Introduction
Patients with ischemic hearts exhibit progressive
myocardial remodeling, which is associated with massive
cardiomyocyte loss, and for which an ideal therapeutic approach has
yet to be identified (1,2). The pathological changes associated
with ischemic hearts primarily involve cardiomyocyte apoptosis and
myocardial fibrosis. The myocardial remodeling process includes
myocardial cell apoptosis, necrosis, subsequent tissue fibrosis
and, ultimately, heart failure, which is an inevitable consequence
when patients are not treated in a timely manner, or if the
patients cannot be revascularized (3). Substitution of apoptotic
cardiomyocytes by collagen and fibrous tissue is an important cause
of heart failure progression and myocardial remodeling. Stem cells
and gene therapy may represent new strategies for blocking cardiac
remodeling before it commences (4). It is advisable to prevent the
progression of heart failure that occurs as a result of myocardial
lesions by altering microRNA (miRNA) expression levels.
miRNAs are endogenous, non-coding small RNAs that
regulate target genes at the post-transcriptional level and have
been found to play an upstream regulatory role in the pathogenesis
of heart failure (5-7). The most abundant miRNA in cardiac
myocytes, and also the first miRNA to be implicated in heart
development, is miRNA-1 (8,9).
Accumulating evidence has demonstrated that cardiomyocyte apoptosis
is closely associated with the abnormal expression of miRNA-1.
Activation of caspases plays a central role in the execution phase
of cell apoptosis (10), and
caspase-3 is considered as the executioner of apoptosis. Previous
studies demonstrated that there is a crucial interrelationship
between caspase-3 activation and functional reserve in
cardiomyocytes (11). It was also
previously reported that miRNAs target caspase-3 in PANC-1 cells
(12).
The expression of the UPS components, such as E2
conjugating enzymes, E3 ubiquitin ligases, or subunits of the
proteasome, are either increased or decreased in cardiac disease.
The expression levels of the UPS components are high in the
impaired myocardium, and may be responsible for the degradation of
the majority of senescent cellular proteins, as well as playing a
key role in DNA repair (13-15). As defective protein degradation in
human cardiomyopathies has been established, recent studies have
started to address the potential underlying mechanisms. The overall
observed increase in the expression of ubiquitination machinery in
failing hearts may be in response to an increased protein burden,
which may be attributed to the increased protein synthesis that
accompanies the hypertrophic response, or to an excess of damaged
or modified proteins to be targeted for proteasomal degradation
(16,17). The association between these two
systems remains to be fully elucidated.
The aim of the present study was to investigate the
expression levels of miRNA-1 in mice with acute heart failure
induced by acute ischemia, elucidate the role of miRNA-1 in
myocardial remodeling and determine whether there is an association
with the ubiquitin-proteasome system (UPS) in the regulation of
heart remodeling following myocardial infarction (MI).
Materials and methods
MI model construction
A total of 48 male C57BL/6 mice, aged 6 weeks and
weighing 25-30 g, were used in the present study. All animals were
housed in an air-conditioned room (22±0.5°C) under a 12/12 h
light-dark cycle with access to standard laboratory food and water
ad libitum. After the mice were anesthetized by inhaling 2%
isoflurane, a skin incision was made over the left chest, followed
by tying a slipknot around one-third of the left anterior
descending artery. The sham group underwent the same surgical
procedure, except the suture placed under the left coronary artery
was not tied. A control vector (30 µl), an miRNA-1 antagomir
(30 µl, 1×109 TU/ml), an miRNA-1 lentiviral
vector (30 µl, 1×109 TU/ml) and bortezomib (30
µl, 0.5 mg/kg) were delivered via intramyocardial
injections, temporarily blanching the left ventricular (LV)
anterior wall. The 48 mice were divided into the following groups
(n=8 per group): i) Sham; ii) MI; iii) MI + control vector; iv) MI
+ miRNA-1 antagomir; v) MI + miRNA-1 lentiviral vector; and vi) MI
+ bortezomib. The protocol of the present study was approved by the
Animal Ethics Committee of Tianjin Union Medical Center.
Antagomir of miRNA-1 and overexpression
of miRNA-1
The antagomir method was used to block the
expression of miRNA-1. A sequence of antisense oligonucleotides was
designed (antagomir); the antisense nucleic acid was allowed to
interact with the target miRNA, and then this was specifically
intervened for downstream regulation. The miRNA-1 gene was
overexpressed using lentiviral vectors. The small RNA gene and its
side sequence were amplified from the genome and then cloned into
lentiviral vectors. The lentivirus was named pPS-EF1-copGFP-LCS,
and the reporter gene was green fluorescent protein (GFP); the
antagomir and lentivirus were injected into the MI area and
surrounding tissues. The antagomir and lentivirus were synthesized
by Shanghai Ji Ma Biotechnology Co., Ltd.
Western blot assay evaluation
Total protein was extracted from tissues to detect
the levels of caspase-3, 19S proteasome, 20S proteasome and
ubiquitin ligase E3 in target tissue via western blotting. The
proteins were separated via 12% SDS-PAGE, blotted and probed with
rabbit anti-caspase-3 (1:500; Sigma-Aldrich; Merck KGaA; cat. no.
C8487), rabbit anti-19S proteasome (1:1,000; Enzo Life Sciences;
cat. no. Q9SEI2), rabbit anti-20S proteasome (1:1,000; Abcam; cat.
no. ab22673) and rabbit anti-enzyme E3 (1:1,000; Abcam; cat. no.
ab84067). A Bradford assay (Bio-Rad Laboratories, Inc.) was used to
quantify protein concentrations. The blots were visualized using a
chemiluminescence system (Amersham Bioscience; GE Healthcare).
Determination of myocardial cell
apoptosis
Myocardial cell apoptosis was determined via
terminal deoxyribonucleotidyl transferase-mediated dUTP-biotin nick
end labeling (TUNEL) staining, as previously described (18,19).
Masson's trichrome staining
Masson's trichrome staining was performed for
histopathological observation. Following inhalation of 2%
isoflurane, the hearts of the mice were isolated, perfused with
normal saline followed by 4% para-formaldehyde for fixation,
dehydrated with ethanol, coronally sectioned into halves along the
long axis, embedded in paraffin blocks, consecutively cut into
5-µm sections, and then treated with commercial reagents for
Masson's trichrome staining. Muscle fibers were stained purple-red,
while collagen fibers were stained green-blue. Collagen volume
fraction, calculated from Masson's trichrome staining images, was
expressed as a percentage of the total LV myocardial volume.
Immunohistochemical staining
The expression of transforming growth factor (TGF)-β
in the process of MI was observed via immunohistochemistry, which
further revealed the degree of myocardial fibrosis and myocardial
remodeling. Paraffin-embedded heart sections were used for the
staining. Briefly, the sections were dewaxed, microwaved to
retrieve antigens, blocked with 5% BSA, incubated overnight at 4°C
with an isotype IgG control antibody, or with a rabbit anti-TGF-β
(1:1,000; Cell Signaling Technology, Inc.; cat. no. 3711), and then
hybridized with horseradish peroxidase-labeled secondary
antibodies, followed by a 3,30-diaminobenzidine (DAB) chromogenic
reaction. The brown DAB deposits were observed under a
microscope.
Measurement of infarct size
After 2 weeks, the heart was frozen at -80°C and
sliced transversely into 1-mmsections. The infarct size area (white
area) and LV area were measured digitally using the Image Pro Plus
software (Media Cybernetics, Inc.). Infarct size was expressed as a
percentage of the white area/LV area.
Echocardiographic examination
After 2 weeks, echocardiography was performed using
a high-resolution ultrasound imaging system. Anesthesia was induced
with isoflurane 2% in 100% oxygen in an induction chamber. The
sedated mice were studied using an echocardiography system (Sequoia
Acuson, 15-MHz linear transducer; Siemens AG). The cardiac
dimensions and function were assessed using M-mode
echocardiography. LV end-diastolic diameter (LVDD) and LV
end-systolic diameter were measured on the parasternal LV long axis
view. LV fractional shortening (LVFS) was measured via the short
axis view of the LV. LV ejection fraction (LVEF) and LV mass were
calculated using computer algorithms. All measurements represent
the mean of 5 consecutive cardiac cycles. All measurements were
performed in a blinded manner.
Statistical analysis
All values and are expressed as mean ± standard
deviation. Comparison between groups was subjected to analysis of
variance followed by Tukey's multiple comparison test. Two-sided
tests were used and P<0.05 was considered to indicate a
statistically significant difference. SPSS software (version 23.0;
IBM Corp.) was used for the data analysis.
Results
Expression levels of miRNA-1 following
miRNA-1 antagomir, miRNA-1 lentiviral vectors and bortezomib
treatment
miRNA-1 antagomir, miRNA-1 lentiviral vector and the
UPS proteasome blocker bortezomib were delivered via three separate
intramyocardial injections. After 2 weeks, the border area tissue
of the myocardial infarct from different groups was obtained in
order to detect the miRNA-1 expression levels. miRNA-1 antagomir
and miRNA-1 lentiviral models were successfully constructed and
assessed via reverse transcription PCR. miRNA-1 expression levels
were found to be signifi-cantly increased in the MI group
(P<0.01) compared with the sham group (Fig. 1B). miRNA-1 expression was
significantly decreased in the miRNA-1 antagomir group (P<0.01),
whereas it was increased in the miRNA-1 lentiviral group
(P<0.01). There was no significant difference between the
bortezomib and control groups (Fig.
1B).
Expression levels of the UPS components
following alterations in miRNA-1 expression and administration of
bortezomib
The expression levels of the main component of the
ubiquitin-proteasome, the 19S proteasome, were significantly
increased in the MI group (P<0.01) compared with the sham group
(Fig. 1A and C). The expression
levels of the 19S proteasome were significantly decreased in the
miRNA-1 antagomir group (P<0.05), increased in the miRNA-1
lentiviral group (P<0.05), and not significantly different in
the bortezomib group when compared with the control group (Fig. 1A and C).
The expression levels of the 20S proteasome in the
MI group were significantly increased (P<0.01) compared with
those in the sham group (Fig. 1A
and D). In addition, the 20S proteasome expression levels were
significantly decreased in the miRNA-1 antagomir group (P<0.01),
increased in the miRNA-1 lentiviral group (P<0.05), and also
decreased in the bortezomib group (P<0.01) when compared with
the control group (Fig. 1A and
D).
The ubiquitin ligase E3 expression levels in the MI
group were significantly increased (P<0.01) compared with those
in the sham group (Fig. 1A and
E). E3 expression was signifi-cantly decreased in the miRNA-1
antagomir group (P<0.01), significantly increased in the miRNA-1
lentiviral group (P<0.01), and not significantly different in
the bortezomib group compared with the control group (Fig. 1A and E).
TUNEL staining following interference
with the miRNA-1 expression levels and inhibition of the UPS
The number of apoptotic cells observed via TUNEL
staining was significantly elevated in the MI group (P<0.01)
compared with that in the sham group (Fig. 2A and B). The extent of apoptosis
was significantly decreased in the miRNA-1 antagomir group
(P<0.05), and there was no significant difference in the
bortezomib group compared with in the control group. The extent of
apoptosis in the miRNA-1 lentiviral group was significantly
increased (P<0.05) compared with the control group (Fig. 2A and B).
Caspase-3 expression following
interference with the miRNA-1 expression levels and inhibition of
the UPS
The expression levels of caspase-3 during MI were
evaluated using western blotting, which further demonstrated the
degree of myocardial cell in the different groups. Caspase-3
expression levels in the MI area were markedly increased in the MI
group (P<0.01) compared with the sham group (Fig. 2C). Caspase-3 expression levels in
the miRNA-1 antagomir group were markedly decreased (P<0.05),
and there was no significant difference in the bortezomib group
compared with the control group. The caspase-3 expression levels in
the miRNA-1 lentiviral group were significantly higher (P<0.05)
compared with those in the control group (Fig. 2C).
Masson's staining following interference
with miRNA-1 expression and administration of bortezomib
Masson's trichrome staining was used to assess the
degree of myocardial fibrosis. Microscopic examination revealed
that the myocardial tissue was purple-red in color, whereas the
fibrous tissue was green-blue. The myocardial fibers in the sham
operation group were arranged in a regular manner; there were no
abnormalities in the myocardial interstitium or myocardial
intravascular region. Collagen hyperplasia, myocardial fiber
disorder, and the proportion of green-blue-stained collagen was
markedly increased in the MI group (P<0.01) compared with those
areas in the sham group (Fig. 3A
and B). Collagen hyperplasia and myocardial fiber disarray were
markedly decreased in the miRNA-1 antagomir (P<0.01) and
bortezomib (P<0.05) groups, but the bortezomib group exhibited
fewer changes. Furthermore, collagen hyperplasia and myocardial
fiber disarray were observed in the miRNA-1 lentiviral group. The
fractional collagen volume in the miRNA-1 lentiviral group was
significantly higher (P<0.01) compared with that in the control
group (Fig. 3A and B).
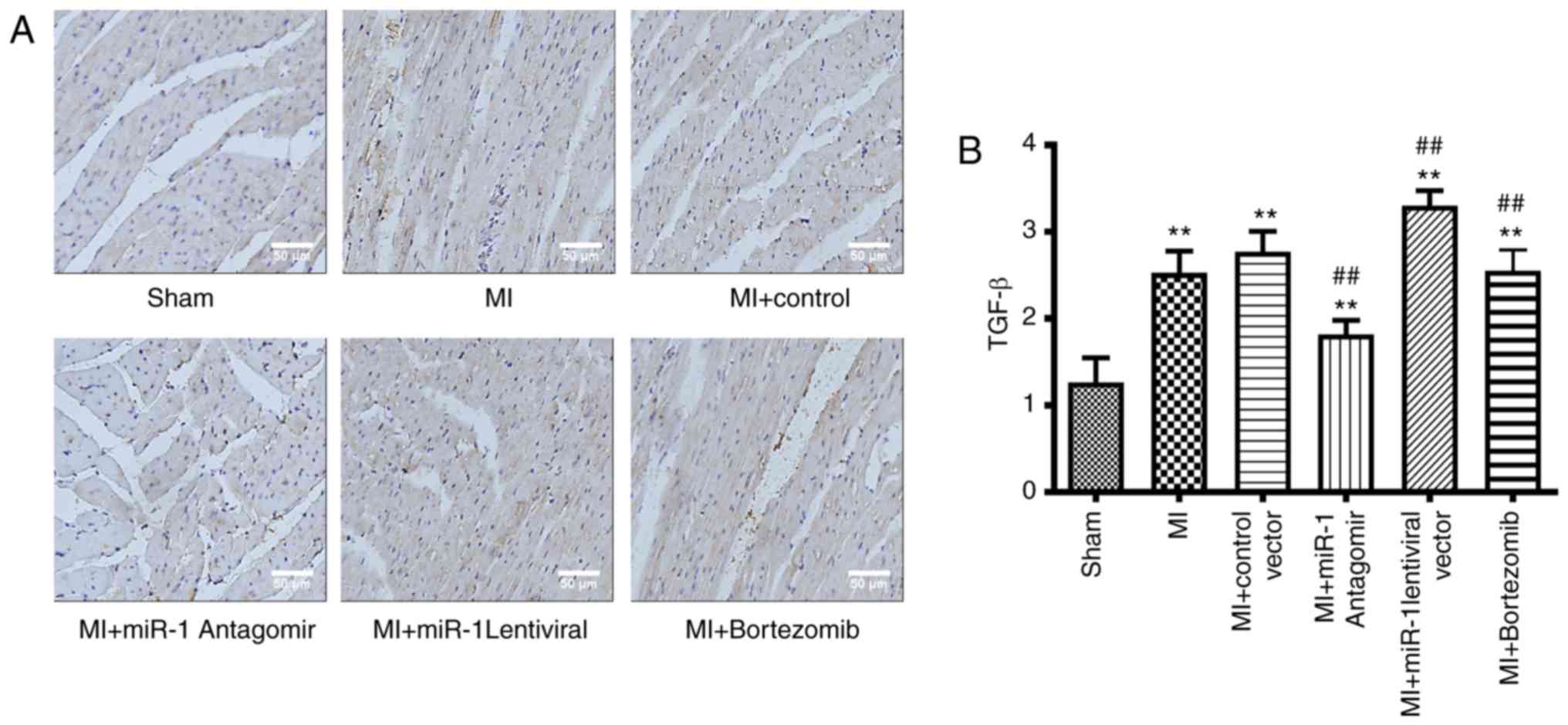
TGF-β expression levels following
interference with miRNA-1 expression levels and inhibition of the
UPS
The expression levels of TGF-β during the MI were
observed via immunohistochemistry, which further demonstrated the
degree of myocardial fibrosis and myocardial remodeling. The TGF-β
expression levels in the MI area were significantly increased in
the MI group (P<0.01) compared with those in the sham group
(Fig. 4A and B). The TGF-β
expression levels in the miRNA-1 antagomir group were significantly
decreased (P<0.01) compared with those in the control group, and
were also significantly decreased in the bortezomib group
(P<0.01). However, the TGF-β expression levels in the miRNA-1
lentiviral vector were significantly higher (P<0.01) compared
with those in the control group (Fig.
4A and B).
miRNA-1 antagomir decreases infarct size
in mice following MI
Infarct size was markedly higher in the MI group
(P<0.01) compared with the sham group (Fig. 5A and B). Infarct size was markedly
decreased in the miRNA-1 antagomir group (P<0.01), but was
significantly larger in the miRNA-1 lenti-viral group (P<0.01).
There was no significant difference in the infarct size between the
bortezomib and control groups (Fig.
5A and B).
Cardiac function changes following
interference with miRNA-1 expression levels and inhibition of the
UPS
The echocardiographic parameters revealed that LVEF
(Fig. 2A and B), LVFS (Fig. 6A and C), LV mass (Fig. 6A and D) and LVDD (Fig. 6A and E) in the MI group exhibited
significant changes (P<0.01) compared with the sham group. LVEF
(Fig. 6A and B) and LVFS
(Fig. 6A and C) were
significantly increased in the miRNA-1 antagomir group (P<0.01),
whereas LVEF (Fig. 6A and B) and
LVFS (Fig. 6A and C) were
decreased in the miRNA-1 lentiviral group (P<0.05), and were not
significantly different in the bortezomib group compared with the
control group.
LV mass (Fig. 6A
and D) and LVDD (Fig. 6A and E)
were significantly decreased in the miRNA-1 antagomir group
(P<0.05), increased in the miRNA-1 lentiviral group (P<0.05),
and mildly decreased in the bortezomib group (P<0.05) compared
with the control group.
Discussion
Previous studies (20-22) support the hypothesis that miRNAs
may play an important role in the upstream regulation of heart
failure progression; however, the role of miRNAs in physiological
and pathophysiological processes in the heart remains elusive.
The most abundant miRNA in cardiac myocytes, and the
first miRNA to be implicated in heart development, is miRNA-1.
miRNA-1 is encoded by two almost identical genes: miRNA-1-1 and
miRNA-1-2, which are located within introns 2 and 12 of the E3
ubiquitin-protein ligase (23).
Mice lacking the miR-1-2 gene develop various heart abnormalities.
Accumulating evidence suggests that cardiomyocyte apoptosis is
associated with abnormal expression of miRNA-1. miRNAs that are
present in the early phases of myocardial ischemia may be
associated with cell death and oxidative stress (24). Transplanting miRNA-1-transfected
embryonic stem cells (miRNA-1-1-ES) into the border zone of the
infarct in mice significantly improved cardiac function (25). Cheng et al (26) revealed that serum miRNA-1 level
increased rapidly following acute MI, reaching a peak value at 6 h,
indicating a strong positive correlation between serum miRNA-1
levels and myocardial infarct size. Besser et al (27) reported that the miR-1/133a
clusters were a prerequisite for maintaining specific functions in
heart electrophysiology and may affect electrical conduction.
The UPS involves two steps: First, covalent
attachment of ubiquitin to a target protein occurs via a cascade of
chemical reactions catalyzed by the ubiquitin-activating enzyme
(E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases
(E3). Second, the combination of ubiquitin proteins is recognized
and degraded by the 26S proteasomes, and this is a principal
pathway for the degradation of certain abnormal intracellular
proteins (14,28,29). The 26S proteasome is composed of a
20S core and two 19S regulatory complexes. The UPS plays an
important role in the removal of damaged proteins involved in the
regulation of inflammation, cell proliferation and differentiation,
signal transduction, transcriptional regulation, apoptosis and DNA
repair, as well as other biological functions (30,31). Bortezomib is a dipeptide boronate
protea-some inhibitor that reversibly binds to and inhibits the 20S
proteasome (32,33).
There are a number of observations suggesting that
certain E3-ligases play a key role in myocardial ischemia, and that
cardiomyocyte apoptosis is closely associated with the abnormal
expression of miRNA-1 (6,17). For example, mice that were
deficient in a co-chaperone with ubiquitin ligase properties,
exhibited an accelerated age-associated pathophysiological
phenotype and increased susceptibility to ischemia/reperfusion
injury (34,35). miRNA-1 is located within introns 2
and 12 of the E3 ubiquitin-protein ligase. It is possible that the
interaction between miRNA and UPS may be involved in the
development of cardiovascular disease and myocardial remodeling
(36-38); however, this interaction and its
possible effects on the cardiovascular system require further
investigation. Studies that assess the effect of miRNA-1 on the UPS
in the development of heart failure and cardiac remodeling are
currently limited.
In the present study, the expression levels of
miRNA-1 increased following MI; in addition, miRNA-1 antagomir
administration inhibited miRNA-1 expression, and miRNA-1 expression
levels increased following injection of the miRNA-1 lentiviral
vector, whereas the UPS proteasome blocker bortezomib was unable to
modulate miRNA-1 expression. The 19S proteasome, 20S proteasome and
ubiquitin ligase E3 in the miRNA-1 antagomir group decreased and
then markedly increased following administration of the miRNA-1
lentiviral vector, but only 20S proteasome expression was decreased
following delivery of the UPS proteasome blocker. These results
demonstrated that miRNA-1 is able to affect all components of the
UPS, while the UPS proteasome blocker bortezomib primarily affects
the 20S proteasome. The miRNA-1 antagomir and bortezomib both
alleviated the ultrastructural impairments, as demonstrated by a
decreased LVDD and LV mass, but the effects of the miRNA-1
antagomir were more noticeable and also increased LVEF and LVFS.
Furthermore, the opposite effect was observed in the miRNA-1
lentiviral vector group.
Cardiomyocyte apoptosis is one of the major
pathological mechanisms underlying MI. Blocking the apoptosis
process may prevent the loss of contractile cells and minimize
cardiac injury. Thus, a TUNEL staining assay was performed and
caspase-3 activity was measured using western blotting to
investigate the underlying molecular mechanisms responsible for the
improvement in cardiac function induced by administration of the
miRNA-1 antagomir. The results indicated that the miRNA-1 antagomir
inhibited cardiomyocyte apoptosis, as demonstrated by a decrease in
TUNEL-positive cardiomyocytes and decreased caspase-3 expression
levels, whereas the UPS proteasome blocker exerted no marked
effect. By contrast, miRNA-1 lentiviral vector administration
increased cardiomyocyte apoptosis and exacerbated myocardial
injury. Masson's trichrome staining and TGF-β expression levels may
be used to assess the degree of myocardial fibrosis and myocardial
remodeling (39). In the present
study, the decreased TGF-β expression levels in the miRNA-1
antagomir group compared with the control group indicate that
miRNA-1 antagomir can inhibit fibrosis after myocardial infarction,
while bortezomib did not exert noticeable effects. However, the
administration of the miRNA-1 lentiviral vector enhanced the
fibrotic responses.
A previous study reported that miRNA-1 levels are
likely increased in the early stages of heart failure, that
overexpression of miRNA-1 aggravates
H2O2-induced cardiomyocyte apoptosis, while
inhibition of miRNA-1 using antisense inhibitory oligonucleotides
results in marked resistance to H2O2
(40). It was previously
hypothesized that miRNA changes that occur in the early phase of
myocardial ischemia may be associated with cell death and oxidative
stress, while those miRNAs altered in the later phase may
contribute to post-infarct remodeling or function as compensatory
mechanisms (41,42). Naga Prasad et al (43) selected a surgical mouse model of
transverse aortic constriction to analyze alterations in eight
miRNAs upon initiation of cardiac dysfunction, and discovered that
the expression of miRNA-1 was decreased in end-stage heart failure.
Profiling miRNAs across various tissues revealed that miRNA-1,
let-7, miR-26a, miR-30c, miR-126-3p and miR-133 are highly
expressed in the murine heart (44). The UPS regulates fundamental cell
functions, including mitosis, DNA replication and repair, cell
differentiation and transcriptional regulation, as well as receptor
internalization, which all play crucial roles in heart biology
(29). A previous study (17) demonstrated that the UPS function
in patients with heart failure is altered, the protein degradation
rate is reduced, and the balance between myocardial protein
synthesis and degradation is disrupted, thus leading to an
accumulation of modified proteins. UPS proteasome blockers were
shown to inhibit nuclear factor-κB and exerted an anti-inflammatory
cardioprotective effect. In addition, inadequate coupling between
ubiquitination and proteasomal degradation appeared to be a major
causative factor behind the UPS functional deficit that contributes
to myocardial ischemic injury (45,46).
The present study indicated that the miRNA-1
antagomir exerted a significant protective effect on heart
function, decreasing cardiomyocyte apoptosis and alleviating
myocardial fibrosis and remodeling. The miRNA-1 antagomir exerted
more prominent effects compared with the UPS proteasome blocker
bortezomib. Studies on the association between miRNA-1 and UPS are
rare. A microRNA/ubiquitin ligase feedback loop was shown to
regulate slug-mediated invasion in breast cancer (47). Another previous study identified a
novel pathophysiological circuit in the heart, which caused
increased miR-199a expression levels and, subsequently, impairment
of the UPS that disrupted the miRNAs present in the early phase of
myocardial ischemia, which may be associated with cardiomyocyte
sarcomere structure and impairment via the release of asymmetric
dimethylarginine (36). It was
also revealed in the present study that miRNA-1 was able to
modulate ubiquitin ligase E3 expression levels. One of the possible
underlying molecular mechanisms that has been proposed is that
miRNA-1 exerts its effects at the regulated transcriptional and
post-transcriptional levels, thus controlling UPS expression and
markedly affecting the various components of the UPS. Furthermore,
the UPS was markedly involved in alleviating collagen hyperplasia
and decreasing apoptosis at the protein level during cardiac
remodeling.
In conclusion, the results of the present study
suggest that the ubiquitin proteasomes may be the most important
mediator of miRNA-1 in the regulation of heart remodeling. miRNA-1
was found to be associated with extensive expression of the UPS
components in the myocardium, resulting primarily in changes in
ubiquitin ligase E3 expression, leading to the accumulation of
proteins, formation of a large number of ubiquitin-positive protein
aggregates, increased myocardial cell apoptosis, and myocardial
fibrosis or remodeling. Conversely, the miRNA-1 antagomir exerted a
significant cardioprotective effect compared with the UPS
proteasome blocker bortezomib. Thus, the present study underlines
the fact that antisense silencing of miRNA-1 sequences that play a
regulatory role in the development of heart failure may be a
potential breakthrough in the treatment of heart failure.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81200158), the
Tianjin Health Bureau Key Project Fund (grant no. 16KG155) and the
Tianjin Chronic Disease Prevention and Treatment Key Project (grant
no. 16ZXMJSY00060).
Availability of materials and data
The datasets used or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LW and XQ designed the experiments. LW and YZ
drafted the manuscript. LW performed the experiments and collected
the data. XS, YL and YX analyzed the data and generated the
figures. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Animal Ethics
Committee of Tianjin Union Medical Center.
Patient consent for publication
Not applicable.
Competing interests
All the authors confirm that they have no competing
interests.
References
|
1
|
Senni M, Paulus WJ, Gavazzi A, Fraser AG,
Díez J, Solomon SD, Smiseth OA, Guazzi M, Lam CS, Maggioni AP, et
al: New strategies for heart failure with preserved ejection
fraction: The importance of targeted therapies for heart failure
phenotypes. Eur Heart J. 35:2797–2815. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Majmudar MD, Keliher EJ, Heidt T,
Leuschner F, Truelove J, Sena BF, Gorbatov R, Iwamoto Y, Dutta P,
Wojtkiewicz G, et al: Monocyte-directed RNAi targeting CCR2
improves infarct healing in atherosclerosis-prone mice.
Circulation. 127:2038–2046. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Varela A, Mavroidis M, Katsimpoulas M,
Sfiroera I, Kappa N, Mesa A, Kostomitsopoulos NG and Cokkinos DV:
The neuropro-tective agent rasagiline mesylate attenuates cardiac
remodeling after experimental myocardial infarction. ESC Heart
Fail. 4:331–340. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharp TE III, Schena GJ, Hobby AR,
Starosta T, Berretta RM, Wallner M, Borghetti G, Gross P, Yu D,
Johnson J, et al: Cortical bone stem cell therapy preserves cardiac
structure and function after myocardial infarction. Circ Res.
121:1263–1278. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagpal V, Rai R, Place AT, Murphy SB,
Verma SK, Ghosh AK and Vaughan DE: MiR-125b is critical for
fibroblast-to-myofibroblast transition and cardiac fibrosis.
Circulation. 133:291–301. 2016. View Article : Google Scholar
|
|
6
|
Hodgkinson CP, Kang MH, Dal-Pra S,
Mirotsou M and Dzau VJ: MicroRNAs and cardiac regeneration. Circ
Res. 116:1700–1711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Lang Y, Guo L, Pei Y, Hao S,
Liang Z, Su G, Shu L, Liu H, Huang C and Xu J: MicroRNA-323a-3p
promotes pressure overload-induced cardiac fibrosis by targeting
TIMP3. Cell Physiol Biochem. 50:2176–2187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee YE, Hong CY, Lin YL and Chen RM:
MicroRNA-1 participates in nitric oxide-induced apoptotic insults
to MC3T3-E1 cells by targeting heat-shock protein-70. Int J Biol
Sci. 11:246–255. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martinez EC, Lilyanna S, Wang P, Vardy LA,
Jiang X, Armugam A, Jeyaseelan K and Richards AM: MicroRNA-31
promotes adverse cardiac remodeling and dysfunction in ischemic
heart disease. J Mol Cell Cardiol. 112:27–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McIlwain DR, Berger T and Mak TW: Caspase
functions in cell death and disease. Cold Spring Harb Perspect
Biol. 5:pp. a0086562013, View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Communal C, Sumandea M, de Tombe P, Narula
J, Solaro RJ and Hajjar RJ: Functional consequences of caspase
activation in cardiac myocytes. Proc Natl Acad Sci USA.
99:6252–6256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park JK, Doseff AI and Schmittgen TD:
MicroRNAs targeting caspase-3 and-7 in PANC-1 cells. Int J Mol Sci.
19:E12062018. View Article : Google Scholar
|
|
13
|
Bozi LHM and Campos JC: Targeting the
ubiquitin proteasome system in diabetic cardiomyopathy. J Mol Cell
Cardiol. 109:61–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Powell SR, Herrmann J, Lerman A, Patterson
C and Wang X: The ubiquitin-proteasome system and cardiovascular
disease. Prog Mol Biol Transl Sci. 109:295–346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmidt M and Finley D: Regulation of
proteasome activity in health and disease. Biochim Biophys Acta.
1843.13–25. 2014.
|
|
16
|
Gilda JE and Gomes AV: Proteasome
dysfunction in cardiomy-opathies. J Physiol. 595:4051–4071. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han QY, Wang HX, Liu XH, Guo CX, Hua Q, Yu
XH, Li N, Yang YZ, Du J, Xia YL and Li HH: Circulating E3 ligases
are novel and sensitive biomarkers for diagnosis of acute
myocardial infarction. Clin Sci (Lond). 128:751–760. 2015.
View Article : Google Scholar
|
|
18
|
Zhang M, Sun D, Li S, Pan X, Zhang X, Zhu
D, Li C, Zhang R, Gao E and Wang H: Lin28a protects against cardiac
ischaemia/reperfu-sion injury in diabetic mice through the
insulin-PI3K-m TOR pathway. J Cell Mol Med. 19:1174–1182. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei L, Sun D, Yin Z, Yuan Y, Hwang A,
Zhang Y, Si R, Zhang R, Guo W, Cao F and Wang H: A PKC-beta
inhibitor protects against cardiac microvascular ischemia
reperfusion injury in diabetic rats. Apoptosis. 15:488–498. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Romaine SP, Tomaszewski M, Condorelli G
and Samani NJ: MicroRNAs in cardiovascular disease: An introduction
for clinicians. Heart. 101:921–928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wong LL, Wang J, Liew OW, Richards AM and
Chen YT: MicroRNA and heart failure. Int J Mol Sci. 17:5022016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bronze-da-Rocha E: MicroRNAs expression
profiles in cardiovascular diseases. Biomed Res Int.
2014.985408:2014.
|
|
23
|
Li J, Dong X, Wang Z and Wu J: MicroRNA-1
in cardiac diseases and cancers. Korean J Physiol Pharmacol.
18:359–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hao YL, Fang HC, Zhao HL, Li XL, Luo Y, Wu
BQ, Fu MJ, Liu W, Liang JJ and Chen XH: The role of microrna-1
targeting of mapk3 in myocardial ischemia-reperfusion injury in
rats undergoing sevoflurane preconditioning via the PI3k/Akt
pathway. . Am J Physiol Cell Physiol. 315:C380–C388. 2018.
View Article : Google Scholar
|
|
25
|
Glass C and Singla DK: MicroRNA-1
transfected embryonic stem cells enhance cardiac myocyte
differentiation and inhibit apoptosis by modulating the PTEN/Akt
pathway in the infarcted heart. . Am J Physiol Heart Circ Physiol.
301:H2038–H2049. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng Y, Tan N, Yang J, Liu X, Cao X, He
P, Dong X, Qin S and Zhang C: A translational study of circulating
cell-free microRNA-1 in acute myocardial infarction. . Clin Sci
(Lond). 119:87–95. 2010. View Article : Google Scholar
|
|
27
|
Besser J, Malan D, Wystub K, Bachmann A,
Wietelmann A, Sasse P, Fleischmann BK, Braun T and Boettger T:
MiRNA-1/133a clusters regulate adrenergic control of cardiac
repolarization. PLoS One. 9:pp. e1134492014, View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Portbury AL, Ronnebaum SM, Zungu M,
Patterson C and Willis MS: Back to your heart: Ubiquitin proteasome
system-regulated signal transduction. . J Mol Cell Cardiol.
52:526–537. 2012. View Article : Google Scholar
|
|
29
|
Pagan J, Seto T, Pagano M and Cittadini A:
Role of the ubiquitin proteasome system in the heart. . Circ Res.
112:1046–1058. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barac YD, Emrich F, Krutzwakd-Josefson E,
Schrepfer S, Sampaio LC, Willerson JT, Robbins RC, Ciechanover A,
Mohr FW, Aravot D and Taylor DA: The ubiquitin-proteasome system: A
potential therapeutic target for heart failure. . J Heart Lung
Transplant. 36:708–714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Divald A, Kivity S, Wang P, Hochhauser E,
Roberts B, Teichberg S, Gomes AV and Powell SR: Myocardial ischemic
preconditioning preserves postischemic function of the 26S
proteasome through diminished oxidative damage to 19S regulatory
particle subunits. . Circ Res. 106:1829–1838. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu N, Liu C, Li X, Liao S, Song W, Yang
C, Zhao C, Huang H, Guan L, Zhang P, et al: A novel proteasome
inhibitor suppresses tumor growth via targeting both 19S proteasome
deubiquitinases and 20S proteolytic peptidases. Sci Rep.
4:52402014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hussain AS, Hari P, Brazauskas R,
Arce-Lara C, Pasquini M, Hamadani M and D'Souza A: Changes in
cardiac biomarkers with bortezomib treatment in patients with
advanced cardiac amyloidosis. . Am J Hematol. 90:E2122015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Calise J and Powell SR: The ubiquitin
proteasome system and myocardial ischemia. . Am J Physiol Heart
Circ Physiol. 304:H337–H349. 2013. View Article : Google Scholar
|
|
35
|
Day SM: The ubiquitin proteasome system in
human cardiomy-opathies and heart failure. . Am J Physiol Heart
Circ Physiol. 304:H1283–H1293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Haghikia A, Missol-Kolka E, Tsikas D,
Venturini L, Brundiers S, Castoldi M, Muckenthaler MU, Eder M,
Stapel B, Thum T, et al: Signal transducer and activator of
transcription 3-mediated regulation of miR-199a-5p links
cardiomyocyte and endothelial cell function in the heart: A key
role for ubiquitin-conjugating enzymes. Eur Heart J. 32:1287–1297.
2011. View Article : Google Scholar
|
|
37
|
Wang H, Lai Y, Mathis BJ, Wang W, Li S, Qu
C, Li B, Shao L, Song H, Janicki JS, et al: Deubiquitinating enzyme
CYLD mediates pressure overload-induced cardiac maladaptive
remodeling and dysfunction via downregulating Nrf2. J Mol Cell
Cardiol. 84:143–153. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang F, Lerman A and Herrmann J:
Dysfunction of the ubiquitin-proteasome system in atherosclerotic
cardiovascular disease. . Am J Cardiovasc Dis. 5:83–100. 2015.
|
|
39
|
Yeh YH, Hsu LA, Chen YH, Kuo CT, Chang GJ
and Chen WJ: Protective role of heme oxygenase-1 in atrial
remodeling. . Basic Res Cardiol. 111:582016. View Article : Google Scholar
|
|
40
|
Tang Y, Zheng J, Sun Y, Wu Z, Liu Z and
Huang G: MicroRNA-1 regulates cardiomyocyte apoptosis by targeting
Bcl-2. . Int Heart J. 50:377–387. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong S, Cheng Y, Yang J, Li J, Liu X, Wang
X, Wang D, Krall TJ, Delphin ES and Zhang C: MicroRNA expression
signature and the role of microRNA-21 in the early phase of acute
myocardial infarction. . J Biol Chem. 284:29514–29525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yin C, Salloum FN and Kukreja RC: A novel
role of microRNA in late preconditioning: Upregulation of
endothelial nitric oxide synthase and heat shock protein 70. . Circ
Res. 104:572–575. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Naga Prasad SV, Duan ZH, Gupta MK,
Surampudi VS, Volinia S, Calin GA, Liu CG, Kotwal A, Moravec CS,
Starling RC, et al: Unique microRNA profile in end-stage heart
failure indicates alterations in specific cardiovascular signaling
networks. J Biol Chem. 284:27487–27499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. . Curr Biol. 12:735–739.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu C, Tian Y, Xu H, Pan B, Terpstra EM, Wu
P, Wang H, Li F, Liu J and Wang X: Inadequate
ubiquitination-proteasome coupling contributes to myocardial
ischemia-reperfusion injury. . J Clin Invest. 128:5294–5306. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Adams B, Mapanga RF and Essop MF: Partial
inhibition of the ubiquitin-proteasome system ameliorates cardiac
dysfunction following ischemia-reperfusion in the presence of high
glucose. . Cardiovasc Diabetol. 14:942015. View Article : Google Scholar
|
|
47
|
Manne RK, Agrawal Y, Bargale A, Patel A,
Paul D, Gupta NA, Rapole S, Seshadri V, Subramanyam D, Shetty P and
Santra MK: A microRNA/Ubiquitin ligase feedback loop regulates
slug-mediated invasion in breast cancer. . Neoplasia. 19:483–495.
2017. View Article : Google Scholar : PubMed/NCBI
|