Introduction
Renal cell carcinoma (RCC) is the most common
malignancy of the kidney and it is estimated that ~65,340 novel
cases and 14,970 cases of mortality are likely to occur in the USA
in 2018 (1). Clear cell RCC is
the most common subtype, accounting for ~70% all RCC cases
(2). In total, >25% patients
have developed metastatic disease on presentation, and reliable
biomarkers for screening RCC have not been established (3). RCC is resistant to conventional
chemoradiotherapy, and surgery remains the primary form of curative
treatment for localized RCC (4).
Despite advances in diagnosis and treatment, the 5-year overall
survival rate of RCC is <50% (4). Therefore, novel diagnostic
approaches and therapeutic strategies for RCC are urgently
required.
MicroRNAs (miRs) are a class of small non-coding
RNAs that suppress gene expression by binding to the
3′-untranslated region of mRNA at the post-transcriptional level
(5). Accumulating evidence
suggested that miRs may be important in the development of
malignancies (6). Previous
studies demonstrated that miRs are involved in a number of cancer
cell biological processes, including metastasis (7), invasion (8), angiogenesis (9), proliferation, apoptosis (10), differentiation (11), metabolism (12) and drug resistance (13). The dysregulation of miRs is
observed in the majority of types of cancer, including RCC
(14). In previous studies,
numerous miRs have been demonstrated to be involved in RCC,
including miR-720 (15), miR-203
(16), miR-204-3p (17) and miR-32-5p (18). miR-222 is associated with
tumorigenesis in gastric cancer (19), glioma (20) and lung cancer (21), among others; however, the role of
miR-222 in RCC remains to be fully elucidated.
In the present study, the expression of miR-222-3p
was investigated in clinical RCC samples and cell lines in order to
determine its effects on the migration, invasion and apoptosis of
RCC cells and to analyze its association with overall survival
rates. The aim of the present study was to determine whether
miR-222-3p may serve as a non-invasive prognostic biomarker and
therapeutic target for patients with RCC.
Materials and methods
Clinical samples
Patients who were initially treated, without
previous surgery, radiotherapy, chemotherapy and other adjuvant
therapy were included in the present study. Patients who had been
diagnosed with tumors other than renal cell carcinoma were
excluded. Tumor tissues and corresponding normal renal tissues were
obtained from 28 patients with RCC at the Department of Urology,
Peking University Shenzhen Hospital (Shenzhen, China) from January
2013 to January 2016. The clinicopathological parameters are
summarized in Table I. In
addition, 42 formaldehyde-fixed paraffin-embedded (FFPE) RCC
samples were obtained from the Department of Pathology, Peking
University Shenzhen Hospital. The clinical and pathological
characteristics are listed in Table
II. Follow-up data was obtained for the 42 FFPE specimens;
however, not for the 28 patients, as no the follow-up data were
available for the 28 patients, which is a limitation of the study.
The present study was approved by the Ethics Committee of Peking
University Shenzhen Hospital, and informed consent was obtained
from all the patients.
 | Table IClinicopathological features in
patients with renal cell carcinoma. |
Table I
Clinicopathological features in
patients with renal cell carcinoma.
| Characteristic | Cases, n |
|---|
| Mean age, years
(range) | 45 (24-87) |
| Sex | |
| Male | 17 |
| Female | 11 |
| Tumor stage | |
| T1 | 17 |
| T2 | 4 |
| T3 + T4 | 7 |
| Fuhrman grade | |
| I | 12 |
| II | 8 |
| III | 7 |
| IV | 1 |
| AJCC clinical
stage | |
| I | 15 |
| II | 3 |
| III+IV | 10 |
 | Table IIAssociation between miR-222-3p status
and clinicopathologic variables in
formaldehyde-fixed-paraffin-embedded renal cell carcinoma tissue
samples. |
Table II
Association between miR-222-3p status
and clinicopathologic variables in
formaldehyde-fixed-paraffin-embedded renal cell carcinoma tissue
samples.
| Variable | Total, n | miR-222-3p status
| P-value |
|---|
| High, n | Low, n |
|---|
| Sex | | | | 0.525a |
| Male | 26 | 14 | 12 | |
| Female | 16 | 7 | 9 | |
| Age, years | | | | 1.000b |
| ≤60 | 33 | 16 | 17 | |
| >60 | 9 | 5 | 4 | |
| Tumor size, cm | | | | 0.116a |
| ≤4.0 | 17 | 6 | 11 | |
| >4.0 | 25 | 15 | 10 | |
| Tumor stage | | | | 0.747a |
| I+II | 27 | 13 | 14 | |
| III+IV | 15 | 8 | 7 | |
RNA isolation and reverse transcription
(RT)
Total RNA was isolated from the tissues and cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and purified using an RNeasy
Maxi kit (Qiagen GmbH, Hilden, Germany) according to the
manufacturer’s protocol. The miRs of the FFPE samples were
separated using the miRNeasy FFPE kit (Qiagen GmbH) according to
the manufacturer’s protocol. Subsequently, the concentration and
quality of RNAs were measured using NanoDrop 2000/2000c (Thermo
Fisher Scientific, Inc.). In total, ~1 µg RNA was used to
synthesize cDNA using the miScript II RT kit (Qiagen GmbH) and the
reaction conditions were as follows: 37°C for 60 min, 95°C for 5
min and 4°C until completion.
RT-quantitative polymerase chain reaction
(qPCR) analysis
The RT-qPCR analysis was performed with the miScript
SYBR® Green PCR kit (Qiagen GmbH). U6 served as an
internal control. The reaction contained 0.5 µl primer, 1
µl cDNA, 5 µl 2x QuantiTect SYBR Green PCR Master Mix
and 3 µl RNase-free water. The reaction was performed in the
Roche Lightcycler 480 Real-Time PCR system (Roche Diagnostics,
Basel, Switzerland) and the reaction conditions were as follows:
95°C for 2 min, followed by 40 cycles of 95°C for 10 sec, 55°C for
30 sec and 72°C for 30 sec. The primers used in the experiment are
summarized in Table III. The
reference gene was U6. The relative expression of miR-222-3p was
evaluated using the comparative Cq and analyzed using the
2−ΔΔCq method (22,23).
 | Table IIISequences of primers and miRs. |
Table III
Sequences of primers and miRs.
| Primer/miR | Sequence |
|---|
| miR-222-3p | Forward:
5′-AGCTACATCTGGCTACTGGGT-3′ |
| Reverse: Universal
primers (miScript SYBR Green PCR kit) |
| U6 | Forward:
5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse:
5′-ACGCTTCACGAATTTGCGT-3′ |
| miR-222-3p
mimic | Forward:
5′-AGCUACAUCUGGCUACUGGGU-3′ |
| Reverse:
5′-CCAGUAGCCAGAUGUAGCUUU-3′ |
| miR-222-3p
inhibitor NC |
5′-ACCCAGUAGCCAGAUGUAGCU-3′ |
| Forward:
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Inhibitor NC | Reverse:
5′-ACGUGACACGUUCGGAGAATT-3′ |
|
5′-CAGUACUUUUGUGUAGUACAA-3′ |
Cell culture and transfection
The 786-O, ACHN, 769-P and Caki-1 RCC cell lines and
the HK-2 normal human renal tubular epithelial cell line were
purchased from the American Type Culture Collection (Manassas, VA,
USA). According to Cellosaurus (https://web.expasy.org/cellosaurus), the ACHN, 769P,
786-O and Caki-1 cell lines appear to be papillary renal cell
carcinoma, renal cell carcinoma, renal cell carcinoma and clear
cell renal cell carcinoma, respectively. The HK-2 cells were
cultured in keratinocyte serum-free medium (ScienCell Research
Laboratories, Inc., Carlsbad, CA, USA) supplemented with 1%
keratinocyte growth supplement (ScienCell Research Laboratories,
Inc.), with 100 µl/ml penicillin and 100 mg/ml streptomycin
sulfate in a humidified incubator with a 5% CO2
atmosphere at 37°C. The ACHN and Caki-1 cells were cultured in
Dulbecco’s modified Eagle’s medium (Gibco; Thermo Fisher
Scientific, Inc.) and McCoy’s 5A (Gibco; Thermo Fisher Scientific,
Inc.) respectively, and supplemented with 10% fetal bovine serum
(FBS, Gibco; Thermo Fisher Scientific, Inc.) and 1% antibiotics
(100 µl/ml penicillin and 100 mg/ml streptomycin sulfate).
The 769-P and 786-O cells were cultured in the same manner in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.). A quick
cell mycoplasma rapid test kit (Shanghai Life iLAB Bio Technology
Co., Ltd., Shanghai, China) was used to detect whether the cells
were contaminated with mycoplasma, according to the manufacturer’s
protocol. The miR-222-3p (miRbase; http://www.mirbase.org/; accession no. MIMAT0000279)
mimics, negative control (NC), miR-222-3p inhibitor and inhibitor
negative control (NC; GenePharma Co., Ltd., Shanghai, China) were
transfected into cells at a concentration of 50 nM using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequences of miRNAs are listed in Table III.
Wound healing assay
The RCC cells (ACHN and 786-O) were seeded into
6-well plates (30×105 cells/well) and incubated in a
humidified chamber supplemented with 5% CO2. The cells
were transfected with miR-222-3p mimics, inhibitor and
corresponding NC when they reached ~90% confluence. Following 6 h
transfection, a wound was created using a sterile 200-µl
pipette tip, followed by washing with PBS three times. Images were
captured using a digital light microscope (magnification, ×100) at
0 and 24 h following introduction of the wound.
Cell migration and invasion assays
The cell migration and invasion capacities were
evaluated using a Transwell assay. Chambers coated with Matrigel
(BD Biosciences, Franklin Lakes, NJ, USA) were used for the
invasion assay, whereas Matrigel was omitted for the migration
assay. Following 24 h transfection, ~100 µl serum-free
medium containing 3×104 cells was added to the upper
chambers, and 500 µl medium supplemented with 10% FBS was
added to the lower chambers. The migration and invasion duration
was 24 and 36 h, respectively. Subsequently, the cells were fixed
in 4% formaldehyde for 15 min at room temperature and stained with
0.1% crystal violet solution for 25 min at room temperature. Images
of those cells that had migrated or invaded to the opposite side of
the membrane were captured using a light microscope (Leica
Microsystems GmbH, Wetzlar, Germany; magnification, ×100).
Apoptosis assay
Flow cytometry was performed to determine the effect
of miR-222-3p on cell apoptosis in vitro. The cells
(~3×105) were added to 6-well plates and cultured in an
incubator for 24 h; subsequently, the cells were treated with
miR-222-3p mimics, miR-222-3p inhibitor or corresponding NC.
Another group of cells (blank group) was treated with
Lipofectamine® 3000 but without mimics, inhibitor or
corresponding NC. Following 48 h transfection, the cells were
collected and washed twice with cold PBS, followed by staining with
Annexin V-fluorescein isothiocyanate/propidium iodide (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer’s
protocol. The apoptotic rate was detected using a flow cytometer
(EPICS, Xl-4; Beckman Coulter, Inc., Brea, CA, USA) and analyzed
using Kaluza Analysis 1.5a (Beckman Coulter, Inc.).
Clinical validation via the cancer genome
atlas (TCGA) dataset
The correlation between the expression of miR-222-3p
and the prognosis of patients with RCC was analyzed using a TCGA
dataset in OncoLnc (www.oncolnc.org). OncoLnc is a tool website that links
TCGA survival data to miRNA expression levels. In OncoLnc, the
input term used was ‘hsa-miR-222-3p’ and the lower and upper
percentile were set to 50. Subsequently, the correlation between
the expression of miR-222-3p and the overall survival rate of the
patients who provided the FFPE samples was analyzed and
Kaplan-Meier curves were constructed.
Bioinformatics and target prediction
analysis
Target prediction was performed for miR-222-3p with
starBase v2.0 (http://starbase.sysu.edu.cn/browseIntersectTargetSite.php),
PicTar (https://pictar.mdc-berlin.de),
TargetScanHuman (www.targetscan.org) and PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html).
Only predictions by at least three programs were included in the
analysis.
Statistical analysis
The results of the experiments were analyzed using
SPSS 19.0 (IBM Corp., Armonk, NY, USA) and are presented as the
mean ± standard deviation or median. Differences between normal
renal tissues and RCC tissues were statistically analyzed using
non-parametric tests and presented as median values. The
differences in the expression level of miR-222-3p among cell lines
was statistically analyzed by one-way analysis of variance and
Dunnett’s post hoc test. Setting the median as a cutoff point, the
expression level of miR-222-3p was classified into higher and lower
groups. The association between the expression of miR-222-3p and
clinical characteristics was evaluated by Pearson’s χ2
test and Fisher’s exact test. Kaplan-Meier overall survival curves
were constructed to evaluate the effect of miR-222-3p on the
prognosis of patients with x RCC. Differences between the curves
were analyzed by the log-rank test. Univariate and multivariate Cox
regression analyses were also performed. The overall survival was
defined as the time between the first surgery for RCC and the
patient succumbing to mortality from any cause. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-222-3p dysregulation in RCC
RT-qPCR analysis was performed to detect the
expression levels of miR-222-3p in the RCC tissues and cell lines.
As presented in Fig. 1, in the 28
paired tissues, miR-222-3p was significantly upregulated in the RCC
tissues compared with the adjacent normal renal tissues (Fig. 1A and B). In addition, miR-222-3p
was upregulated in the RCC cell lines compared with the HK-2 cells
(Fig. 1C). Therefore, the results
suggested that miR-222-3p may serve as an onco-miR in RCC.
The 786-O and ACHN cell lines were selected for use
in the subsequent experiments. The 786-O cells are RCC cells and
ACHN cells are papillary RCC cells. By transfecting with miR-222-3p
mimics, miR-222-3p inhibitor and corresponding NC, miR-222-3p was
overexpressed or knocked down. The 24-h transfection efficiency was
detected by RT-qPCR analysis (Fig.
1D).
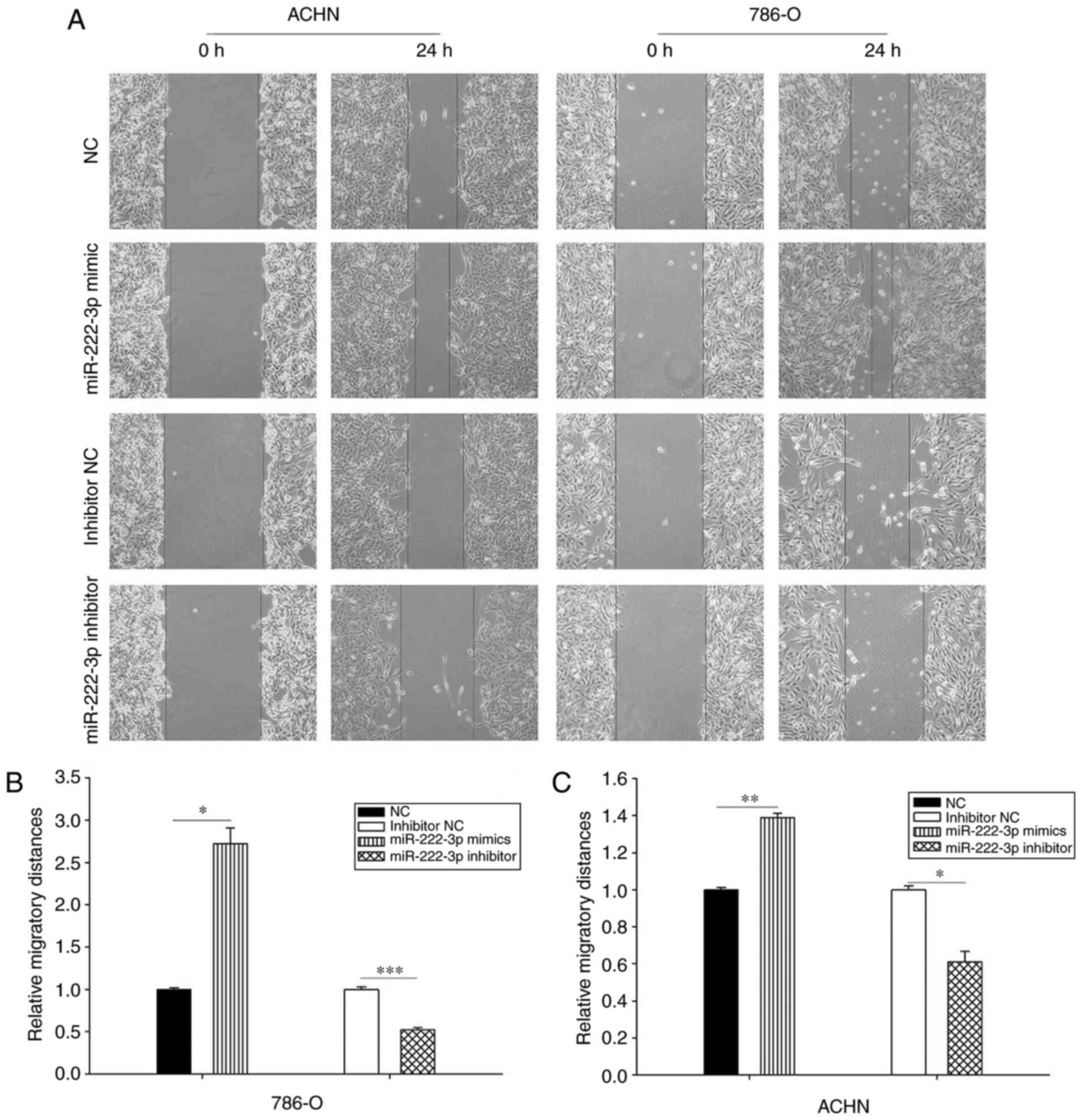
miR-222-3p accelerates cell mobility
Wound healing and Transwell assays were performed to
evaluate the effect of miR-222-3p on cell mobility. The results
demonstrated that the overexpression of miR-222-3p promoted cell
migration. As presented in Fig.
2, cells transfected with miR-222-3p mimics migrated faster
compared with those transfected with NC; by contrast, cells
transfected with miR-222-3p inhibitor migrated more slowly compared
with those transfected with inhibitor NC. The results of the
Transwell assay demonstrated that the migration ability of the two
cell lines transfected with miR-222-3p mimics was enhanced compared
with those transfected with NC, and the inhibition of miR-222-3p
repressed cell migration ability compared with the inhibitor
control group. In addition, the results of the Transwell assay
demonstrated that the overexpression of miR-222-3p facilitated the
invasion ability of cells compared with those in the NC group. The
invasion ability of the two cell lines was suppressed in the
miR-222-3p inhibitor group compared with the inhibitor NC group
(Fig. 3). These results
demonstrated that miR-222-3p promotes RCC cell migration and
invasion in vitro.
miR-222-3p inhibits apoptosis in RCC cell
lines
The results of the apoptosis assay demonstrated that
the apoptotic rate of the 786-O cells was lower in the miR-222-3p
mimics group compared with the NC group (Fig. 4A and B), with similar results
observed in the ACHN cell line (Fig.
4A and C). The apoptotic rates of the two cell lines were
higher in the inhibitor groups compared with those in the inhibitor
NC group (Fig. 4). These results
suggested that miR-222-3p partly decreased the apoptosis of RCC
cells.
miR-222-3p is a potential prognostic
marker for RCC
The expression level of miR-222-3p in 42 FFPE
samples was detected by RT-qPCR analysis. No significant
association was observed between the expression level of miR-222-3p
and sex (P=0.525), age (P=1.000), tumor size (P=0.116) or tumor
stage (P=0.747; Table II).
However, patients with a higher expression of miR-222-3p exhibited
a statistically significant shorter overall survival rate, compared
with patients with a lower expression of miR-222-3p [hazard ratio
(HR)=5.120; 95% confidence interval (CI)=1.113-23.539; P=0.036].
When controlling for age, sex, tumor size and tumor stage in the
multivariate analysis, patients with a higher expression of
miR-222-3p retained the statistically significant decrease in
overall survival rate compared with patients with a lower
expression of miR-222-3p (HR=5.636; 95% CI=1.181-26.882; P=0.030;
Table IV). Furthermore, the
Kaplan-Meier survival curves demonstrated that patients with higher
expression levels of miR-222-3p exhibited significantly lower
overall survival rates compared with patients with lower levels of
miR-222-3p (P=0.020; Fig. 5). The
results from the database analysis in OncoLnc of 506 RCC samples
additionally demonstrated that patients with a higher expression of
miR-222-3p exhibited significantly lower overall survival rates
compared with patients with a lower expression of miR-222-3p
(P<0.01; Fig. 6). These
results suggested that miR-222-3p may serve as a potential
prognostic biomarker for patients with RCC.
 | Table IVExpression of microRNR-222-3p and
patient survival rates. |
Table IV
Expression of microRNR-222-3p and
patient survival rates.
A, Univariate
analysis
|
|---|
| Variable
P-value | Overall survival
|
|---|
| H R (9 5 % C
I) | |
|---|
| Low | 1 | |
| High | 5.120
(1.113-23.539) | 0.036 |
| Age | 2.943
(0.932-9.293) | 0.066 |
| Sex | 0.988
(0.313-3.117) | 0.983 |
| Tumor size | 2.113
(0.571-7.818) | 0.262 |
| Tumor stage | 4.872
(1.462-16.241) | 0.010 |
B, Multivariate
analysisa
|
|---|
| Variable
P-value | Overall
survival |
|---|
| H R (9 5 % C
I) | |
|---|
| Low | 1 | |
| High | 5.636
(1.181-26.882) | 0.030 |
| Age | 2.502
(0.513-12.202) | 0.257 |
| Sex | 1.630
(0.341-7.788) | 0.540 |
| Tumor size | 1.316
(0.341-5.076) | 0.690 |
| Tumor stage | 4.346
(1.233-15.313) | 0.022 |
Potential targets of miR-222-3p
In order to examine the potential involvement of
miR-222-3p in the tumorigenesis of RCC, numerous databases were
searched. The results demonstrated that 37 target genes were
significantly correlated with miR-222-3p (Table V). These genes will be the focus
of future investigations.
 | Table VTarget genes of microRNA-222-3p
determined using starBase v2.0. |
Table V
Target genes of microRNA-222-3p
determined using starBase v2.0.
| No. | Gene | R | Rank | P-value |
|---|
| 1 | SNX4 | −0.39727 | 15,268 | <0.0001 |
| 2 | MYLIP | −0.38758 | 16,842 | <0.0001 |
| 3 | PAIP2 | −0.34117 | 26,749 | <0.0001 |
| 4 | PLEKHA2 | −0.30646 | 37,313 | <0.0001 |
| 5 | CDKN1B | −0.30500 | 37,827 | <0.0001 |
| 6 | PPP1R15B | −0.29684 | 40,824 | <0.0001 |
| 7 | ARNT | −0.28919 | 43,721 | <0.0001 |
| 8 | ANKHD1 | −0.21829 | 80,889 | 0.0001 |
| 9 | ATXN1 | −0.16837 | 121,356 | 0.0034 |
| 10 | FNDC3A | −0.14729 | 142,969 | 0.0106 |
| 11 | POGZ | −0.14009 | 151,019 | 0.0152 |
| 12 | MIA3 | −0.12970 | 163,365 | 0.0247 |
| 13 | HIPK1 | −0.12861 | 164,708 | 0.0259 |
| 14 | TMCC1 | −0.12025 | 175,357 | 0.0374 |
| 15 | MIER3 | −0.11226 | 185,905 | 0.0521 |
| 16 | MEGF9 | −0.09859 | 205,420 | 0.0883 |
| 17 | RAB1A | −0.09235 | 214,787 | 0.1104 |
| 18 | CTCF | −0.07860 | 236,696 | 0.1745 |
| 19 | QKI | −0.07410 | 244,326 | 0.2006 |
| 20 | HECTD2 | −0.06762 | 255,577 | 0.2429 |
| 21 | MIDN | −0.06706 | 256,576 | 0.2469 |
| 22 | VAPB | −0.03828 | 309,841 | 0.5089 |
| 23 | TRPS1 | −0.03086 | 324,412 | 0.5944 |
| 24 | UBE2J1 | −0.03054 | 325,054 | 0.5983 |
| 25 | ANKRD52 | −0.01265 | 361,316 | 0.8272 |
| 26 | DYRK1A | 0.00371 | 379,893 | 0.9489 |
| 27 | TCF12 | 0.00799 | 370,998 | 0.8904 |
| 28 | DCUN1D1 | 0.00866 | 369,617 | 0.8812 |
| 29 | TLE3 | 0.01128 | 364,187 | 0.8457 |
| 30 | ARID1A | 0.04000 | 306,486 | 0.4901 |
| 31 | BCL2L11 | 0.06746 | 255,858 | 0.2440 |
| 32 | MEX3A | 0.07353 | 245,319 | 0.2041 |
| 33 | BMF | 0.11406 | 183,484 | 0.0484 |
| 34 | VGLL4 | 0.11958 | 176,213 | 0.0385 |
| 35 | AMMECR1 | 0.16296 | 126,609 | 0.0047 |
| 36 | ARF4 | 0.24646 | 63,505 | <0.0001 |
Discussion
RCC is the most common type of renal malignancy and,
despite advances in therapeutic approaches, its prognosis remains
poor (24). Novel approaches in
RCC diagnosis and treatment are required. miRs have been reported
to be involved in a number of biological processes of tumorigenesis
(6), and miR replacement therapy
has been considered to be promising in cancer treatment (14).
Previous studies demonstrated that miR-222 is
critical in the pathogenesis of a number of diseases. The
upregulation of miR-222 in response to increased extracellular
signal-regulated kinase 1/2 activity exacerbates neointimal
hyperplasia in diabetes mellitus (25). Zhao et al (26) demonstrated that long non-coding
RNA Gas5 suppresses glioma malignancy by downregulating miR-222,
and the knockdown of miR-222 was correlated with tumor size and
survival rate in mice. A previous study performed by Tan et
al (19) demonstrated that
miR-222-3p promoted tumor cell proliferation and invasion and
inhibited apoptosis by targeting homeodomain-interacting protein
kinase 2 in gastric cancer. In colorectal cancer, miR-222 was
identified to promote cell migration and invasion through targeting
MIA3 (27). In addition, Zhang
et al (28) observed that
miR-222 inhibited tumor cell migration and invasion by
downregulating guanine nucleotide binding protein, a inhibiting
activity polypeptide 3 in hepatocellular carcinoma (28). Furthermore, miR-222 was identified
to serve as a biomarker in a number of types of cancer, including
lung (29), pancreatic (30), breast (31), bladder (32) and oral (33) cancer. miR-222 is involved in
promoting cancer and suppressing cancer in different tumors, and
its mechanism may be caused by different target genes in the
downstream. According to Kafshdooz et al (34), overexpressed miRs may function as
onco-miRs by downregulating tumor suppressor genes, whereas
downregulated miRs may serve as tumor suppressors by negatively
regulating oncogenes.
miR-222 has already been characterized as a
discriminator miR for RCC subtypes (35) and miR-222-3p has been demonstrated
to offer potential in distinguishing between normal tissues and RCC
subtypes (36). However, in the
present study, miR-222-3p was identified to promote the progression
of RCC. These results demonstrated that miR-222 has an important
function in RCC.
In the present study, miR-222-3p was observed to be
upregulated in RCC tissues and cell lines, compared with adjacent
normal renal tissues and the HK-2 cell line. The overexpression of
miR-222-3p promoted cell migration and invasion and suppressed
cellular apoptosis in RCC cell lines. Survival analysis
demonstrated that a higher expression of miR-222-3p was correlated
with poor prognosis in patients with RCC. However, a lack of target
gene investigation was a limitation of the present study, which is
to be performed in future investigations. Brodaczewska et
al(37) demonstrated that RCC
cells established from primary or metastatic disease may express
different molecules or the same molecules; however, in different
quantities. As 786-O and ACHN cells are different RCC cell lines,
the apoptotic rate between the two cell lines was somewhat
inconsistent. This may be caused by the different expression levels
of apoptosis-associated proteins. However the exact mechanism
leading to this difference remains to be fully elucidated, which
was a limitation of the present study.
In conclusion, the present study demonstrated that
miR-222-3p serves as an onco-miR in RCC, and a high expression of
miR-222-3p was associated with poor prognosis in patients with RCC.
These results suggested that miR-222-3p may serve as a biomarker
and therapeutic target in patients with RCC. However, there were
limitations to the present study, including the number of patients
included being insufficient, and further investigations are
required to elucidate the mechanism of miR-222-3p in RCC.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Basic
Research Project of Peking University Shenzhen Hospital (grant nos.
JCYJ2017001, JCYJ2017004, JCYJ2017005, JCYJ2017006, JCYJ2017007 and
JCYJ2017012; Shenzhen, China), the Clinical Research Project of
Peking University Shenzhen Hospital (grant no. LCYJ2017001), the
Science and Technology Development Fund Project of Shenzhen (grant
no. JCYJ20170307111334308; Shenzhen, China) and the Clinical
Research Project of Shenzhen Health Commission (grant no.
SZLY2018023; Shenzhen, China).
Availability of data and materials
The analyzed datasets generated during the present
study are available from the corresponding author on reasonable
request.
Authors’ contributions
YL, SY, YG and YC designed the experiments. LZ, JQ,
ZL, XP and JW performed the experiments, and analyzed and
interpreted the data. JX, WX and XG collected the renal cell
carcinoma specimens and follow-up data of formaldehye-fixed
paraffin-embedd specimens. HL analyzed the experimental data and
wrote the manuscript. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Peking University Shenzhen Hospital (Shenzhen, China),
and written informed consent for the use of the specimens for
investigation purposes was provided by all patients prior to
surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brugarolas J: Molecular genetics of
clear-cell renal cell carcinoma. J Clin Oncol. 32:1968–1976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rossi SH, Klatte T, Usher-Smith J and
Stewart GD: Epidemiology and screening for renal cancer. World J
Urol. 36:1341–1353. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karbasforooshan H, Roohbakhsh A and Karimi
G: SIRT1 and microRNAs: The role in breast, lung and prostate
cancers. Exp Cell Res. 367:1–6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kian R, Moradi S and Ghorbian S: Role of
components of microRNA machinery in carcinogenesis. Exp Oncol.
40:2–9. 2018.PubMed/NCBI
|
|
7
|
Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang
Y, Bao X, Du Q, Luo G, Liu K, et al: miR-122 promotes metastasis of
clear-cell renal cell carcinoma by downregulating Dicer. Int J
Cancer. 142:547–560. 2018. View Article : Google Scholar
|
|
8
|
Kabir TD, Ganda C, Brown RM, Beveridge DJ,
Richardson KL, Chaturvedi V, Candy P, Epis M, Wintle L, Kalinowski
F, et al: A microRNA-7/growth arrest specific 6/TYRO3 axis
regulates the growth and invasiveness of sorafenib-resistant cells
in human hepatocellular carcinoma. Hepatology. 67:216–231. 2018.
View Article : Google Scholar
|
|
9
|
Fang L, Deng Z, Shatseva T, Yang J, Peng
C, Du WW, Yee AJ, Ang LC, He C, Shan SW and Yang BB: MicroRNA
miR-93 promotes tumor growth and angiogenesis by targeting
integrin-β8. Oncogene. 30:806–821. 2011. View Article : Google Scholar
|
|
10
|
Jia YJ, Liu ZB, Wang WG, Sun CB, Wei P,
Yang YL, You MJ, Yu BH, Li XQ and Zhou XY: HDAC6 regulates
microRNA-27b that suppresses proliferation, promotes apoptosis and
target MET in diffuse large B-cell lymphoma. Leukemia. 32:703–711.
2018. View Article : Google Scholar
|
|
11
|
Ferreira AF, Calin GA, Picanco-Castro V,
Kashima S, Covas DT and de Castro FA: Hematopoietic stem cells from
induced pluripotent stem cells-considering the role of microRNA as
a cell differentiation regulator. J Cell Sci. 131:pii:
jcs2030182018. View Article : Google Scholar
|
|
12
|
Kim S, Lee E, Jung J, Lee JW, Kim HJ, Kim
J, Yoo HJ, Lee HJ, Chae SY, Jeon SM, et al: microRNA-155 positively
regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast
cancer. Oncogene. 37:2982–2991. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu W, Tan C, He Y, Zhang G, Xu Y and Tang
J: Functional miRNAs in breast cancer drug resistance. Onco Targets
Ther. 11:1529–1541. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hosseinahli N, Aghapour M, Duijf PHG and
Baradaran B: Treating cancer with microRNA replacement therapy: A
literature review. J Cell Physiol. 233:5574–5588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bhat NS, Colden M, Dar AA, Saini S, Arora
P, Shahryari V, Yamamura S, Tanaka Y, Kato T, Majid S and Dahiya R:
MicroRNA-720 regulates E-cadherin-αE-catenin complex and promotes
renal cell carcinoma. Mol Cancer Ther. 16:2840–2848. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dasgupta P, Kulkarni P, Majid S, Shahryari
V, Hashimoto Y, Bhat NS, Shiina M, Deng G, Saini S, Tabatabai ZL,
et al: MicroRNA-203 inhibits long noncoding RNA HOTAIR and
regulates tumorigenesis through epithelial-to-mesenchymal
transition pathway in renal cell carcinoma. Mol Cancer Ther.
17:1061–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han Z, Zhang Y, Sun Y, Chen J, Chang C,
Wang X and Yeh S: ERβ-mediated alteration of circATP2B1 and
miR-204-3p signaling promotes invasion of clear cell renal cell
carcinoma. Cancer Res. 78:2550–2563. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang M, Sun Y, Xu J, Lu J, Wang K, Yang
DR, Yang G, Li G and Chang C: Preclinical studies using miR-32-5p
to suppress clear cell renal cell carcinoma metastasis via altering
the miR-32-5p/TR4/HGF/Met signaling. Int J Cancer. 143:100–112.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan X, Tang H, Bi J, Li N and Jia Y:
MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2
to promote cell proliferation, invasion, and inhibits apoptosis in
gastric cancer. J Cell Biochem. 119:5153–5162. 2018. View Article : Google Scholar
|
|
20
|
Li Q, Shen K, Zhao Y, He X, Ma C, Wang L,
Wang B, Liu J and Ma J: MicroRNA-222 promotes tumorigenesis via
targeting DKK2 and activating the Wnt/β-catenin signaling pathway.
FEBS Lett. 587:1742–1748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamashita R, Sato M, Kakumu T, Hase T,
Yogo N, Maruyama E, Sekido Y, Kondo M and Hasegawa Y: Growth
inhibitory effects of miR-221 and miR-222 in non-small cell lung
cancer cells. Cancer Med. 4:551–564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Barata PC and Rini BI: Treatment of renal
cell carcinoma: Current status and future directions. CA Cancer J
Clin. 67:507–524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lightell DJ Jr, Moss SC and Woods TC:
Upregulation of miR-221 and -222 in response to increased
extracellular signal-regulated kinases 1/2 activity exacerbates
neointimal hyperplasia in diabetes mellitus. Atherosclerosis.
269:71–78. 2018. View Article : Google Scholar
|
|
26
|
Zhao X, Wang P, Liu J, Zheng J, Liu Y,
Chen J and Xue Y: Gas5 exerts tumor-suppressive functions in Human
Glioma cells by targeting miR-222. Mol Ther. 23:1899–1911. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao H, Cong X, Zhou J and Guan M:
MicroRNA-222 influences migration and invasion through MIA3 in
colorectal cancer. Cancer Cell Int. 17:782017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Yao J, Huan L, Lian J, Bao C, Li
Y, Ge C, Li J, Yao M, Liang L and He X: GNAI3 inhibits tumor cell
migration and invasion and is post-transcriptionally regulated by
miR-222 in hepatocellular carcinoma. Cancer Lett. 356:978–984.
2015. View Article : Google Scholar
|
|
29
|
Lv S, Xue J, Wu C, Wang L, Wu J, Xu S,
Liang X and Lou J: Identification of A panel of serum microRNAs as
biomarkers for early detection of lung adenocarcinoma. J Cancer.
8:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee C, He H, Jiang Y, Di Y, Yang F, Li J,
Jin C and Fu D: Elevated expression of tumor miR-222 in pancreatic
cancer is associated with Ki67 and poor prognosis. Med Oncol.
30:7002013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han SH, Kim HJ, Gwak JM, Kim M, Chung YR
and Park SY: MicroRNA-222 expression as a predictive marker for
tumor progression in hormone receptor-positive breast cancer. J
Breast Cancer. 20:35–44. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Puerta-Gil P, Garcia-Baquero R, Jia AY,
Ocaña S, Alvarez-Múgica M, Alvarez-Ossorio JL, Cordon-Cardo C, Cava
F and Sánchez-Carbayo M: miR-143, miR-222, and miR-452 are useful
as tumor stratification and noninvasive diagnostic biomarkers for
bladder cancer. Am J Pathol. 180:1808–1815. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang YA, Weng SL, Yang SF, Chou CH, Huang
WC, Tu SJ, Chang TH, Huang CN, Jong YJ and Huang HD: A
Three-MicroRNA signature as a potential biomarker for the early
detection of oral cancer. Int J Mol Sci. 19:pii: E7582018.
View Article : Google Scholar
|
|
34
|
Kafshdooz L, Pourfathi H, Akbarzadeh A,
Kafshdooz T, Razban Z, Sheervalilou R, Ebrahimi Sadr N, Khalilov R,
Saghfi S, Kavetskyy T, et al: The role of microRNAs and
nanoparticles in ovarian cancer: A review. Artif Cells Nanomed
Biotechnol. 1–7. 2018.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meo AD, Saleeb R, Wala SJ, Khella HW, Ding
Q, Zhai H, Krishan K, Krizova A, Gabril M, Evans A, et al: A
miRNA-based classification of renal cell carcinoma subtypes by PCR
and in situ hybridization. Oncotarget. 9:2092–2104. 2017.
|
|
36
|
Youssef YM, White NM, Grigull J, Krizova
A, Samy C, Mejia-Guerrero S, Evans A and Yousef GM: Accurate
molecular classification of kidney cancer subtypes using microRNA
signature. Eur Urol. 59:721–730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brodaczewska KK, Szczylik C, Fiedorowicz
M, Porta C and Czarnecka AM: Choosing the right cell line for renal
cell cancer research. Mol Cancer. 15:832016. View Article : Google Scholar : PubMed/NCBI
|