Introduction
Systemic lupus erythematosus (SLE) is a chronic
systemic autoimmune disease characterized by multiple
autoantibodies production, immune complex deposits and multiple
organ damage (1,2). Early diagnosis and appropriate
treatment may prevent serious clinical manifestations in patients
with SLE. Although the effects of therapeutic regimens for SLE have
made remarkable progress in recent years, the majority of patients
experience relapse, due to the ambiguity of pathogenesis (3). Therefore, efforts into investigating
the genetic and molecular abnormities of SLE are urgently required
and likely to be crucial for identifying new biomarkers for SLE
diagnosis.
Circular RNAs (circRNAs) are a unique form of RNAs,
which are composed primarily of transcripts from the exons
(4). Compared to linear RNAs,
circRNAs have a remarkable characteristic of non-canonical splicing
without free 3′or 5′ ends (5).
This characteristic enables circRNAs to resist degradation by RNase
R. In addition to having more stable characteristics, circRNAs
often exhibit tissue/developmental stage-specific expression
(6-9), which make them more appropriate to
be biomarkers compared with linear RNAs (6,10,11). Previous studies have identified
that circRNAs may serve as 'microRNA (miRNA) sponges' by
sequestering target microRNAs (miRNAs) and regulating RNA-binding
proteins to control gene transcription (12,13). Accumulating evidence has indicated
that circRNAs may be involved in neurological disorders,
atherosclerotic vascular disease risk, prion diseases, cancer and
autoimmune disease (14-17), supporting the hypothesis that
circRNAs have potential to be new diagnostic and prognostic
biomarkers, and novel therapy targets of diseases (18-20). However, little is known about
circRNAs in peripheral blood mononuclear cells (PBMCs) in human
SLE. The present study aimed to determine whether circRNAs in PBMCs
may be used as novel diagnosis biomarkers for SLE.
Materials and methods
Patient variables
A total of 79 patients with SLE were admitted from
the First Affiliated Hospital of Nanchang University from November
2016 to August 2018. All cases fulfilled the revised American
College of Rheumatology criteria for SLE (21). Disease activity was assessed by
the SLE disease activity index (SLEDAI) (22). Patients with SLE were classified
into inactive (SLEDAI, 0-9) and active (SLEDAI, ≥10) groups,
according to SLEDAI score. In the same time period, 62 healthy
subjects who had no inflammatory or autoimmune diseases and
genetically unrelated to the patients with SLE were selected as
healthy controls (HC). A total of 30 rheumatoid arthritis (RA)
patients admitted to the First Affiliated Hospital of Nanchang
University from November 2016 to August 2018 were used as disease
controls. All patients with RA fulfilled the revised American
College of Rheumatology criteria for RA (23).
The demographic characteristics of the study
population are demonstrated in Table
I. In the discovery set, 4 patients with new-onset SLE and 4
sex- and age-matched HCs were registered for microarray analysis.
An additional 30 patients with relapsed SLE and 20 HCs were
included in a validation testing set for the validation of
differentially expressed circRNAs and diagnostic model
construction. An independent cohort consisting of 45 SLE patients
(13 patients with new-onset SLE), 30 patients with RA and 38 HCs
were enrolled in a double validation testing set for clinical
evaluation of SLE diagnosis. Among the total 79 patients with SLE,
17 were newly diagnosed SLE patients with no history of
immunosuppressive drugs or corticosteroids treatment prior to
recruitment. The other patients with SLE were those with relapsed
SLE that had been previously diagnosed with SLE and had received
treatment prior to recruitment. The characteristics of patients
with new-onset and relapsed SLE are summarized in Table SI. In addition, the
characteristics of the patients with SLE and HCs in the validation
testing set and the characteristics of the patients with SLE, HCs
and patients with RA in the double validation testing set are
summarized in Tables SII and
SIII. The study was approved by
the Ethics Committee of the First Affiliated Hospital of Nanchang
University (approval no. 2014003) and was performed according to
the Declaration of Helsinki. Written informed consent was obtained
from all participants.
 | Table IDemographic characteristics of the
study population. |
Table I
Demographic characteristics of the
study population.
| Study set | Categories | SLE | HC | RA |
|---|
| Discovery | n | 4 | 4 | - |
| Females, n (%) | 4 (100.00) | 4 (100.00) | - |
| Age, mean (SD),
years | 30.50±17.53 | 34.00±7.21 | - |
| SLEDAI score, mean
(SD) | 10.25±6.76 | - | - |
| Validation
testing | n | 30 | 20 | - |
| Females, n (%) | 29 (96.67) | 17 (85.00) | - |
| Age, mean (SD),
years | 40.47±11.78 | 35.15±9.03 | - |
| SLEDAI score, mean
(SD) | 7.43±6.50 | - | - |
| Double validation
testing | n | 45 | 38 | 30 |
| Females, n (%) | 40 (89.79) | 32 (85.71) | 25 (83.33) |
| Age, mean (SD),
years | 36.08±16.77 | 35.94±8.48 | 53.04±12.41 |
| SLEDAI score, mean
(SD) | 9.75±6.50 | - | - |
PBMCs preparation and total RNA
extraction
PBMCs were isolated from EDTA anticoagulated blood
samples from each patient using Ficoll-Hypaque density gradients
(Sigma-Aldrich; Merck KGaA) at 25°C. The cells were frozen in
TRIzol® (Thermo Fisher Scientific, Inc.) at a
concentration of 106/ml and stored at −80°C. Following
extraction of total RNA from PMBCs with TRIzol® reagent
according to the manufacturer's protocol, the determination of RNA
quantity and quality was assessed by absorbance spectrometry,
measuring the absorbance ratios of A260/A280 and A260/A230 using a
NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.).
Microarray analysis
Sample labeling and array hybridization were
executed according to the manufacturer's protocol (Arraystar Inc.).
The specific protocol was described previously by Ouyang et
al (24). The microarray
analysis was performed by KangChen BioTech, Co., Ltd.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
cDNA was acquired by reverse transcription using a
PrimeScript™ RT reagent kit (Takara Bio, Inc.). The relative
expression of circRNAs was determined on an ABI 7500 Real-time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
SYBR® Premix Ex Taq™ II (Takara Bio, Inc.), with the
following PCR thermocycler protocol: Initial denaturation step at
95°C for 5 min, followed by 40 cycles of 95°C for 15 sec
(denaturation), 60°C for 1 min (annealing and elongation) and 72°C
for 2 min (final extension). The primers sequences are listed in
Table SIV. β-actin was used as
an internal control and the relative expression of circRNAs were
analyzed using the 2−∆∆Cq method (25) normalized to the internal control,
with ∆Ct=Cttarget-Ctreference.
Serum immunoglobulin G (IgG), complement
3 (C3), complement 4 (C4), C-reactive protein (CRP), autoantibody,
erythrocyte sedimentation rate (ESR), and routine urine and blood
analysis
The levels of serum C3, IgG, C4 and CRP were
detected by nephelometry methods according to the manufacturer's
protocol (IMMUNE800; Beckman Coulter, Inc.). Anti-extractable
nuclear antigens (ENAs) antibodies including
anti-Sjögren's-syndrome-related antigen A (anti-SSA),
anti-Sjögren's-syndrome-related antigen B (anti-SSB),
anti-tripartite motif-containing protein 21 (anti-Ro52), anti-Smith
(anti-Sm), anti-nuclear ribonuclear protein/Smith (anti-nRNP/Sm),
anti-ribosomal protein P (anti-RIB-P), and anti-nucleosome antibody
were determined using an immunoenzyme dot assay (Euroimmun AG)
according to the manufacturer's protocol. The results of the
anti-ENAs detection were presented as negative (−) and positive (+;
++; +++) by EuroBlot One (Euroimmun AG).
The serum levels of anti-dsDNA were determined using
a commercially available anti-dsDNA Enzyme Immunoassay Test kit
(cat. no. ED180401; Shanghai Kexin Biotech Co., Ltd.) with the
following protocol: Blood samples (5 ml) were collected from
patients with SLE in a tube without anticoagulant and centrifuged
at 2,000 × g for 10 min at normal temperature. The supernatants
were carefully collected and stored at 80°C until use.
Subsequently, the serum was used to detect the level of anti-dsDNA
according to the manufacturer's protocol. Sodium citrate
anti-coagulated blood, urine and K2-EDTA anti-coagulated blood were
used to detect ESR, and analyse routine urine and blood parameters
with a LBY-XC40 analyzer (Beijing Pulisheng Biotech Co., Ltd.),
URIT-500B analyzer (Guilin Youlite Biotech Co., Ltd.) and Sysmex
XE-2100 analyzer (Sysmex Corporation), respectively. Urine routine
parameters included pH, nitrite, glucose, vitamin C, specific
gravity, occult blood, proteinuria, bilirubin, urobilinogen, ketone
body, leucocyte lipase, cylindruria, hematuresis, pyuria and
crystalluria. Blood routine parameters included white blood cell
count, red blood cell count, hemoglobin, hematocrit, mean
corpuscular volume, mean corpuscular hemoglobin, mean corpuscular
hemoglobin concentration, red blood cells distribution width,
platelet count, mean platelet volume, plateletcrit, platelet
distributing width, numbers of lymphocytes, lymphocytes percentage,
numbers of moncytes, moncytes percentage, numbers of neutrophils,
neutrophils percentage, numbers of eosinophils, eosinophils
percentage, numbers of basophil, basophils percentage and platelet
large cell ratio.
Annotation for circRNA/miRNA
interaction
The circRNA/miRNA interaction was predicted using
Arraystar's home-made miRNA target prediction software based on
TargetScan (26) and miRanda
(27), and the differentially
expressed circRNAs within all the comparisons were annotated in
detail with the circRNA/miRNA interaction information.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism v5.0 software (GraphPad Software, Inc.) and SPSS
version 16.0 software (SPSS, Inc.). Differences in circRNAs
expression between two groups were analyzed using the Student's
t-test or nonparametric Mann-Whitney test. Kruskal-Wallis test
followed by Dunn's multiple comparison test was used for
statistical analysis between three groups. The correlation analyses
were performed using the nonparametric Spearman method. Receiver
operating characteristic (ROC) curves were performed to evaluate
the diagnostic value of circRNAs. P<0.05 was considered to
indicate a statistically significant difference.
Results
Dysregulated circRNAs expression
profiling
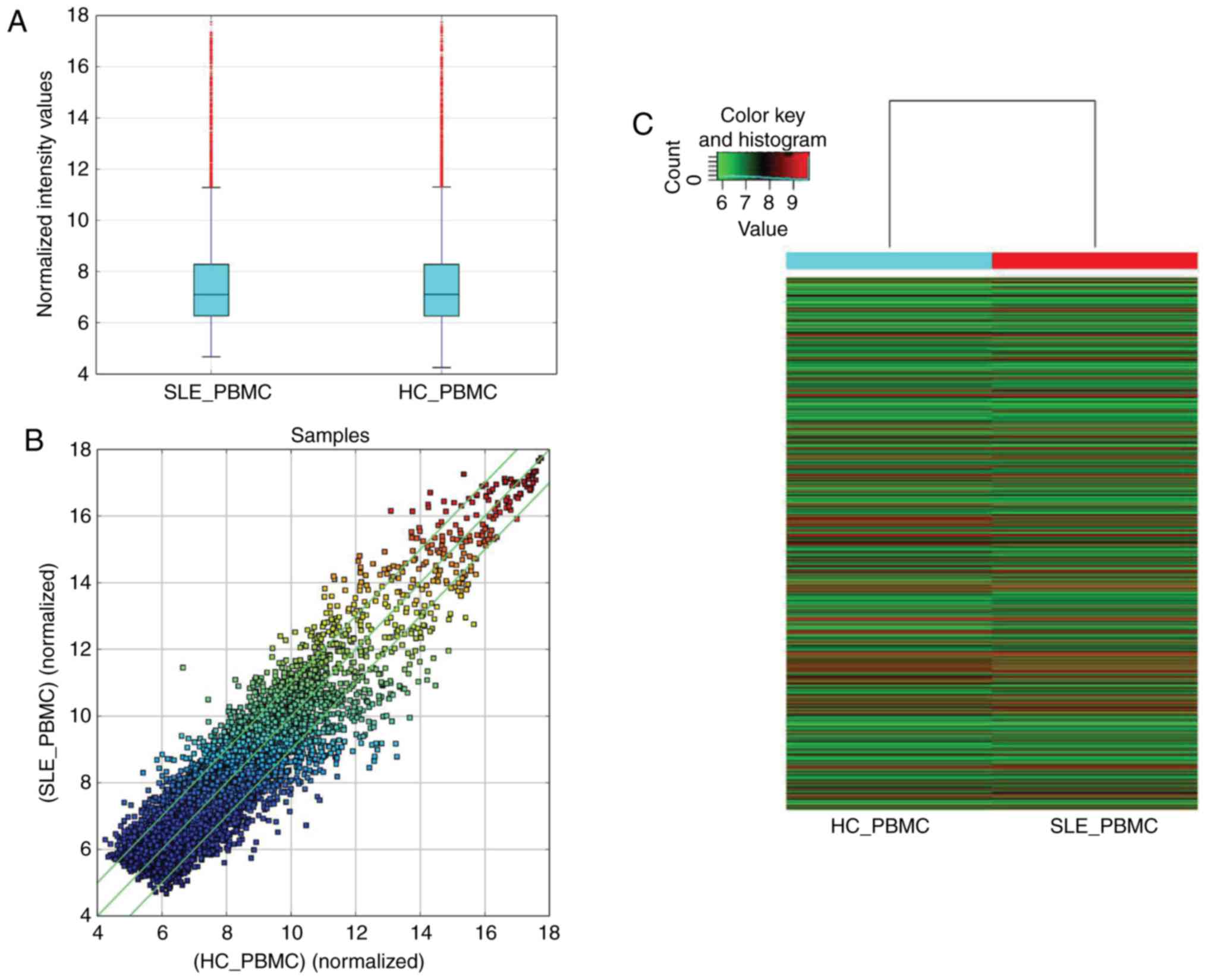
To explore the differentially expressed circRNAs in
SLE, PBMCs samples from 4 patients with SLE and 4 age- and
sex-matched HC were selected to perform microarray analysis using
an Arraystar Human circRNAs Microarray version 2.0. Based on the
criteria of a fold change >2.0 and P<0.05 (Fig. 1A and B), 1,603 circRNAs were
differentially expressed between the patients with SLE and HCs, of
which the top 30 differently expressed circRNAs (15 were
upregulated and 15 were downregulated) are listed in Table SV. Of the 1,603 differentially
expressed circRNAs, 838 circRNAs were upregulated and 765 circRNAs
were downregulated in the patients with SLE. A heatmap was
constructed to group the circRNAs based on their expression levels
among the samples (Fig. 1C).
Validation of circRNAs expression
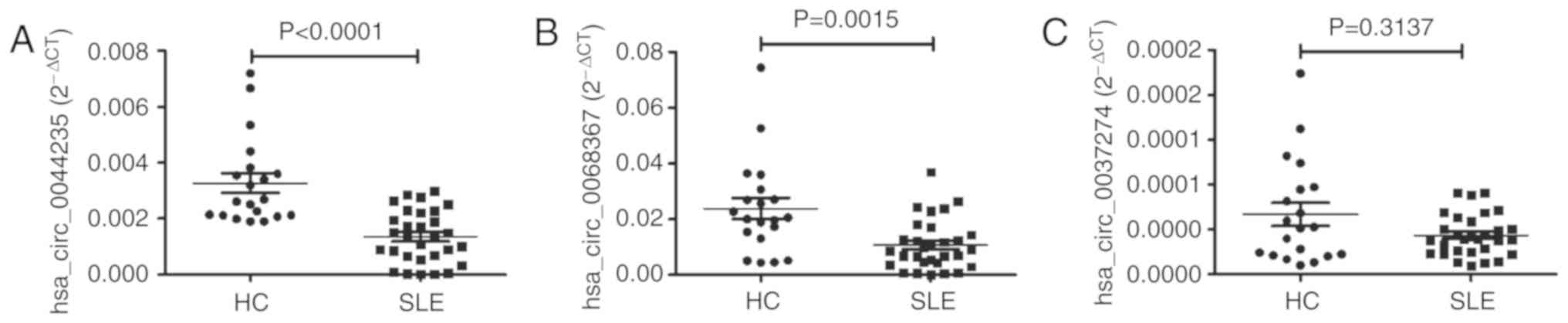
To verify the microarray data, 3 circRNAs
(hsa_circ_0068367; hsa_circ_0037274; and hsa_circ_0044235) were
selected for validation via RT-qPCR in a training set with 30
patients with SLE and 20 HCs. The 3 selected circRNAs were listed
in the top 45 (fold change >6) downregulated circRNAs, and had
been demonstrated to be downregulated in peripheral blood from
patients with SLE in our previous research (Luo et al,
unpublished data). Consistent with the results of the microarray
analysis, the levels of hsa_ circ_0044235 and hsa_circ_0068367 in
the PBMCs of patients with SLE were significantly decreased
compared with those of the HCs, while the expression levels of
hsa_circ_0037274 did not exhibit any remarkable differences between
patients with SLE and the HCs (Fig.
2).
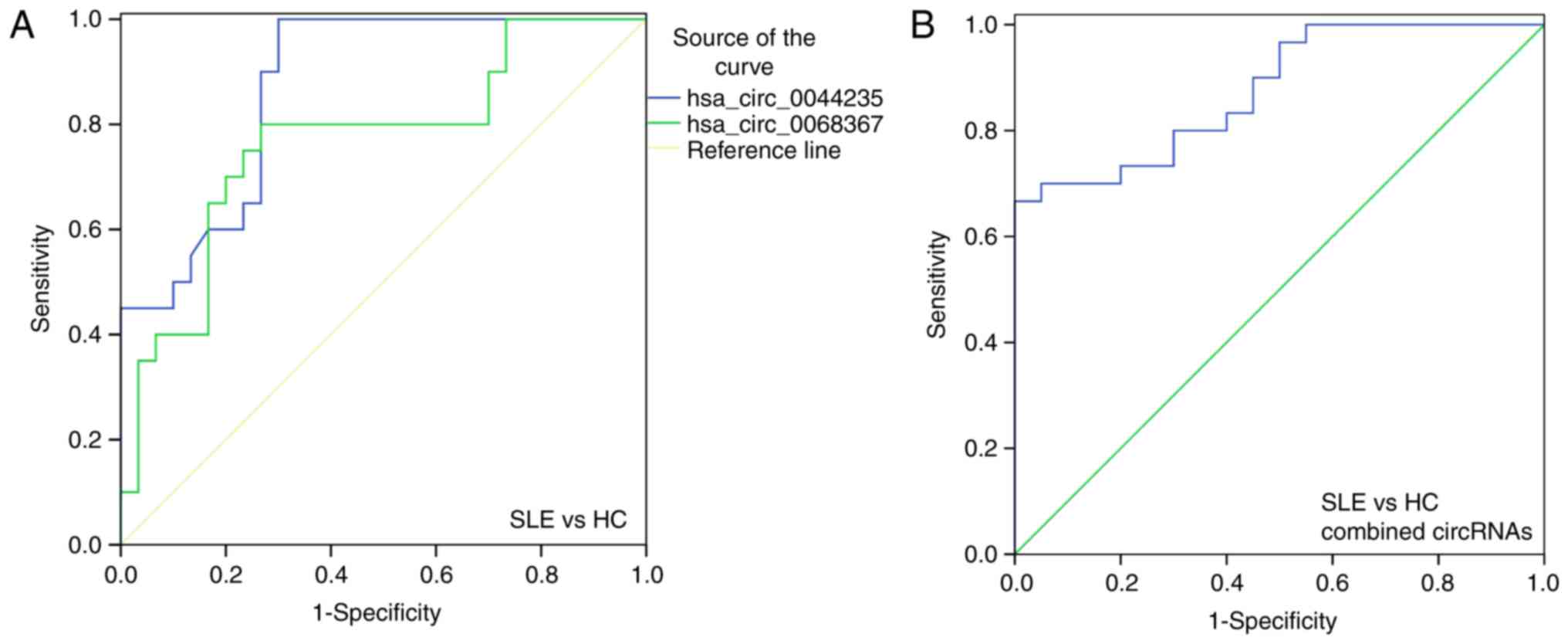
ROC curve analysis
To additionally evaluate the potential of these two
differentially expressed circRNAs (hsa_circ_0044235 and
hsa_circ_0068367) in SLE diagnosis, ROC curve analysis was
performed. ROC curves indicated that the levels of hsa_circ_0044235
and hsa_circ_0068367 in PBMCs were able to distinguish between
patients with SLE and the HCs. The highest area under the curve
(AUC) was hsa_circ_0044235 [AUC = 0.873; 95% confidence interval
(CI), 0.778-0.967; P<0.0001; sensitivity =70.00%, specificity
=100.00%; Fig. 3A], followed by
hsa_circ_0068367 (AUC =0.768; 95% CI, 0.629-0.907; P=0.0014;
sensitivity =73.33%; specificity =80.00%; Fig. 3A). The combined AUC of
hsa_circ_0044235 and hsa_circ_0068367 (AUC =0.876; 95% CI,
0.778-0.967; P<0.0001; sensitivity =70.00%; specificity
=100.00%; Fig. 3B) was identical
to the individual hsa_circ_0044235 AUC value (AUC =0.873).
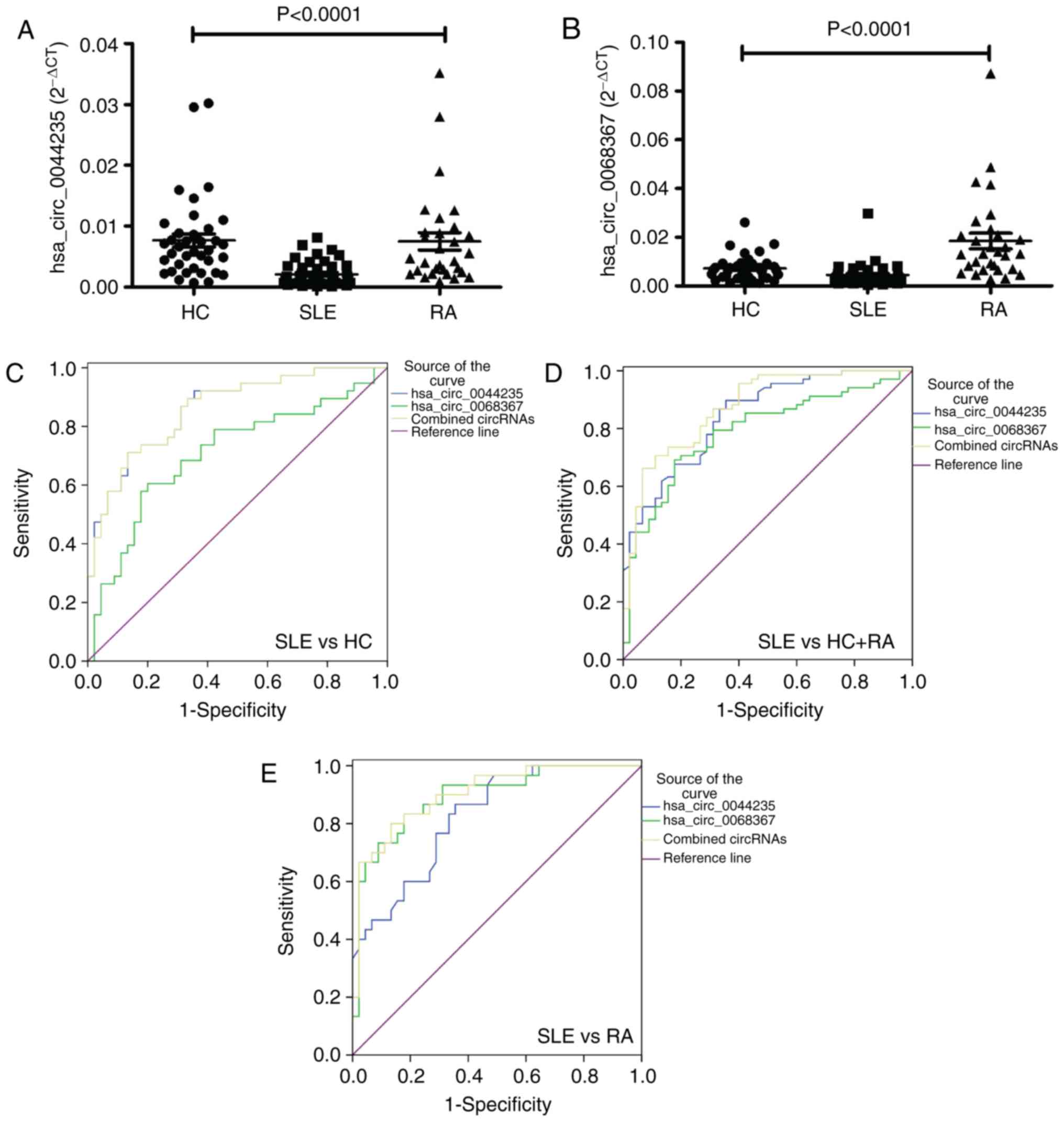
Double validation of the diagnostic value
of differentially expressed circRNAs
To additionally evaluate the value of the
aforementioned hsa_circ_0044235-only, hsa_ circ_0068367-only and
hsa_circ_0044235-hsa_circ_0068367 combination models in SLE
diagnosis, an independent validation testing set consisting of 45
patients with SLE, 30 patients with RA and 38 HCs were enrolled and
their circRNAs levels were determined. Similar to the training set,
the results demonstrated that the levels of hsa_circ_0044235 and
hsa_circ_0068367 were all significantly decreased in the patients
with SLE compared with the patients with RA and HCs (all
P<0.0001) (Fig. 4A and B). In
addition, ROC curves from the patients with SLE and HCs indicated
that the AUC values of hsa_circ_0044235, hsa_circ_0068367 and
hsa_circ_0044235-hsa_circ_0068367 were 0.861 (95% CI, 0.783-0.939;
P<0.0001; sensitivity =86.67%; specificity =71.05%), 0.707 (95%
CI, 0.592-0.822; P=0.0010; sensitivity =80.00%; specificity
=60.53%) and 0.860 (95% CI, 0.781-0.938; P<0.0001; sensitivity
=86.67%; specificity =71.05%), respectively (Fig. 4C).
A risk score analysis based on hsa_circ_0044235 and
hsa_circ_0068367 was subsequently performed in patients with SLE
and all controls (HCs and patient with RA) (Fig. 4D). The AUC for the risk score
based on hsa_circ_0044235 was 0.845 (95% CI: 0.774-0.916;
P<0.0001; sensitivity =66.44%; specificity =89.71%). The AUC for
the risk score based on hsa_circ_0068367 was 0.789 (95% CI,
0.705-0.874; P<0.0001; sensitivity =82.22%; specificity
=69.12%), and the AUC for the risk score based on the combination
of hsa_circ_0044235 and hsa_circ_0068367 was 0.874 (95% CI,
0.809-0.939; P<0.0001; sensitivity =93.33%; specificity
=66.18%). The risk score also significantly distinguished between
the patients with SLE and the disease controls (patients with RA)
(Fig. 4E); the AUC based on
hsa_circ_0044235 was 0.825 (95% CI, 0.734-0.916; P<0.0001;
sensitivity =64.44%; specificity =86.67%); the AUC based on
hsa_circ_0068367 was 0.893 (95% CI, 0.818-0.968; P<0.0001;
sensitivity =82.22%; specificity =83.33%); and the AUC based on the
combination of hsa_circ_0044235 and hsa_circ_0068367 was 0.903 (95%
CI, 0.834-0.972; P<0.0001; sensitivity =86.67%; specificity
=80.00%).
Association of hsa_circ_0044235 and
hsa_circ_0068367 levels in PBMCs with SLE clinical
characteristics
To determine whether the hsa_circ_0044235 and
hsa_circ_0068367 levels in the PBMCs from patients with SLE
reflected the severity of the disease, analysis was performed to
assess the correlation between the clinical features of SLE and the
levels of circRNAs (hsa_circ_0044235 and hsa_circ_0068367) in the
double validation testing set. The data suggested that the
expression levels of all confirmative circRNAs in the PBMCs from
patients with SLE were not associated with SLEDAI, CRP, ESR, C3 or
C4, which are established biomarkers used to measure the severity
and activity of SLE (Table II).
However, the level of hsa_circ_0044235 was negatively correlated
with the numbers of monocytes (rs =−0.33; P=0.02), and
the level of hsa_circ_0044235 was correlated with the level of
hsa_circ_0068367 (rs=0.42; P<0.01; Table II). Following the addition of the
data from the validation testing set, the level of hsa_circ_0044235
was identified to be correlated with the numbers of monocytes
(rs=−0.35; P<0.01) and the level of hsa_circ_0068367
(rs=0.52; P<0.01; Table II).
 | Table IIAssociations of hsa_circ_0044235 and
hsa_circ_0068367 levels in PBMCs with SLE clinical
characteristics. |
Table II
Associations of hsa_circ_0044235 and
hsa_circ_0068367 levels in PBMCs with SLE clinical
characteristics.
A, Double
validation testing set
|
|---|
hsa__circ_0044235
| hsa_circ_0068367
|
|---|
| Categories | rs | P-value | categories | rs | P-value |
|---|
| ESR | 0.04 | 0.79 | ESR | 0.07 | 0.64 |
| CRP | −0.12 | 0.46 | cRP | −0.13 | 0.41 |
| IgG | 0.16 | 0.29 | IgG | 0.07 | 0.64 |
| c3 | −0.23 | 0.14 | c3 | −0.17 | 0.27 |
| c4 | −0.27 | 0.07 | c4 | −0.12 | 0.46 |
| wbc | −0.16 | 0.30 | wbc | −0.06 | 0.69 |
| rbc | −0.07 | 0.64 | rbc | 0.02 | 0.90 |
| HGB | −0.09 | 0.56 | HGB | 0.02 | 0.91 |
| hct | −0.07 | 0.64 | hct | 0.03 | 0.84 |
| PLT | −0.12 | 0.45 | PLT | −0.07 | 0.64 |
| Neutrophils | −0.09 | 0.57 | Neutrophils | −0.12 | 0.45 |
| Lymphocytes | −0.19 | 0.22 | Lymphocytes | 0.02 | 0.91 |
| Monocytes | −0.33 | 0.02 | Monocytes | −0.21 | 0.17 |
| SLEDAI | −0.01 | 0.95 | SLEDAI | 0.10 | 0.52 |
|
hsa_circ_0068367 | 0.42 | <0.01 |
hsa_circ_0044235 | 0.42 | <0.01 |
B, Double
validation testing and validation testing sets
|
|---|
hsa_circ_0044235
| -
| -
| -
|
|---|
| categories | rs | P-value | - | - | - |
|---|
| Monocytes | −0.35 | <0.01 | - | - | - |
|
hsa_circ_0068367 | 0.52 | <0.01 | - | - | - |
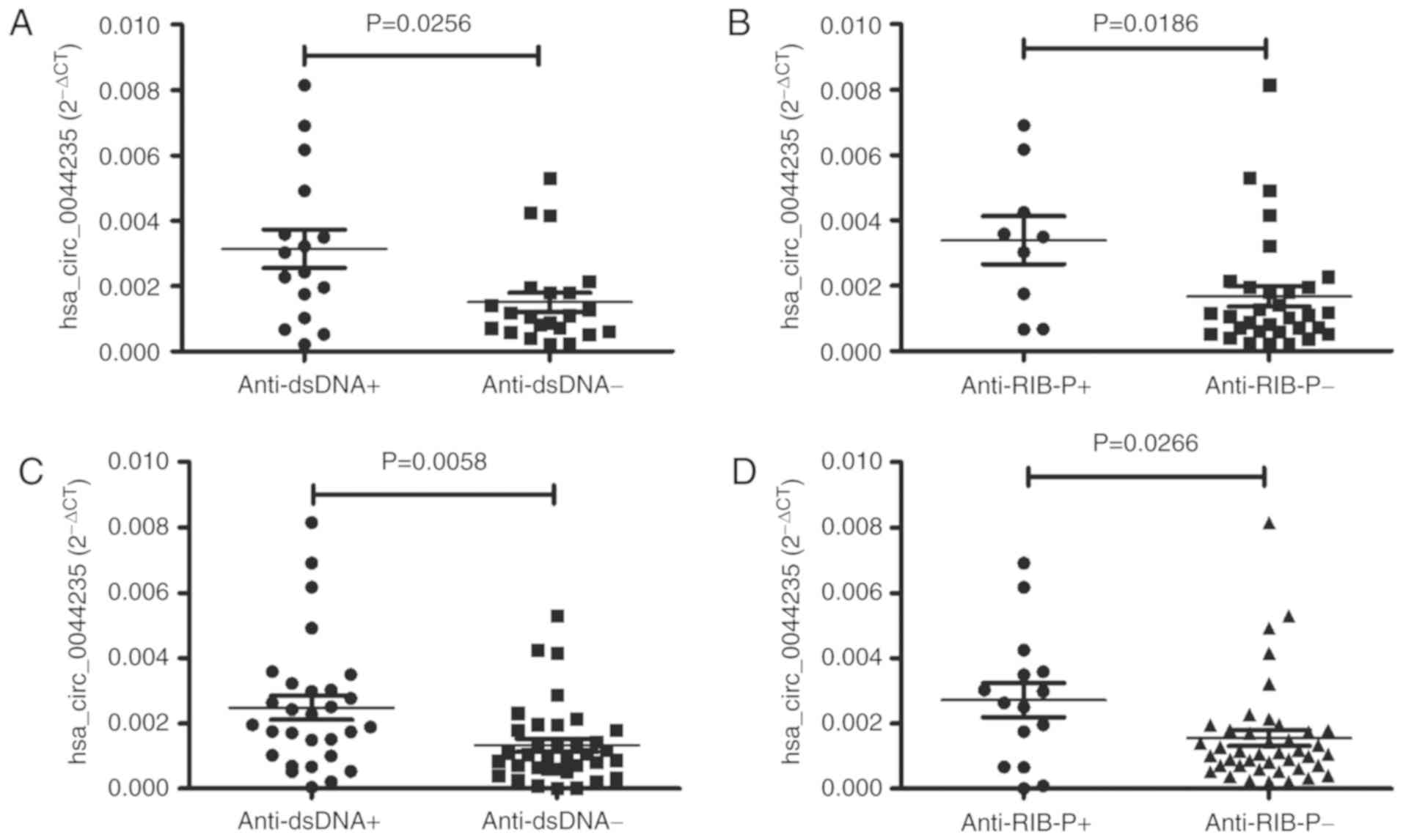
SLE is a prototypical systemic autoimmune disease
characterized by the production of autoantibodies, including
anti-dsDNA and anti-ENAs. Therefore, the correlations between the
levels of circRNAs (hsa_circ_0044235 and hsa_circ_0068367) and
anti-dsDNA, anti-Sm, anti-SSB, anti-Ro52, anti-SSA, anti-nRNP/Sm,
anti-nucleosome and anti-RIB-P were examined in patients with SLE
in the double validation testing set. These analyses demonstrated
that the levels of hsa_circ_0044235 were significantly increased in
the patients with SLE who were positive for anti-dsDNA and
anti-RIB-P antibodies compared with patients who were negative for
anti-dsDNA and anti-RIB-P antibodies (P=0.0256 and P=0.0186,
respectively; Fig. 5A and B). No
marked correlation was observed between the levels of circRNAs
(hsa_circ_0044235 and hsa_circ_0068367) and other autoantibodies
(data not shown). Following the addition of the data from the
validation testing, the levels of hsa_circ_0044235 remained
significantly increased in patients with SLE who were positive for
anti-dsDNA and anti-RIB-P antibodies compared with patients who
were negative for anti-dsDNA and anti-RIB-P antibodies (P=0.0058
and P=0.0266, respectively; Fig. 5C
and D).
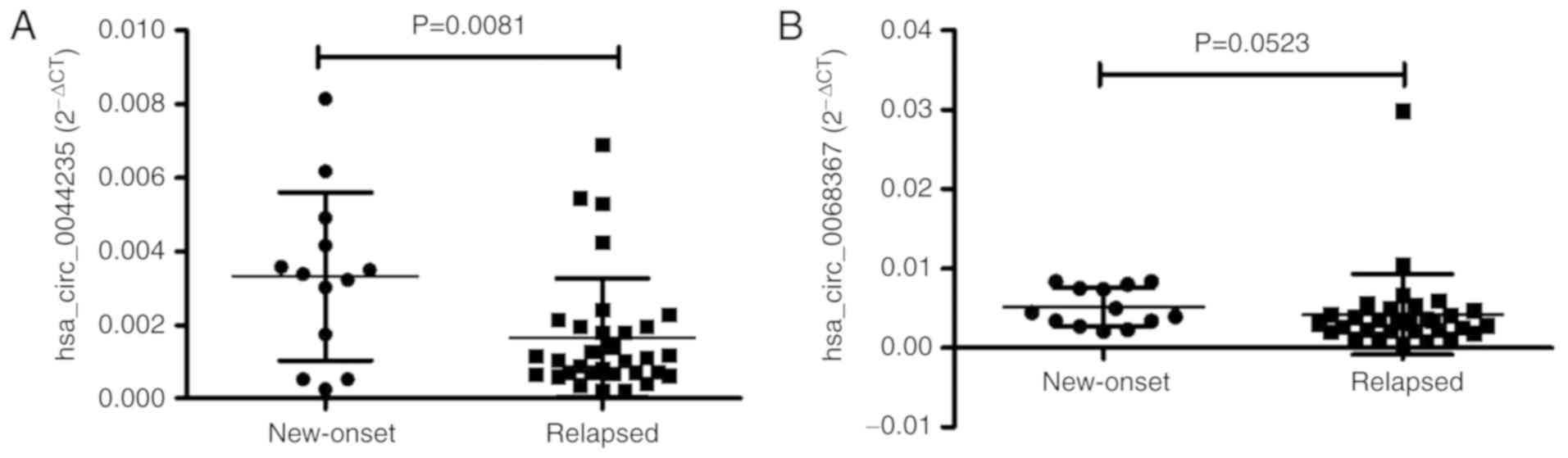
The levels of circRNAs (hsa_circ_0044235 and hsa_
circ_0068367) between patients with new-onset and relapsed SLE were
also compared. The results demonstrated that the level of
hsa_circ_0044235 was significantly increased in patients with
new-onset disease compared with patients with relapsed SLE
(P=0.0081; Fig. 6A), and the
level of hsa_circ_0068367 were slightly increased in patients with
new-onset disease, but the difference was not statistically
significant (P=0.0523; Fig.
6B).
Next, the clinical features of patients with SLE,
including lupus nephritis, cutaneous manifestations, alopecia,
arthritis, effusion, neuropathic lupus and fever were recorded, and
the correlations between these features and the levels of circRNAs
(hsa_circ_0044235 and hsa_circ_0068367) were analyzed. The results
indicated that no correlation was identified between the clinical
features of patients with SLE and the levels of circRNAs
(hsa_circ_0044235 and hsa_circ_0068367) (data not shown).
Target miRNA prediction of
hsa_circ_0044235 and hsa_ circ_0068367
The field of circRNAs research is relatively new,
and to the best of our knowledge, no studies have definitively
demonstrated the bio-function of hsa_circ_0044235 and
hsa_circ_0068367. It has been suggested that the miRNA response
elements (MREs) of circRNAs may bind target miRNA, and thereby
decrease miRNA-mediated post-transcriptional repression. To
determine the potential functions of hsa_circ_0044235 and
hsa_circ_0068367, target miRNA were predicted in the present study
by aligning prospective target miRNA with the MREs of
differentially expressed circRNAs using Arraystar's home-made miRNA
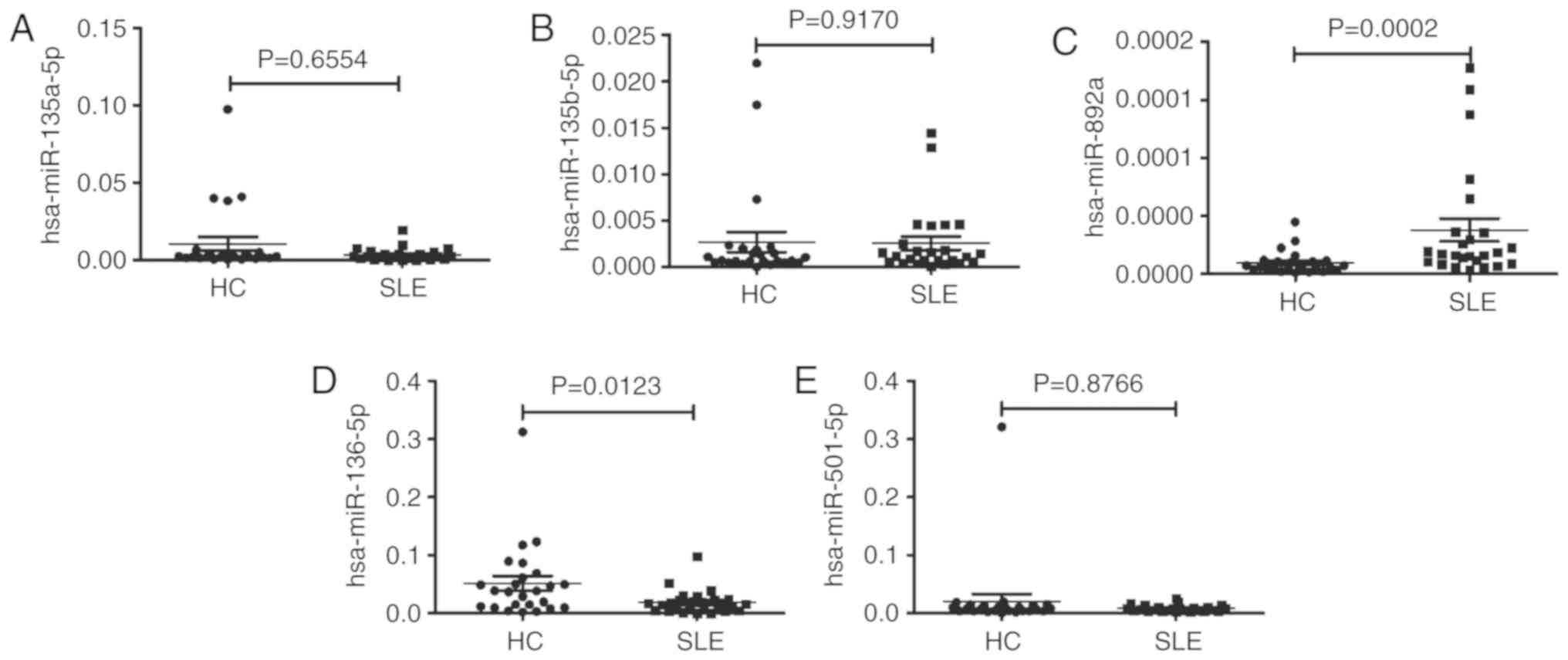
target prediction software. For hsa_circ_0044235, 3 putative miRNAs
targets (hsa-miR-892a; hsa-miR-135b-5; and hsa-miR-135a-5p) were
identified. For hsa_circ_006837, 3 putative miRNAs targets
(hsa-miR-136-5p; hsa-miR-501-5p; and hsa-miR-5696) were identified.
These putative miRNAs targets of hsa_circ_0044235 and
hsa_circ_0068367 were detected in PBMCs from SLE and HC. It was
identified that the levels of hsa-miR-135a-5p and hsa-miR-135b-5p
did not exhibit any remarkable differences between patients of SLE
and HC (Fig. 7A and B), while the
level of hsa-miR-892a was increased significantly in PBMCs from
patients of SLE compared with HC (Fig. 7C), suggesting that
hsa_circ_0044235 may serve a role in SLE by interacting with
hsa-miR-892a. In addition, the present study identified that the
level of hsa-miR-136-5p was decreased significantly in PBMCs from
the patients with SLE compared with the HCs (Fig. 7D), whereas the level of
hsa-miR-501-5p was not remarkably different between the patients
with SLE and the HCs (Fig. 7E).
However, the level of hsa-miR-5696 in PBMCs from the patients with
SLE was not detectable. Due to the decreased level of hsa_
circ_0068367 in the patients with SLE, hsa_circ_0068367 may serve a
role in SLE, by not as hsa-miR-136-5p, hsa-miR-501-5p or
hsa-miR-5696 sponges.
Discussion
SLE is a systemic autoimmune disease with indistinct
etiology. The clinical manifestations in patients with SLE are
heterogeneous and their disease progressions are unpredictable,
which has prompted studies to explore novel biomarkers to achieve
improved diagnoses and prognostic monitoring. CircRNAs are a
special class of endogenous RNAs and generally expressed in human
cells. It has been demonstrated that the expression levels of
circRNAs may be ≥10-fold compared with their corresponding linear
isomers (11). In addition,
circRNAs are more stable compared with long noncoding RNAs and
miRNAs in mammalian cells (28).
These characteristics give circRNAs the potential to be well-suited
biomarkers for human diseases. Shang et al (29) identified that hsa_circ_0005075 may
be used as a diagnostic biomarker for hepatocellular carcinoma and
that the expression level of hsa_circ_0005075 was correlated with
tumor size. Zhang et al (30) suggested that circ_101222 may be
used as a predictive biomarker for pre-eclampsia. Ouyang et
al (17) described 12
circRNAs that were differentially expressed in PBMCs from patients
with RA, and demonstrated that circRNA_104871 may be used to
diagnose RA. Li et al (31) described the comprehensive
expression profiles of circRNAs in plasma from patients with SLE
and initiated the development of circRNAs as novel non-invasive
biomarkers for SLE disease. However, the global expression profile
of patients with SLE and the clinical significance of circRNAs in
PBMCs from patients with SLE remain unknown.
To the best of our knowledge, the present study was
the first to characterize the expression profiles of circRNA by
comparing the transcriptome profiles of PBMCs from patients with
SLE and those from HCs using circRNAs microarray analysis. When
compared with HCs, the microarray data indicated a total of 1,603
circRNAs that were significantly dysregulated in patients with SLE.
By using circRNAs sequencing, Wang et al (32) and Miao et al (33) also identified a number of
differently expressed circRNAs in the PBMCs of patients with SLE.
These data may assist future patho-physiology studies in SLE and
assist in determining whether circRNAs in PBMCs may be used as
novel non-invasive biomarkers for SLE diagnosis.
In the present study, hsa_circ_0044235 and
hsa_circ_0068367 were confirmed to be significantly down-regulated
in the PBMCs of patients with SLE. It was also identified that the
levels of hsa_circ_0044235 and hsa_circ_ 0068367 in the PBMCs of
the patients with SLE were not correlated with SLEDAI, CRP, ESR, C3
or C4, indicating that these circRNAs may not be relevant
biomarkers for disease severity, disease activity or systemic
inflammation in SLE. However, it was revealed that the level of
hsa_circ_0044235 were associated with anti-dsDNA and anti-RIB-P.
Notably, the level of hsa_circ_0044235 was significantly increased
in patients with new-onset disease compared with patients with
relapsed conditions, indicating that hsa_circ_0044235 may serve a
role in SLE pathogenesis. However, it was demonstrated that the
level of hsa_circ_0044235 was correlated with the level of
hsa_circ_0068367 in patients with SLE, but no correlation was
identified between the levels of hsa_circ_0044235 and
hsa_circ_0068367 in HC. Therefore, we hypothesize that
hsa_circ_0044235 may interact with hsa_circ_0068367 directly or
indirectly in patients with SLE, although future studies are
required to verify this hypothesis.
Evidence from Zhang et al (34) have indicated that circRNAs in
PBMCs may be potential biomarkers for SLE. To explore whether
hsa_circ_0044235 and hsa_circ_0068367 in PBMCs may be used as
diagnostic biomarkers for SLE, their expression levels were
confirmed in a validation testing set and a double validation
testing set by ROC curves. In the validation testing set,
hsa_circ_0044235 had an AUC value of 0.873, hsa_circ_0068367 had an
AUC value of 0.768 for SLE diagnosis, and the combination of
hsa_circ_0044235 and hsa_circ_0068367 had an AUC value of 0.876. In
the double validation testing set, these circRNAs yielded parallel
AUC values in discriminating between the patients with SLE and the
HCs, indicating that the levels of hsa_circ_0044235 and
hsa_circ_0068367 in PBMCs have potential diagnostic value for SLE.
These diagnostic efficacies for SLE were quite comparable to those
previously described circRNAs (24,32,34). In addition, the efficacies of
hsa_circ_0044235 and hsa_ circ_0068367 in distinguishing SLE from
other autoimmune disease (RA) were assessed. The risk score
demonstrated that hsa_circ_0044235 and hsa_circ_0068367 also
distinguished between the patients with SLE and patients with
RA.
The principal types of PBMCs are lymphocytes and
monocytes. In the present study, the numbers of lymphocytes and
monocytes were compared between the patients with SLE and the HCs,
and it was identified that the numbers of lymphocytes were
significantly decreased in SLE group (P<0.05); no difference in
the numbers of monocytes between SLE and HCs groups was observed.
In addition, the correlation between the levels of circRNAs and the
numbers of monocytes and the numbers of lymphocytes was determined.
The results indicated that the level of hsa_circ_004235 was
negatively correlated with the number of monocytes, suggesting that
the difference in circRNAs expression was not the result of changes
in the number of lymphocytes or monocytes in the PBMCs of patients
with SLE.
Increasing evidence has demonstrated that circRNAs
may perform biological functions by binding to target miRNAs. Li
et al (35) suggested that
hsa_circ_0045272 served an important role in pathogenesis of SLE by
regulating the level of miR-6127 via directly binding. The present
study revealed that hsa_circ_0044235 and hsa_circ_0068367 were
significantly decreased in PBMCs of patients with SLE. To explore
the potential roles of hsa_circ_0044235 and hsa_circ_0068367 in
SLE, bioinformatics was employed in the present study to predict
the potential target miRNAs, and 3 putative target miRNAs were
obtained respectively for both hsa_circ_0044235 and
hsa_circ_0068367. Among these 6 putative target miRNAs,
hsa-miR-136-5p and hsa-miR-501-5p, putative targets of
hsa_circ_006837, have been demonstrated to be associated with SLE
(36,37). The functions of the other 4 miRNAs
(hsa-miR-892a; hsa-miR-135b-5p; hsa-miR-135a-5p; and hsa-miR-5696)
have not been described previously, to the best of our knowledge.
The levels of these putative target miRNAs in the PBMCs from
patients with SLE and HCs were then detected and compared. The data
demonstrated that the level of hsa-miR-892a in PBMCs was increased
significantly in the patients with SLE, suggesting that
hsa_circ_0044235 may serve a role in SLE by interacting with
hsa-miR-892a. As the levels of hsa-miR-136-5p and hsa_circ_0068367
in PBMCs were both decreased in patients with SLE, the role of
hsa_circ_0068367 in SLE may not be achieved by binding
hsa-miR-136-5p. Nevertheless, the exact mechanisms of
hsa_circ_0044235 and hsa_circ_0068367 in SLE require additional
investigation.
Several limitations of the present study should be
acknowledged. Firstly, the sample size of the patients with
new-onset SLE was relatively small, and the samples were sourced
from only one hospital, which may limit the universality of the
results. Secondly, circRNAs were not detected in the serum or
plasma of patients with SLE. Previous data have indicated that
exonic circRNAs in serum are not stable, with a half-life of less
than 15 sec (13), and that the
circRNAs in serum constitute only a fraction of the total RNA.
Thirdly, the specific role of circRNAs in SLE pathogenesis were not
fully explored.
In conclusion, differentially expressed circRNAs
were detected in PBMCs of patients with SLE. The data indicated
that the levels of hsa_circ_0044235 and hsa_circ_0068367 levels in
the PBMCs of patients with SLE were decreased and may represent
novel biomarkers for SLE diagnosis. However, the molecular
mechanisms and specific functions of these circRNAs in SLE require
additional investigation.
Supplementary Data
Abbreviations:
|
anti-dsDNA
|
anti-double-stranded DNA
|
|
anti-ENA
|
anti-extractable nuclear antigen
|
|
anti-nRNP/Sm
|
anti-nuclear ribonuclear
protein/Smith
|
|
anti-RIB-P
|
anti-ribosomal protein P
|
|
anti-Ro52
|
anti-tripartite motif-containing
protein 21
|
|
anti-SSA
|
anti-Sjögren's-syndrome-related
antigen A
|
|
anti-SSB
|
anti-Sjögren's-syndrome-related
antigen B
|
|
anti-Sm
|
anti-Smith
|
|
AUC
|
area under the curve
|
|
C3
|
complement 3
|
|
C4
|
complement 4
|
|
CRP
|
C-reactive protein
|
|
circRNAs
|
circular RNAs
|
|
ESR
|
erythrocyte sedimentation rate
|
|
HCs
|
healthy controls
|
|
IgG
|
immunoglobulin G
|
|
miRNA
|
microRNA
|
|
PBMCs
|
peripheral blood mononuclear cells
|
|
RA
|
rheumatoid arthritis
|
|
RT-qPCR
|
reverse transcription quantitative
polymerase chain reaction
|
|
ROC
|
receiver operating characteristic
|
|
SLE
|
systemic lupus erythematosus
|
|
SLEDAI
|
SLE disease activity index
|
Acknowledgments
We would like to acknowledge the help from Dr Rui Wu
from the Department of Rheumatology, The First Affiliated Hospital
of Nanchang University (Nanchang, China).
Funding
The present study was supported by the Key Research
and Development Plan Project of Jiangxi Province (grant. no.
20181BBG70013), the Science and Technology Plan Project of the
Education Department of Jiangxi Province (grant. no. GJJ170008),
the National Natural Science Foundation of China (grant. nos.
81360459 and 81660277), Jiangxi Provincial Natural Science
Foundation of China (grant. nos. 20151BAB215031 and
20171BAB205113), the Science and Technology Project of Health and
Family Planning Commission of Jiangxi Province of China (grant. no.
20165094) and the Foundation for Distinguished Young Scientists of
Jiangxi Province of china (grant. no. 20171BCB23087).
Availability of data and materials
The data used to support the results of this study
are available from the corresponding author upon request.
Authors' contributions
QL, LZ and XL performed the experiments. BF, YG, ZH
and JL analyzed and interpreted the data. QL and JL made
substantial contributions to the design supervision of the present
study, and wrote the manuscript. All authors reviewed the results
and approved the final version of the manuscript.
Ethics approval and consent to
participate
This study was authorized by the Ethics Committee of
the First Affiliated Hospital of Nanchang University. All
participants provided written informed consent prior to the
initiation of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rosenbaum JT and Silverman GJ: The
microbiome and systemic lupus erythematosus. N Engl J Med.
378:2236–2237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
D'Cruz DP, Khamashta MA and Hughes GR:
Systemic lupus erythematosus. Lancet. 369:587–596. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lerang K, Gilboe IM, Steinar Thelle D and
Gran JT: Mortality and years of potential life loss in systemic
lupus erythematosus: A population-based cohort study. Lupus.
23:1546–1552. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hentze MW and Preiss T: Circular RNAs:
Splicing's enigma variations. EMBO J. 32:923–925. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
10
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Szabo L, Morey R, Palpant NJ, Wang PL,
Afari N, Jiang C, Parast MM, Murry CE, Laurent LC and Salzman J:
Statistically based splicing detection reveals neural enrichment
and tissue-specific induction of circular RNA during human fetal
development. Genome Biol. 16:1262015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumar L, Shamsuzzama, Haque R, Baghel T
and Nazir A: Circular RNAs: The emerging class of non-coding RNAs
and their potential role in human neurodegenerative diseases. Mol
Neurobiol. 54:7224–7234. 2017. View Article : Google Scholar
|
|
15
|
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu
X, Li L, Yang B, Chen J, Chen S, et al: Identification of circular
RNA Hsa_ circ_0001879 and Hsa_circ_0004104 as novel biomarkers for
coronary artery disease. Atherosclerosis. 286:88–96. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R
and Li H: The emerging functions and roles of circular RNAs in
cancer. Cancer Lett. 414:301–309. 2018. View Article : Google Scholar
|
|
17
|
Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R,
Lou A, Zhu D, Shi GP and Yang M: Microarray expression profile of
circular RNAs in peripheral blood mononuclear cells from rheumatoid
arthritis patients. Cell Physiol Biochem. 42:651–659. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang F, Nazarali AJ and Ji S: Circular
RNAs as potential biomarkers for cancer diagnosis and therapy. Am J
Cancer Res. 6:1167–1176. 2016.PubMed/NCBI
|
|
19
|
Shao Y and Chen Y: Roles of circular RNAs
in neurologic disease. Front Mol Neurosci. 9:252016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu D and Xu AD: Mini review: Circular RNAs
as potential clinical biomarkers for disorders in the central
nervous system. Front Genet. 7:532016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hochberg MC: Updating the American College
of Rheumatology revised criteria for the classification of systemic
lupus erythema-tosus. Arthritis Rheum. 40:17251997. View Article : Google Scholar
|
|
22
|
Bombardier C, Gladman DD, Urowitz MB,
Caron D and Chang CH: Derivation of the SLEDAI. A disease activity
index for lupus patients. The Committee on Prognosis Studies in
SLE. Arthritis Rheum. 35:630–640. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS,
et al: The American Rheumatism Assocaition 1987 revised criteria
for the classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP
and Yang M: Using plasma circRNA_002453 as a novel biomarker in the
diagnosis of lupus nephritis. Mol Immunol. 101:531–538. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KG and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Enright A, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar
|
|
27
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that hsa_
circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular crcinoma development. Medicine (Baltimore).
95:e38112016. View Article : Google Scholar
|
|
30
|
Zhang YG, Yang HL, Long Y and Li WL:
Circular RNA in blood corpuscles combined with plasma protein
factor for early prediction of pre-eclampsia. BJOG. 123:2113–2118.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li H, Li K, Lai W, Li X, Wang H, Yang J,
Chu S, Wang H, Kang C and Qiu Y: Comprehensive circular RNA
profiles in plasma reveals that circular RNAs can be used as novel
biomarkers for systemic lupus erythematosus. Clin Chim Acta.
480:17–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X, Zhang C, Wu Z, Chen Y and Shi W:
CircIBTK inhibits DNA demethylation and activation of AKT signaling
pathway via miR-29b in peripheral blood mononuclear cells in
systemic lupus erythematosus. Arthritis Res Ther. 20:1182018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miao Q, Zhong Z, Jiang Z, Lin Y, Ni B,
Yang W and Tang J: RNA-seq of circular RNAs identified circPTPN22
as a potential new activity indicator in systemic lupus
erythematosus. Lupus. 28:520–528. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang MY, Wang JB, Zhu ZW, Li LJ, Liu RS,
Yang XK, Leng RX, Li XM, Pan HF and Ye DQ: Differentially expressed
circular RNAs in systemic lupus erythematosus and their clinical
significance. Biomed Pharmacother. 107:1720–1727. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu Li LJ, Zhao ZW, Tao W, Li SS, Xu BZ,
Wang SZ, Zhang JB, Wu MY, Leng JRX, et al: Circular RNA expression
profile and potential function of hsa_circ_0045272 in systemic
lupus erythematosus. Immunology. 155:137–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu D, Zhao H, Zhao S and Wang X: MicroRNA
expression profiles of peripheral blood mononuclear cells in
patients with systemic lupus erythematosus. Acta Histochem.
116:891–897. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hewagama A, Gorelik G, Patel D,
Liyanarachchi P, McCune WJ, Somers E, Gonzalez-Rivera T, Michigan
Lupus Cohort, Strickland F and Richardson B: Overexpression of
X-linked genes in T cells from women with lupus. J Autoimmun.
41:60–71. 2013. View Article : Google Scholar : PubMed/NCBI
|