Introduction
Epithelial ovarian cancer (EOC) is the most lethal
gynecological malignancy and has a number of histological subtypes
(1). An advanced stage at the
time of diagnosis and relapse due to chemoresistance are the
principal reasons for its poor prognosis (1,2),
and the 5-year survival rate is only ~30% (3). Therefore, there is an urgent need to
elucidate the underlying mechanisms of EOC and apply the knowledge
obtained to the development of novel treatments, including targeted
therapy, to improve patient survival.
Various covalent modifications of histone tails,
including acetylation, ubiquitination, phosphorylation and
methylation, may modulate the chromatin structure and serve pivotal
roles in the regulation of DNA repair, gene transcription, cell
differentiation, cell cycle progression and embryonic development
(4,5). Histone acetylation is generally
associated with transcriptional activation, and histone methylation
is associated with transcriptional activation and repression. For
example, methylation of histone H3 at the lysine 9, 20 or 27
residues (H3K9, H3K20 or H3K27, respectively) leads to
transcriptional gene silencing, whereas methylation at H3K4, H3K36
or H3K79 is correlated with chromatin opening and transcriptional
activation (6).
Histone H3K4 methylation is one of the most
prominent epigenetic modifications associated with gene activation
(5,7). As the major histone H3K4
methyltransferases in mammals, histone-lysine N-methyltransferase
SETD1A (SET1)/histone-lysine N-methyltransferase 2A (MLL) complexes
comprise SET1, histone-lysine N-methyltransferase SETD1B, MLL, and
histone-lysine N-methyltransferase 2B, 2C or 2D as the catalytic
subunit, and WD repeat-containing protein 5 (WDR5),
retinoblastoma-binding protein 5 (RbBp5), Set1/Ash2 histone
methyltransferase complex subunit ASH2 (ASH2L) and protein dpy-30
homolog (DPY30) as integral core subunits exerting methylation
activity (8-10). DPY30, a common member of the human
SET1/MLL complexes, has been reported to be required for complete
SET1/MLL methyltransferase activity (11,12). DPY30 catalyzes histone H3K4
methylation, through which it regulates gene expression, cell
proliferation and differentiation, and therefore affects tissue
development. Furthermore, dysfunction of DPY30 may lead to the
occurrence of cancer (11,12).
Ovarian carcinogenesis entails the progressive
accumulation of various genetic and epigenetic alterations that
lead to gains of function in oncogenes and loss of function in
tumor suppressor genes. Since gene transcriptional activation is
affected by the chromatin structure, abnormal histone methylation,
which alters the chromatin structure, is commonly associated with
tumor progression and prognosis (13). Although alterations in histone
methylation have been well described in various types of cancer,
any alterations in histone methylation in EOC remain poorly
characterized (14,15). The present study aimed to examine
the role of DPY30 in EOC by analyzing DPY30 expression in EOC
tissues and cell lines. Correlations between the
clinicopathological characteristics of EOC cases and the survival
rate among patients with EOC were analyzed. Furthermore, the
effects of DPY30 on EOC cell proliferation, migration and invasion
were investigated. Finally, the mechanism of action of DPY30 was
further elucidated by identifying its association with
epithelial-mesenchymal transition (EMT).
Materials and methods
Clinical specimens
The present study was approved by the ethics
committee of Liaocheng People’s Hospital (Liaocheng, China).
Written informed consent was obtained from all participants prior
to surgical treatment.
Overall, 60 patients who were diagnosed with EOC and
underwent cytoreductive surgery at Liaocheng People’s Hospital
between January 2009 and December 2011 were included in the study.
The clinicopathological data of the enrolled patients were
recorded. Patients were grouped by age, histological type,
International Federation of Gynecology and Obstetrics (FIGO) stage,
pathological grade and lymph node metastasis. The postoperative
follow-up period was 5 years. An additional 20 patients with a
benign ovarian epithelial tumor with a median age of 35 years
(range, 18-50 years) and 15 perimenopausal patients who underwent
ovariectomy due to a uterine fibroid with a median age of 54 years
(range, 48-60 years) were included as controls.
Separately, 40 fresh ovarian carcinoma tissues and
adjacent normal ovarian tissues were obtained from patients who
underwent initial hysterectomy at Liaocheng People’s Hospital
between May 2014 and October 2016. All specimens were stored frozen
at −80°C.
Cell culture
The ovarian cancer cell lines SKOV3, OVCAR3, A2780
and IOSE80 were obtained from the American Type Culture Collection
(Manassas, VA, USA). SKOV3 cells were cultured in McCoy’s 5A Medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the
OVCAR3 and A2780 lines were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.). IOSE80 cells were cultured in
Dulbecco’s modified Eagle’s medium (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA). All medium was supplemented with 10%
fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc.). Cells were maintained at 37°C with 5% CO2 in a
humidified incubator.
Immunohistochemistry
Immunohistochemical staining for DPY30 expression in
EOC tissue specimens was performed as follows. The specimens were
fixed in 10% neutral formalin at room temperature for 48 h. The
collected paraffin-embedded tissues were sectioned to a thickness
of 4 µm. The slides were deparaffinized in xylene,
rehydrated in graded alcohol solutions, and boiled in citrate
buffer for 2.5 min in an autoclave. The slides were treated with
0.3% hydrogen peroxide for 10 min at room temperature to inhibit
endogenous peroxidase activity. The slides were incubated at 4°C
overnight with an anti-DPY30 primary antibody (cat. no. ab214010;
1:100; Abcam, Cambridge, UK). To each slide was added 100 µl
horseradish peroxidase-labeled goat anti-rabbit IgG complex (cat.
no. PV-6001; OriGene Technologies, Inc., Beijing, China), which was
incubated for 20 min at room temperature. The peroxidase reaction
was developed with 3,3′-diaminobenzi-dine (DAB), and slides were
counterstained for 2 min at room temperature with hematoxylin
staining buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
DAB was obtained from OriGene Technologies, Inc. The
immunohistochemical evaluation was performed by two experienced
pathologists who had no knowledge of the clinical status of the
patients. Using a light microscope (BX53; Olympus Corporation,
Tokyo, Japan), the digital images were processed using Image-Pro
plus 6.0 software (Media Cybernetics, Inc.). Nuclear expression of
DPY30 was regarded as positive. The status of DPY30 protein
expression was assessed by an evaluation of the intensity of
staining and the percentage of stained tumor cells.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cell lines and tissues was extracted
using TRIzol® (Thermo Fisher Scientific, Inc.),
according to the manufacturer’s protocol. RT was performed using
the PrimeScript RT Master Mix Perfect Real Time (Takara Bio, Inc.,
Otsu, Japan) for DPY30 (forward, 5′-ACT CGT GCC TAC CTG GAT CA-3′
and reverse, 5′-CGA TCT TCA AAC TGT GCC TTG T-3′), and GAPDH
(forward, 5′-GGA GCG AGA TCC CTC CAA AAT-3′ and reverse, 5′-GGC TGT
TGT CAT ACT TCT CAT GG-3′) was used as an internal loading control.
The reaction conditions were as follows: 37°C for 15 min and 85°C
for 5 sec, 4°C for 10 min. qPCR was performed using the RNA PCR kit
(Takara Bio, Inc.), and SYBR-Green qPCR Master mix (Takara Bio,
Inc.) was added to a 20-µl reaction volume. Amplification
was conducted using a Applied Biosystems 7500 Fast Real-Time PCR
System (Thermo Fisher Scientific, Inc.) under the following
conditions: Initial denaturation for 1 cycle at 95°C for 30 sec,
followed by denaturation at 95°C for 5 sec, and amplification at
60°C for 34 sec for a total of 40 cycles, followed by a
dissociation stage. Finally, the relative mRNA expression levels of
the target genes were calculated following normalization to GAPDH
mRNA expression using the 2−ΔΔCq method (16).
Stable cell line establishment by
lentiviral transfection
The plasmid vector LV-pLKO-1-EGFP-puro carrying
either DPY30 short hairpin (sh)RNA (LV-sh-DPY30) or control
oligonucleotide (LV-sh-DPY30-NC) was purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). Lentiviruses were produced
according to the instructions from GenePharma. The cells were
infected with 30 µl of each lentivirus (108
particles/ml) with 4 µg/ml polybrene (Shanghai GenePharma
Co., Ltd.) for ~24 h to establish an anti-DPY30-expressing stable
cell line (SKOV3/sh-DPY30) and a control cell line
(SKOV3/shDPY30-NC). The DPY30 expression levels in the established
cell lines were examined by RT-qPCR using GAPDH as an endogenous
control.
Protein extraction and western
blotting
Total protein was extracted using
radioimmunoprecipitation assay buffer (Vazyme, Piscataway, NJ, USA)
with phenylmethylsulfonyl fluoride (Roche Diagnostics, Basel,
Switzerland). Western blotting was performed according to the
standard protocol. The concentration of protein in the supernatant
was determined with a Bicinchoninic Acid Protein Assay kit
(Wanleibio Co., Ltd., Shanghai, China), according to the
manufacturer’s protocol. A total of 30 µg protein was
separated by SDS-PAGE (12% gel) and transferred to polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA). Following
blocking with 5% skimmed milk at room temperature for 1 h, the
membranes were incubated at 4°C overnight with the following
primary antibodies: DPY30 (cat. no. ab214010; 1:1,000; Abcam),
vimentin (cat. no. 5741; 1:1,000; Cell Signaling Technology, Inc.,
Danvers, MA, USA), E-cadherin (cat. no. 3195; 1:1,000; Cell
Signaling Technology, Inc.), N-cadherin (cat. no. 13116; 1:1,000;
Cell Signaling Technology, Inc.), zinc finger protein SNAI1 (Snail;
cat. no. 3879; 1:1,000; Cell Signaling Technology, Inc.),
trimethylated histone H3K4 (H3K4me3; cat. no. 9751; 1:1,000; Cell
Signaling Technology, Inc.), total histone H3 (cat. no. 9728;
1:1,000; Cell Signaling Technology, Inc.) and β-actin (cat. no.
T4014; 1:3,000; Abmart, Shanghai, China), which served as a loading
control. Subsequently, the membranes were incubated at room
temperature for 60 min with anti-rabbit IgG horseradish peroxidase
secondary antibody (cat. no. WLA023a; 1:3,000; Wanleibio Co.,
Ltd.), in blocking buffer. Protein bands were visualized using an
enhanced chemiluminescence system (ProteinSimple, San Jose, CA,
USA) and analyzed by Quantity One software version 4.0.1 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The experiments were
performed in triplicate.
Functional study
Cell proliferation
Cell proliferation was evaluated using a
water-soluble tetrazolium salt assay and counted via a Cell
Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). Cells (5×103/well) were seeded on
96-well culture plates in triplicate and incubated for 3 days at
37°C with 5% CO2 in a humidified incubator. The numbers
of viable cells were quantified every 24 h by measuring the
absorbance at an optical density of 450 nm using a microplate
reader (Epoch; BioTek Instruments, Inc., Winooski, VT, USA).
Wound-healing assay
Cells were seeded on six-well plates, and upon
reaching 70-80% confluence, the cell monolayer was scratched using
a sterilized 10-µl pipette tip. Detached cells were removed,
and the plates were incubated at 37°C with McCoy’s 5A containing 1%
FBS. Images of the scratches were captured every 24 h (0, 24 and 48
h total) for the assessment of cell migration. Images of at least
five independent scratches were recorded, and the experiments were
repeated three times.
Transwell invasion assays
Cell invasion and migration were assessed using cell
culture inserts coated with or without basement membrane matrix (BD
Biosciences, Franklin Lakes, NJ, USA), according to the
manufacturer’s protocol. A total of ~5×105 cells in 100
µl serum-free culture medium were placed in the upper
chamber of triplicate wells and incubated at 37°C in a 5%
CO2 humidified incubator, and medium containing 20% FBS
was placed in the lower chamber of these wells. After 24 h in
culture, the cells in the upper chamber were gently removed with a
cotton swab, and the cells on the bottom of the insert were stained
with 1% crystal violet for 20 min at room temperature. Cells in
five random fields were counted, and the relative extents of
invasion and migration were interpreted as the average cell number
± standard deviation per field.
Cell cycle analysis
Cells (1×106/ml) were fixed in 75%
ethanol at 4°C overnight, washed with cold PBS, and then treated
with RNaseI in a 37°C water bath for 30 min, followed by propidium
iodide (Wanleibio Co., Ltd.) staining for 30 min in darkness. Cell
cycle analysis was performed via flow cytometry (BD FACSAriaII; BD
Biosciences), according to the manufacturer’s protocol.
Chromatin immunoprecipitation
(ChIP)
ChIP was performed using an EZ-Magna ChIP kit (EMD
Millipore, Billerica, MA, USA), according to the manufacturer’s
protocol. Anti-H3K4me3 antibody (cat. no. 9751; 1:50) was obtained
from Cell Signaling Technology, Inc., and anti-DPY30 antibody (cat.
no. ab214010; 1:50) was obtained from Abcam. Anti-IgG (cat. no.
3900; Cell Signaling Technology, Inc.) was used as the control
antibody. Vimentin promoter primers (forward, 5′-GCT GTA AGT TGG
TAG CAC TGA-3′ and reverse, 5′-TTC TGT CGA GGG ACC TAA CG-3′) were
used in this experiment (17).
Statistical analysis
All statistical analyses were performed using SPSS
version 22.0 software (IBM Corp., Armonk, NY, USA). Differences
between two groups were assessed using the Student’s t-test.
Differences among three or more groups were evaluated using one-way
analysis of variance. Data are presented as the mean ± standard
deviation from three independent experiments. Survival analysis was
performed using a log-rank test and generating Kaplan-Meier plots.
P<0.05 was considered to indicate a statistically significant
difference.
Results
DPY30 is upregulated in EOC tissues and
cell lines
DPY30 expression in primary ovarian tissues from 95
patients was examined by immunohistochemical staining, and the
associations between DPY30 expression and the clinicopathological
factors of EOC were examined. The results demonstrated that the
DPY30 positive staining rate was significantly increased in EOC
tissues (75%) compared with benign ovarian tumors (10%) and normal
tissues (6.67%; both P<0.05; Table
I; Fig. 1). Furthermore,
DPY30 positive staining was observed to be associated with FIGO
stage, pathological grade and lymph node metastasis in patients
with EOC, although no association with histological type or age was
found (Table II).
 | Table IDPY30 expression in different ovarian
tissues. |
Table I
DPY30 expression in different ovarian
tissues.
| Tissue type | n | DPY30 expression, n
(%)
|
|---|
| − (%) | + (%) |
|---|
| Normal ovarian
tissue | 15 | 14 (93.3) | 1 (6.67) |
| Benign ovarian
epithelial tumor | 20 | 18 (90) | 2 (10) |
| Epithelial ovarian
cancer | 60 | 15 (25) | 45 (75)a,b |
 | Table IICorrelation between DPY30 expression
and the clinicopathological features of ovarian cancer. |
Table II
Correlation between DPY30 expression
and the clinicopathological features of ovarian cancer.
| Clinical
pathology | n | DPY30 expression, n
(%)
| χ2 | P-value |
|---|
| − (%) | + (%) |
|---|
| Age, years | | | | | |
| ≤50 | 14 | 4 (28.6) | 10 (71.4) | | |
| >50 | 46 | 11 (23.9) | 35 (76.1) | 0.134 | 0.734 |
| Histological
type | | | | | |
| Serous | 35 | 8 (22.9) | 27 (77.1) | | |
| Endometrioid | 25 | 7 (28.0) | 18 (72.0) | 0.206 | 0.765 |
| FIGO stage | | | | | |
| I + II | 18 | 8 (44.4) | 10 (55.6) | | |
| III + IV | 42 | 7 (16.7) | 35 (83.3) | 5.185 | 0.048 |
| Pathological
grade | | | | | |
| G1 | 11 | 6 (54.5) | 5 (45.5) | | |
| G2+G3 | 49 | 9 (18.4) | 40 (81.6) | 6.271 | 0.021 |
| Lymph node
metastasis | | | | | |
| Yes | 45 | 8 (17.8) | 37 (82.2) | 5.007 | 0.039 |
| No | 15 | 7 (46.7) | 8 (53.3) | | |
DPY30 expression was further evaluated by RT-qPCR
and western blotting in 40 EOC tissues and adjacent normal ovarian
tissues as controls. Compared with normal controls, DPY30
expression at the RNA level in EOC tissues was significantly
elevated (Fig. 2A). In addition,
DPY30 expression at the RNA and protein levels in three EOC cell
lines was higher compared with that in a normal ovarian cell line
(Fig. 2B and C), and DPY30
expression was highest in SKOV3 cells among the three EOC cell
lines. The upregulation of DPY30 expression in EOC suggested that
it may serve an important role in EOC development.
DPY30 promotes EOC cell proliferation,
migration and invasion in vitro
To further examine the role of DPY30 in the genesis
and development of EOC, SKOV3 cells, which exhibited a high level
of DPY30 expression (Fig. 2B and
C) were transfected with LV-DPY30 shRNA to establish
SKOV3/shDPY30 stable clones (Fig.
3). The relative control clones (SKOV3/sh-DPY30-NC) were also
generated. The RT-qPCR results indicated that DPY30 expression was
significantly lower in SKOV3/sh-DPY30 cells compared with
SKOV3/sh-DPY30-NC cells (Fig.
3A). The cell transfection efficacy was verified by RT-qPCR,
and invasion and migration assays were subsequently performed.
As presented in Fig.
3B and D, SKOV3/sh-DPY30 cells exhibited diminished migratory
and invasion capacities compared with SKOV3/sh-DPY30-NC cells
(P<0.05), indicating that DPY30 expression may promote the
invasion and migration of EOC cells in vitro.
According to the results for cell proliferation
obtained from the CCK-8 assay, SKOV3/sh-DPY30 cells exhibited
markedly inhibited proliferation compared with SKOV3/shDPY30-NC
cells (Fig. 3C). Cell cycle
analysis was conducted, and SKOV3/sh-DPY30 cells exhibited
increased populations at the G0/G1 phase and reduced populations at
the G2/M phase, suggesting that DPY30 expression may promote the
proliferation of EOC cells in vitro (Fig. 3E).
DPY30 promotes EMT
The EMT process is critical to the acquisition of
malignant traits during cancer progression (18-20). Considering the high DPY30
expression in EOC cells and its association with increased cell
migration and invasion in vitro, it was hypothesized that
DPY30 may be involved in the EMT process in EOC cells. The in
vitro experiments demonstrated that compared with
SKOV3/sh-DPY30-NC control cells, the expression of E-cadherin, an
epithelial cell marker, in SKOV3/sh-DPY30 cells was significantly
increased, and the expression levels of the mesenchymal cell
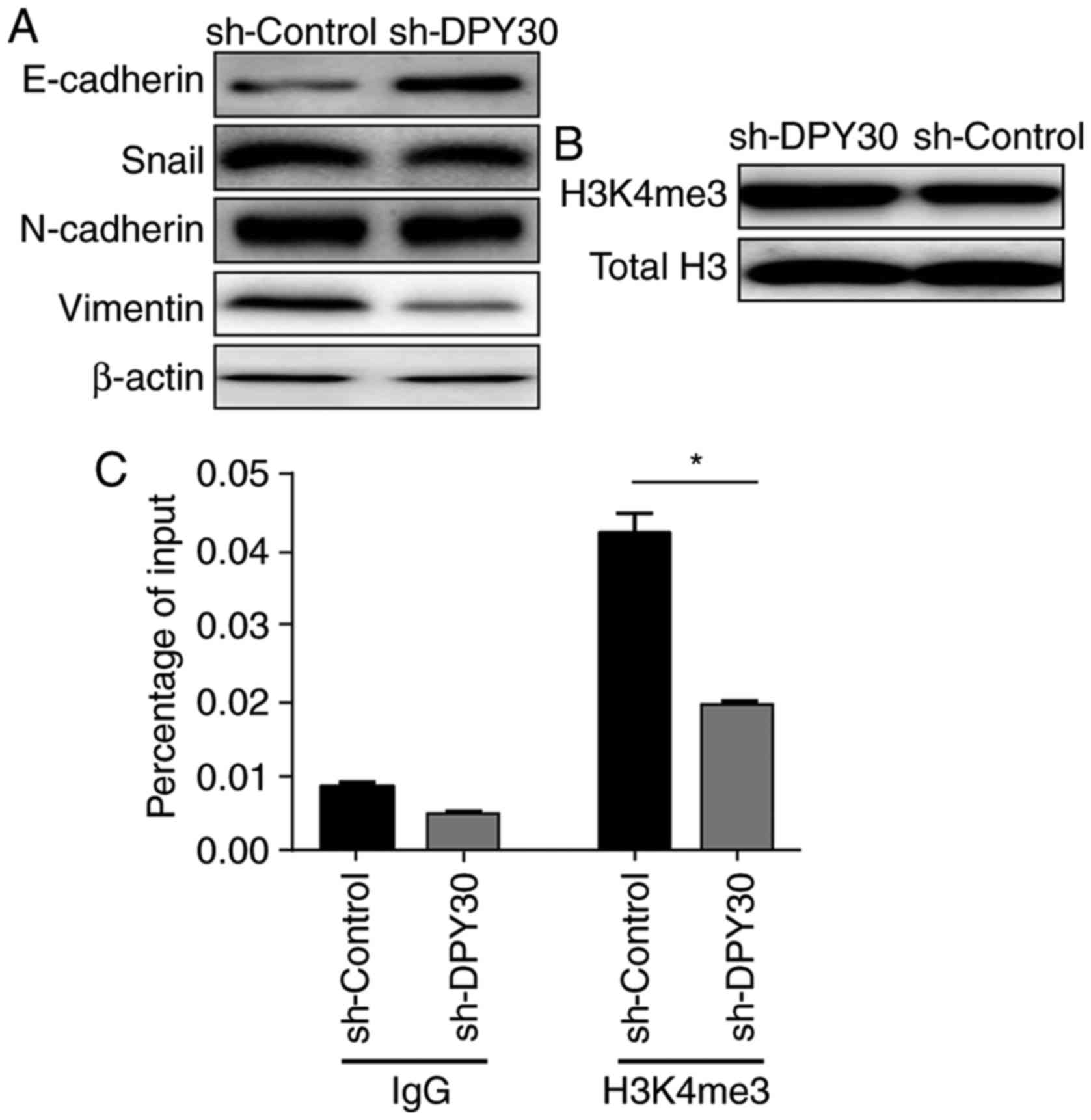
markers vimentin, N-cadherin and Snail were decreased (Fig. 4A). These findings supported the
hypothesis that DPY30 may promote EMT in EOC cells.
DPY30 regulates vimentin expression
through histone H3K4me3 modification
As mentioned above, DPY30, as a member of the human
SET1/MLL complexes, is able to catalyze the methylation of histone
H3K4. Methylation at H3K4, H3K36 or H3K79 is associated with
chromatin opening and gene transcription activation (6), and DPY30 is primarily required for
H3K4me3 (8,12,21).
E-cadherin expression was increased upon knockdown
of DPY30 expression in SKOV3 cells, whereas the expression levels
of vimentin, N-cadherin and Snail were decreased, with the greatest
reduction observed for vimentin expression. In stable
SKOV3/sh-DPY30 cells, the global H3K4me3 level was upregulated upon
DPY30 depletion (Fig. 4B). To
investigate whether DPY30 promotes vimentin expression through
H3K4me3 methylation, ChIP was performed using antibodies against
DPY30 and H3K4me3, with IgG as a control. It was observed that in
SKOV3/sh-DPY30 cells, the expression level of H3K4me3 was decreased
more significantly at the vimentin promoter region compared with
SKOV3/shDPY30-NC control cells (Fig.
4C). These data suggested that DPY30 may regulate histone H3K4
modification at the vimentin promoter and thus enhance vimentin
expression.
High DPY30 expression is associated with
poor survival of patients with EOC
A survival analysis was performed in order to
investigate the association between DPY30 expression levels and the
survival of patients with EOC. The Kaplan-Meier method was used to
estimate overall survival. From our analysis, high DPY30 expression
was significantly associated with a poor prognosis in patients with
EOC (P<0.05; Fig. 5), which
suggested that high DPY30 expression may affect patient survival in
EOC, likely by promoting tumor metastasis.
Discussion
EOC remains a leading cause of cancer-associated
mortality among women, and much research has been devoted to
pursuing an effective treatment for EOC through the discovery of
novel therapeutic targets. The present study focused on DPY30, a
common member of the human SET1/MLL complexes that is required for
complete SET1/MLL methyltransferase activity, a key process in
cancer development (11,12). Previously, DPY30 was reported to
be essential for the differentiation and proliferation of
hematopoietic progenitor cells (12) and was implicated in the
differentiation potential of embryonic stem cells along the
neuronal lineage (11). Research
has demonstrated that depletion of DPY30 leads to a senescent-like
state in cells and upregulated cyclin-dependent kinase 4 inhibitor
B and cyclin-dependent kinase inhibitor 2A expression levels, which
are directly associated with cell senescence (13). Notably, DPY30 was recently
reported to be important for gastric cancer progression, suggesting
that DPY30 may be a therapeutic target in gastric cancer (22).
The results of the present study indicated that
DPY30 may serve important roles in EOC. The majority of ovarian
cancer tissues exhibited high expression of DPY30, and DPY30
expression was positively associated with FIGO stage, pathological
grade and lymph node metastasis. DPY30 expression was higher in the
advanced stages (III-IV) of EOC compared with the early stages
(I-II), higher in less-differentiated carcinomas compared with
well-differentiated tissues, and higher in cases with lymph node
metastasis compared with those without lymph node metastasis.
Therefore, the present results indicated a strong association
between DPY30 and EOC development and progression.
The functional experiments further revealed that
DPY30 knockdown was able to regulate the proliferation, migration
and invasion of EOC cells. Importantly, DPY30 induced G0/G1 arrest
in SKOV3/sh-DPY30 cells, which was further supported by the fact
that DPY30 knockdown in SKOV3 cells increased the cell population
at the G0/G1 phase and therefore restrained cell proliferation.
The present study also indicated that DPY30 promoted
EMT, a process that is important for tumor progression and
metastasis (18,23). DPY30 knockdown in SKOV3 cells
induced increased expression of E-cadherin and decreased expression
of vimentin, N-cadherin and Snail, demonstrating a potential
tumorigenic effect of DPY30 in EOC.
The underlying molecular mechanisms of the
cancer-promoting effects of DPY30 have been examined in previous
studies, and a number of hypotheses have been proposed (22). One hypothesis is that DPY30
overexpression leads to oncogene overexpression by increasing the
methylation of histone H3 lysine 4 methyltransferase (H3K4MT).
ASH2L or DPY30 depletion has been observed to lead to decreased
H3K4me3 expression (8,11). Notably, RbBp5 and WDR5 are crucial
for the methylation of all three H3K4 subtypes, whereas DPY30 is
primarily required for H3K4me3 (5,19,21). In another hypothesis,
overexpression of DPY30 alone increases H3K4MT methylation activity
(11). Since H3K4me2/3 expression
is an indicator of transcriptional activity (6,24),
increased H3K4MT activity may directly upregulate the expression of
oncogenes or downregulate the expression of tumor suppressors
indirectly. Previous research found that ASH2L, another crucial
component of the SET1/MLL complexes, functions as an oncoprotein
(25,26), which strongly supports this
hypothesis. A third hypothesis is that DPY30 is able to directly
activate the expression of inhibitor of DNA binding proteins via
H3K4 methylation (10,27). This is supported by the present
finding that DPY30 promotes vimentin expression via H3K4me3
methylation at the vimentin promoter. Overall, the present study
along with previous work suggested that DPY30 may promote EOC
development via multiple pathways.
The results of the present study revealed important
roles for DPY30 in EOC, with DPY30 acting as an oncogene and
promoting EOC cell proliferation, migration and invasion capacity.
The present data establish a possible mechanism through which DPY30
may promote cancer metastasis in EOC cells. DPY30 was able to
promote EMT in EOC, and DPY30 promoted vimentin expression through
H3K4me3 methylation at the vimentin promoter. Therefore, DPY30 may
represent a therapeutic target and prognostic marker in EOC.
Funding
The present study was supported by the Natural
Science Foundation of Shandong Province (grant nos. ZR2017PH029 and
ZR2015YL047) and Projects of the Medical and Health Technology
Development Program in Shandong Province (grant nos. 2017WS641 and
2015WS0157).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
LZ, ShuZ and ShiZ contributed to the conception of
the work and designing the study. AL and LC searched the literature
and collated the data. LZ, LC and AZ performed the experiments. LZ
analyzed the data and drafted the manuscript. ShiZ made substantial
contributions to the analysis and interpretation of data, and
revised the manuscript critically for important intellectual
content. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Liaocheng People’s Hospital (Liaocheng, China).
Written informed consent was obtained from all participants prior
to surgical treatment.
Patient consent for publication
Consent for publication was obtained from the
participants.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to acknowledge the continuous
support and valuable guidance of Dr Haiying Chen and Dr Shaoda Ren
of the Central Laboratory of Liaocheng People’s Hospital
(Liaocheng, China).
References
|
1
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck
A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R,
Etemadmoghadam D, et al: Rethinking ovarian cancer: Recommendations
for improving outcomes. Nat Rev Cancer. 11:719–725. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gao L, Ye X, Ma RQ, Cheng HY, Han HJ, Cui
H, Wei LH and Chang XH: Low programmed cell death 5 expression is a
prognostic factor in ovarian cancer. Chin Med J (Engl).
128:1084–1090. 2015. View Article : Google Scholar
|
|
3
|
Kim MK, James J and Annunziata CM:
Topotecan synergizes with CHEK1 (CHK1) inhibitor to induce
apoptosis in ovarian cancer cells. BMC Cancer. 15:1962015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhaumik SR, Smith E and Shilatifard A:
Covalent modifications of histones during development and disease
pathogenesis. Nat Struct Mol Biol. 14:1008–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berger SL: The complex language of
chromatin regulation during transcription. Nature. 447:407–412.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Martin C and Zhang Y: The diverse
functions of histone lysine methylation. Nat Rev Mol Cell Biol.
6:838–849. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee
JW, Verdine GL, Allis CD and Roeder RG: Regulation of MLL1 H3K4
methyltransferase activity by its core components. Nat Struct Mol
Biol. 13:713–719. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shilatifard A: Molecular implementation
and physiological roles for histone H3 lysine 4 (H3K4) methylation.
Curr Opin Cell Biol. 20:341–348. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shilatifard A: The COMPASS family of
histone H3K4 methylases: Mechanisms of regulation in development
and disease pathogenesis. Annu Rev Biochem. 81:65–95. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang H, Shukla A, Wang X, Chen WY,
Bernstein BE and Roeder RG: Role for Dpy-30 in ES cell-fate
specification by regulation of H3K4 methylation within bivalent
domains. Cell. 144:513–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Z, Augustin J, Chang C, Hu J, Shah K,
Chang CW, Townes T and Jiang H: The DPY30 subunit in SET1/MLL
complexes regulates the proliferation and differentiation of
hematopoietic progenitor cells. Blood. 124:2025–2033. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Simboeck E, Gutierrez A, Cozzuto L,
Beringer M, Caizzi L, Keyes WM and Di Croce L: DPY30 regulates
pathways in cellular senescence through ID protein expression. EMBO
J. 32:2217–2230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Varier RA and Timmers HT: Histone lysine
methylation and demethylation pathways in cancer. Biochim Biophys
Acta. 1815.75–89. 2011.
|
|
15
|
Chervona Y and Costa M: Histone
modifications and cancer: Biomarkers of prognosis. Am J Cancer Res.
2:589–597. 2012.
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Liu Y, Wang Y, Chen C, Zhang J, Qian W,
Dong Y, Liu Z, Zhang X, Wang X and Zhang Z: LSD1 binds to HPV16 E7
and promotes the epithelial-mesenchymal transition in cervical
cancer by demethylating histones at the Vimentin promoter.
Oncotarget. 8:11329–11342. 2017.
|
|
18
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Steward MM, Lee JS, O’Donovan A, Wyatt M,
Bernstein BE and Shilatifard A: Molecular regulation of H3K4
trimethylation by ASH2L, a shared subunit of MLL complexes. Nat
Struct Mol Biol. 13:852–854. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee YJ, Han ME, Baek SJ, Kim SY and Oh SO:
Roles of DPY30 in the proliferation and motility of gastric cancer
cells. PLoS One. 10:e013–1863. 2015.
|
|
23
|
Knösel T, Schlüns K, Stein U, Schwabe H,
Schlag PM, Dietel M and Petersen I: Chromosomal alterations during
lymphatic and liver metastasis formation of colorectal cancer.
Neoplasia. 6:23–28. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi Y and Whetstine JR: Dynamic regulation
of histone lysine methylation by demethylases. Mol Cell. 25:1–14.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
South PF, Fingerman IM, Mersman DP, Du HN
and Briggs SD: A conserved interaction between the SDI domain of
Bre2 and the Dpy-30 domain of Sdc1 is required for histone
methylation and gene expression. J Biol Chem. 285:595–607. 2010.
View Article : Google Scholar :
|
|
26
|
Takahashi YH, Westfield GH, Oleskie AN,
Trievel RC, Shilatifard A and Skiniotis G: Structural analysis of
the core COMPASS family of histone H3K4 methylases from yeast to
human. Proc Natl Acad Sci USA. 108:20526–20531. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ohtani N, Zebedee Z, Huot TJ, Stinson JA,
Sugimoto M, Ohashi Y, Sharrocks AD, Peters G and Hara E: Opposing
effects of Ets and Id proteins on p16INK4a expression during
cellular senescence. Nature. 409:1067–1070. 2001. View Article : Google Scholar : PubMed/NCBI
|