Introduction
Uterine leiomyomas are benign smooth muscle cell
tumors originating from the myometrium (1). During growth, a leiomyoma compresses
the surrounding structures (the myometrium and connective tissue),
causing the progressive formation of a sort of pseudocapsule, rich
in collagen fibers (2),
characterized by an abnormal extracellular matrix (ECM) and high
interstitial fluid pressure (3).
Several factors, such as growth factors, retinoic acid, ovarian
hormones and vitamin D, are associated with tumor development
(4,5).
Protein phosphorylation is a common
post-translational modification (6) and usually occurs in serine,
threonine or tyrosine residues (7). In uterine leiomyomas, altered
protein phosphorylation is associated with the inhibition of
apoptosis and the promotion of cell survival (8). Several receptors, such as estrogen
receptor α (9) and receptor
tyrosine kinases (10), exhibit
increased phosphorylation levels in leiomyoma compared with the
myometrium, subsequently promoting tumor growth. Other receptors,
such as transforing growth factor β, mediate gene expression via
the phosphorylation of Smad proteins (11).
Oxidative stress is associated with several
gynecological disorders, such as uterine fibroids and endometriosis
(12). Fletcher et al
(13) reported that leiomyoma is
characterized by an impaired antioxidant cellular system,
suggesting a role of oxidative stress in its pathogenesis. In
tumors, the increase in reactive oxygen species (ROS) is balanced
by the upregulation of antioxidant systems (14). Pyruvate dehydrogenase kinase 1 is
a ROS sensor activated by the rise of oxidative stress, leading to
detoxification and cell survival (15). Oxidative stress stimulates
phosphorylation of mixed-lineage protein kinase 3 by ERK1/2,
enhancing the invasion of cancer cells (16). Identification of changes in
protein phosphorylation levels in leiomyoma associated with
oxidative stress may be useful for the understanding of the
physiopathology of the tumor.
The present study used immobilized metal affinity
chromatography (IMAC), two-dimensional gel electrophoresis (2-DE)
and mass spectrometry (MS) to analyze leiomyoma tissues. The aim
was to identify differentially phosphorylated proteins involved in
the suppression of oxidative stress and synthesis of ROS leading to
tumor growth.
Materials and methods
Patients
Tissues samples were obtained from ten premenopausal
patients who underwent hysterectomy for symptomatic uterine
leiomyomas. All procedures conformed with the Declaration of
Helsinki and were approved by the Review Board of the Institute for
Maternal and Child Health-IRCCS 'Burlo Garofolo' (Trieste, Italy).
All subjects involved signed a written informed consent. The median
age of patients was 44 years, with a range of 36-48 years. The
patients were recruited from January to Febuary 2019 at the
Institute for Maternal and Child Health-IRCCS 'Burlo Garofolo'
(Trieste, Italy), where all hysterectomies took place. Oncologic
patients, HIV, HBV, HCV-seropositive patients, and patients with
adenomyosis were excluded from the study. The patients had not
received hormonal therapy in the three months prior to surgery.
Tissue samples
Two samples were collected from each patient: One
from the central area of the leiomyoma and one from the unaffected
myometrium. All leiomyomas were confirmed histologically as benign
ordinary leiomyomas. Samples were stored at −80°C until proteomic
analysis was performed.
Phosphoprotein isolation and 2-DE
One hundred mg of myometrium and leiomyoma tissue
from ten patients were used for phosphoprotein isolation using the
Phosphoprotein Enrichment kit (Thermo Fisher Scientific, Inc.).
Tissues were homogenized in buffer [1% NP-40, 50 mM Tris-HCl (pH
8.0), NaCl 150 mM] with 1X Phosphatase Inhibitor Cocktail Set II
(EMD Millipore), 2 mM PMSF and 1 mM benzamidine. The concentration
of the supernatant was determined by Bradford assay. Tissue
homogenates were then diluted to a final concetration of 0.5 mg/ml
in lysis buffer provided by the Phosphoprotein Enrichment kit. Six
ml of final samples were used for isolation of the phosphoproteins,
according to the manufacturer's instructions.
For 2-DE analysis, a single patient-single gel
strategy was adopted, and the analyzes were performed as previously
described (17). For 2-DE
analysis, 250 µg of proteins from each sample were denatured
in 315 µl of dissolution buffer [7 M urea, 2 M thiourea, 4%
3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, 40 mM
Tris, 65 mM dithiothreitol (DTT) and 0.24% Bio-Lyte 3/10 (Bio-Rad
Laboratories, Inc.)] with a trace of bromophenol blue. ReadyStrip™
pH 4-7 18-cm immobilized pH gradient (IPG) strips (Bio-Rad
Laboratories, Inc.) were rehydrated in dissolution buffer at 50 V
for 12 h at 20°C, and isoelectric focusing (IEF) was performed in a
PROTEAN IEF Cell (Bio-Rad Laboratories, Inc.). After the IEF,
serial incubations were performed: first, the IPG strips were
equilibrated for 15 min in an equilibration buffer [6 M urea, 2%
SDS, 50 mM Tris-HCl (pH 8.8), 30% glycerol and 1% DTT] and then in
another equilibration buffer containing 4% iodoacetamide instead of
DTT. For the second dimension, the equilibrated IPG strips were
transferred to a 12% polyacrylamide gel.
After electrophoresis, gels were fixed in 40%
methanol and 10% acetic acid for 1 h, and then stained for 16 h
with SYPRO Ruby (Bio-Rad Laboratories, Inc.). 2-DE gels were
scanned with a Molecular Imager PharosFX System (Bio-Rad
Laboratories, Inc.). Double experimental replicates were performed
per sample. For all gels, molecular weights were determined by
comparison with Precision Plus Protein Prestained Standards
(Bio-Rad Laboratories, Inc.) covering a range 10-250 kDa, and
analyzed using the Proteomweaver 4.0 software (Bio-Rad
Laboratories, Inc.).
Western blotting
Phosphoprotein extracts (20 µg) from IMAC
columns used for 2-DE were separated by 12% SDS-PAGE and then
transferred to a nitrocellulose membrane. The western blotting
procedure for phosphoproteins was conducted as previously described
(18). The membrane was blocked
with 5% BSA in TBS/0.05% Tween-20 (TBST) for 2 h at room
temperature. After BSA saturation, the membrane was incubated
overnight at 4°C with primary rabbit polyclonal antibodies against
peroxiredoxin 2 (PRDX2; 1:800; cat. no. SAB2101878), protein
disulfide isomerase family A member 3 (PDIA3; 1:400; cat. no.
SAB2107799) and peroxiredoxin 4 (PDRX4; 1:500; cat. no. SAB4301759;
all Sigma-Aldrich; Merck KGaA). The membrane was washed three times
in TBST for 10 min, and then incubated for 90 min at 4°C with a
horseradish peroxidase-conjugated anti-rabbit immunoglobulin G
antibody (1:3,000; cat. no. G4018; Sigma-Aldrich; Merck KGaA).
Protein expression was visualized by chemiluminescence (SuperSignal
West Pico Chemiluminescent Substrate; Thermo Fisher Scientific,
Inc.), and the intensity of the signals was quantified by VersaDoc
Imaging System (Bio-Rad Laboratories, Inc.). The intensities of the
immunostained bands were normalized with the protein intensities
measured with Red Ponceau (Bio-Rad, Laboratories, Inc.) from the
same blot.
Trypsin digestion and MS analysis
Spots from 2-DE were digested and analyzed by MS, as
described by Ura et al (19). After 2-DE gel excision, the spots
were washed four times with 50 mM NH4HCO3 and
acetonitrile (ACN; Sigma-Aldrich; Merck KGaA) alternatively, and
dried under vacuum in a SpeedVac system. Three µl of 12.5
ng/µl sequencing grade modified trypsin (Promega
Corporation) in 50 mM NH4HCO3 were added to
gel spots for digestion (overnight at 37°C). Peptide extraction was
achieved by three changes of 50% ACN/0.1% formic acid (FA; Fluka).
Peptide mixtures were dried under vacuum and dissolved in 10
µl of 5% ACN/0.1% FA and 5 µl of each sample were
analyzed by liquid chromatography with tandem mass spectrometry
(LC-MS/MS) on a 6520 Q-TOF mass spectrometer (Agilent Technologies,
Inc.) coupled to a chip-based chromatographic interface.
Raw data files were converted into Mascot Generic
Format (MGF) files with MassHunter Qualitative Analysis Software
version B.03.01 (Agilent Technologies, Inc.) and searched with
Mascot Search Engine (version 2.2.4; Matrix Science) through the
Proteome Discoverer Software interface (version 1.4; Thermo Fisher
Scientific, Inc.). Spectra were searched against the human section
of the UniProt database (version July 2018, 95,057 sequences) using
the following parameters: Enzyme specificity was set to trypsin
with one missed cleavage allowed, while precursor and fragment ions
tolerance were set to 20 ppm and 0.05 Da, respectively.
Carbamidomethyl cysteine and oxidation of methionine were set as
fixed modification and variable modification, respectively. MS/MS
spectra containing <5 peaks or with a total ion count <50
were discarded. Proteins were considered as positive hits if, for
each protein, at least two unique peptides were identified with
high confidence (FDR <0.01%). For protein spots that did not
return any significant hit, a Peptide Mass Fingerprint (PMF) was
also performed with Mascot. All identified proteins were verified
to have phosphorylated residues in PhosphoSitePlus database
(www.phosphosite.org).
PANTHER and Ingenuity Pathway
analyses
Phosphorylated proteins were analyzed by PANTHER
11.0 (Protein Analysis Through Evolutionary Relationships;
http://www.pantherdb.org) and Gene Ontology
(http://amigo.geneontology.org/rte).
Proteins were then classified according to their protein class.
Since the majority of the proteins identified were involved in
multiple processes, only the most relevant ones were reported.
These proteins were analyzed by Ingenuity Pathway
Analysis (IPA; Qiagen GmbH), as previously described (20). Selected genes were used to
generate bio-functions. For the filter summary, only high
confidence associations (predicted) or that had been experimentally
observed were considered.
Statistical analysis
Statistical analyses were performed with the
non-parametric Wilcoxon signed-rank test for matched samples for
both 2-DE and western blot data. P<0.05 was considered to
indicate a statistically significant difference. All analyses were
conducted with Stata/IC 14.1 for Windows (StataCorp LLC).
Results
Enrichment by IMAC
For phosphoproteome enrichment, an IMAC column was
used. This column, although non-specific, allowed us to efficiently
enrich the phosphoproteome of both the leiomyoma and the myometrium
tissues. An average of 1,800 spots were detected on gels for both
types of the enriched phosphoproteome. The analysis revealed 26
protein spots (Table I) with a
significantly different abundance (Fig. 1) in leiomyoma tissue compared with
myometrium tissues. The correlation of gel-pairs performed well,
with an average matching efficiency of ~80%. In the present study,
only spots corresponding to putative phosphoproteins were
considered, with the following criteria: fold change in %V (where V
indicates the spot volume) ≥1.5 or ≤0.6 in intensity and P<0.05.
Of these spots, 25 were significantly upregulated (>1.5-fold)
while one was significantly downregulated (<0.6-fold).
 | Table IList of putative phosphoproteins with
a significantly different abundance in leiomyoma compared with
myometrium tissues. |
Table I
List of putative phosphoproteins with
a significantly different abundance in leiomyoma compared with
myometrium tissues.
| Accession number | Spot number | Protein
description | Gene symbol | Fold change | Protein class |
|---|
| A0A087WU08 | 7 | Haptoglobin | HP | 4.25 | Hydrolase |
| P11021 | 17 | 78 kDa
glucose-regulated protein | HSPA5 | 3.61 | Chaperone |
| H9KV75 | 19 | α-actinin 1 | ACTN1 | 3.2 | Cytoskeletal |
| | | | | protein |
| A0A0C4DGB6 | 21 | Albumin | ALB | 3.1 | Transfer/carrier
protein |
| E9PFZ2 | 18 C | eruloplasmin | CP | 3 | Oxidoreductase |
| Q5JRR6 | 20 | Ubiquitin-like
modifier-activating enzyme 1 | UBA1 | 3 | Ligase |
| H7C3T4 | 3 | Peroxiredoxin 4 | PRDX4 | 3 | Peroxidase |
| Q3BDU5 | 8 | Prelamin A/C | LMNA | 3 | Cytoskeletal
protein |
| H7BZ94 | 26 | Protein
disulfide-isomerase | P4HB | 2.9 | Oxidoreductase |
| P30101 | 11 | Protein
disulfide-isomerase family A member 3 | PDIA3 | 2.75 | Disulfide
oxidoreductase |
| P10809 | 25 | 60 kDa heat shock
protein | HSPD1 | 2.63 | Chaperone |
| P18206-2 | 24 | Isoform 1 of
vinculin | VCL | 2.54 | Cytoskeletal
protein |
| P02790 | 13 | Hemopexin | HPX | 2.45 |
Metalloprotease |
| P08238 | 16 | Heat shock protein
HSP 90β | HSP90AB1 | 2.32 | Chaperone |
| P04792 | 3 | Heat shock protein
β1 | HSPB1 | 2.3 | Chaperone |
| E9PN50 | 9 | 26S protease
regulatory subunit 6A | PSMC3 | 2.2 | Hydrolase |
| Q8NBS9 | 10 | Thioredoxin
domain-containing protein 5 | TXNDC5 | 2 | Isomerase |
| P08603 | 22 | Complement factor
H | CFH | 1.93 | Complement control
protein |
| P01023 | 23 |
α2-macroglobulin | A2M | 1.9 | Defence immunity
protein |
| B7ZAR1 | 14 | T-complex protein 1
subunit epsilon | CCT5 | 1.84 | Chaperone |
| P28070 | 2 | Proteasome subunit
β type 4 | PSMB4 | 1.8 | Defence immunity
protein |
| Q13409-2 | 15 | Isoform 2B of
Cytoplasmic dynein 1 intermediate chain 2 | DYNC1H1 | 1.78 | Cytoskeletal
protein |
| P01023 | 19 | Peroxiredoxin
2 | PRDX2 | 1.7 | Oxidoreductase |
| P01024 | 21 | Complement C3 | CO3 | 1.61 | Complement control
protein |
| Q16555 | 15 |
Dihydropyrimidinase-related protein 2 | DPYSL2 | 1.5 |
Metalloprotease |
| P17661 | 5 | Desmin | DES | 0.07 | Cytoskeletal
protein |
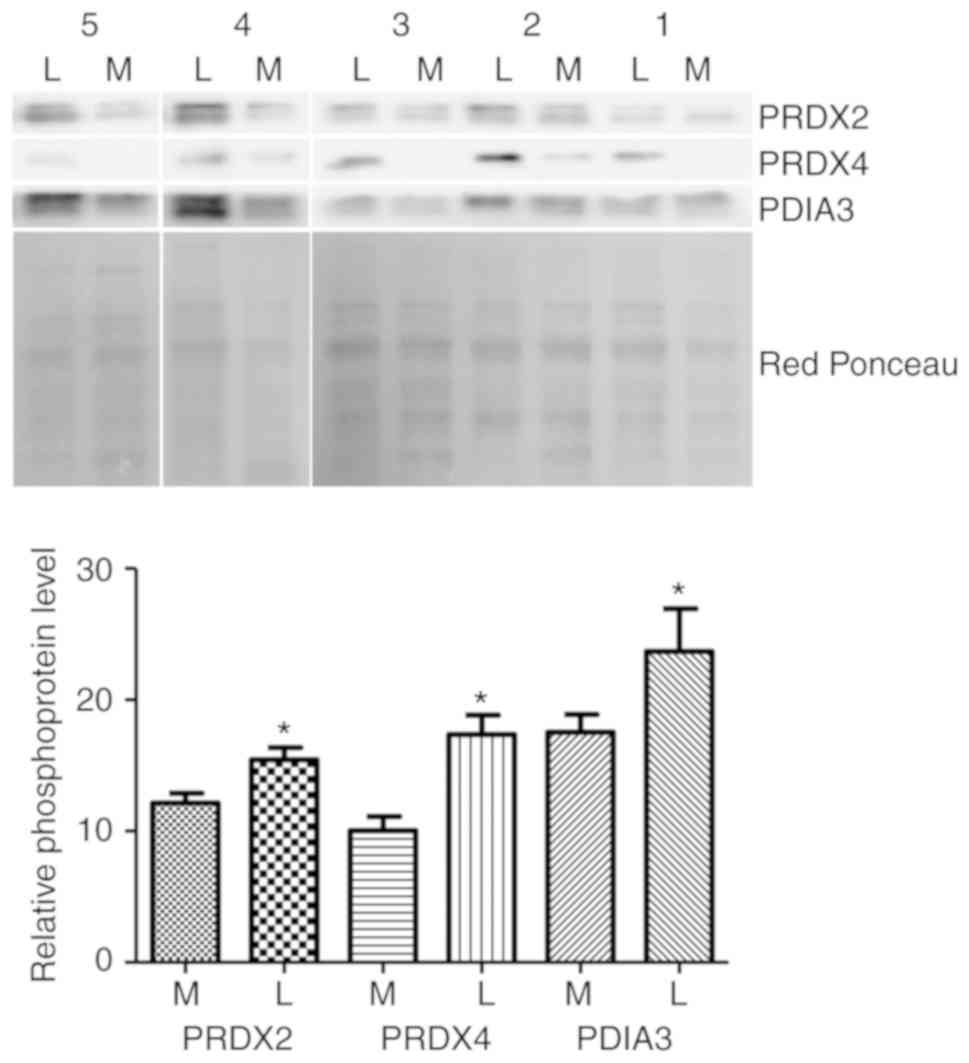
Western blotting validation
Western blot analysis was used to confirm the
alteration of three putative phosphorylated proteins: PRDX2, PDIA3
and PDRX4. The abundance of the propsphorylated forms of these
proteins in five leiomyoma samples was compared to the matched
normal myometrial tissue samples (same samples as used in 2-DE
analysis) by western blotting (Fig.
2). The results demonsatrted that, in the five patients tested,
phosphorylation levels of PRDX2, PDIA3 and PRDX4 were markedly
higher in the leiomyoma tissues compared with the myometrium
tissues, consistent with the 2-DE results.
Bioinformatic analysis
A PANTHER analysis of the identified proteins was
conducted. Based on the PANTHER classification system, the results
revealed that these proteins could be grouped into: Chaperone,
enzyme modulator, transfer/carrier protein, cytoskeletal protein,
signaling molecule and in three enzyme classes, hydrolase, ligase
and oxidoreductase.
The in silico data analysis was then
explanded by using the core analysis in the IPA software to
construct a network in which these proteins were involved. The top
networks in which these proteins were involved corresponded to:
migration of cells, synthesis of ROS, cell movement, oxidative
stress and cell proliferation of tumor cell lines (Fig. 3). Thirteen putative
phosphoproteins were involved in the 'migration of cells' network,
two were involved in the 'oxidative stress' network, five in the
'synthesis of ROS' network, 14 in the 'cell movement' network,
while 13 were involved in the 'cell survival' network. Of these,
four proteins [heat shock protein β1 (HSPB1), PRDX4, haptoglobin
(HP) and PRDX2] suppress oxidative stress and synthesis of reactive
oxygen.
Discussion
Phosphorylation of membrane proteins and kinases is
fundamental for leiomyoma development (9,21).
In the present study, by combining the IMAC column, 2-DE and MS, 26
putative phosphoproteins were identified to be differentially
phosphorylated/expressed in leiomyoma tissues compared with
myometrium tissues. By using western blotting, the differential
levels of three putative phosphorylated proteins were confirmed:
PRDX2, PDIA3 and PDRX4. All identified proteins have already been
reported to be phosphorylated in vivo in the PhosphoSitePlus
database, suggesting that the IMAC column was effective in
isolating mainly phosphorylated proteins. Unfortunately, the
validation experiments could not be performed using antibodies
against the specific phosphorylated sites of these proteins,
because these are not commercially available.
PRDX4 is a thiol-specific peroxidase that catalyzes
the reduction of hydrogen peroxide. This enzyme protects against
oxidative stress by detoxifying peroxides (22). The increase of this enzyme in
tumors is associated with the protection of the cell from oxidative
stress (23,24).
HSPB1 is a heat shock protein which maintains
denatured proteins in a folding-competent state (25). Phosphorylation of HSPB1 inhibits
apoptosis by protecting the cell from oxidative stress (26). The present study identified this
putative protein as differently phosphorylated in leiomyoma,
suggesting a possible association of phosphorylated HSPB1 with the
inhibition of oxidative stress and tumor growth.
PRDX2 is a thiol-specific peroxidase that serves a
role in cell protection against oxidative stress by detoxifying
peroxides, and as a sensor of hydrogen peroxide-mediated signaling
events (27). This enzyme has an
important role in cell survival by modulating the signaling
involved in apoptosis and the phosphorylation of JNK, and by
blocking the synthesis of ROS (28).
HP binds free hemoglobin (Hb), prevents oxidative
stress and acts as a potent immunoreactive modulator in the acute
phase (29). Fedorovych et
al (30) found that the
function of the HP/Hb complex in sera of patients with lung cancer
was to neutralize superoxidative products (30). A similar mechanism may also be
present in leiomyoma, in which the upregulation of phosphorylated
HP may contribute to cell protection from reactive oxygen
species.
In conclusion, the present study identified several
putative phosphoproteins involved in oxidative stress in leiomyoma
tissues. Further studies are needed to understand the role of
phosphorylation in oxidative stress. The current data represented a
step forward in the understanding of the mechanism involving
oxidative stress in tumor growth.
Acknowledgments
We greatly appreciate the help of the obstetrics and
gynecologigy operating room personnel of the Institute for Maternal
and Child Health-IRCCS 'Burlo Garofolo'.
Funding
The work was funded with current research funds
(grant no. 27/17) from our institute (Institute for Maternal and
Child Health-IRCCS 'Burlo Garofolo', Trieste, Italy).
Availability of data and materials
The datasets generated and analyzed during the
current study are not publicly available due to restrictions
imposed by the Regional Bioethics Committee of Friuli-Venezia
Giulia (Comitato Etico Unico Regionale), but can be available from
the corresponding author prior to approval of the research protocol
by the Review Board of the Institute for Maternal and Child
Health-IRCCS 'Burlo Garofolo' (Trieste, Italy).
Authors' contributions
Conceived and designed the experiments: GR and FS.
Performed the experiments: BU, GA, BG and DL. Analyzed the data:
LM, BU and GA. Contributed to data analysis: GDL and FR. Wrote the
paper: GDL, FR, BU, LM and GA. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Procedures incolving the use of human tissue were
approved by the Review Board of the Institute for Maternal and
Child Health-IRCCS 'Burlo Garofolo' (Trieste, Italy). All subjects
involved signed a written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ura B, Scrimin F, Arrigoni G, Aloisio M,
Monasta L and Ricci G: Dysregulated chaperones associated with cell
proliferation and negative apoptosis regulation in the uterine
leiomyoma. Oncol Lett. 15:8005–8010. 2018.PubMed/NCBI
|
|
2
|
Okolo S: Incidence, aetiology and
epidemiology of uterine fibroids. Best Pract Res Clin Obstet
Gynaecol. 22:571–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ura B, Di Lorenzo G, Romano F, Monasta L,
Mirenda G, Scrimin F and Ricci G: Interstitial fluid in gynecologic
tumors and its possible application in the clinical practice. Int J
Mol Sci. 19:2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moravek MB, Yin P, Ono M, Coon JS V, Dyson
MT, Navarro A, Marsh EE, Chakravarti D, Kim JJ, Wei JJ and Bulun
SE: Ovarian steroids, stem cells and uterine leiomyoma: Therapeutic
implications. Hum Reprod Update. 21:1–12. 2015. View Article : Google Scholar
|
|
5
|
Ura B, Scrimin F, Monasta L, Radillo O and
Ricci G: Association between up-regulated expression proteins and
circulating steroidal hormones in leiomyoma. Med Hypotheses.
85:5152015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ardito F, Giuliani M, Perrone D, Troiano G
and Lo Muzio L: The crucial role of protein phosphorylation in cell
signaling and its use as targeted therapy (Review). Int J Mol Med.
40:271–280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nishi H, Shaytan A and Panchenko AR:
Physicochemical mechanisms of protein regulation by
phosphorylation. Front Genet. 5:2702014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ura B, Monasta L, Arrigoni G, Battisti I,
Licastro D, Di Lorenzo G, Romano F, Aloisio M, Peterlunger I,
Stabile G, et al: Phosphoproteins involved in the inhibition of
apoptosis and in cell survival in the leiomyoma. J Clin Med.
8:2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kovács KA, Oszter A, Göcze PM, Környei JL
and Szabó I: Comparative analysis of cyclin D1 and oestrogen
receptor (alpha and beta) levels in human leiomyoma and adjacent
myometrium. Mol Hum Reprod. 7:1085–1091. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu L, Saile K, Swartz CD, He H, Zheng X,
Kissling GE, Di X, Lucas S, Robboy SJ and Dixon D: Differential
expression of receptor tyrosine kinases (RTKs) and IGF-I pathway
activation in human uterine leiomyomas. Mol Med. 14:264–275. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chegini N, Luo X, Ding L and Ripley D: The
expression of Smads and transforming growth factor beta receptors
in leiomyoma and myometrium and the effect of gonadotropin
releasing hormone analogue therapy. Mol Cell Endocrinol. 209:9–16.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fletcher NM, Abusamaan MS, Memaj I, Saed
MG, Al-Hendy A, Diamond MP and Saed GM: Oxidative stress: A key
regulator of leiomyoma cell survival. Fertil Steril.
107:1387.e1–1394.e1. 2017. View Article : Google Scholar
|
|
13
|
Fletcher NM, Saed MG, Abu-Soud HM,
Al-Hendy A, Diamond MP and Saed GM: Uterine fibroids are
character-ized by an impaired antioxidant cellular system:
Potential role of hypoxia in the pathophysiology of uterine
fibroids. J Assist Reprod Genet. 30:969–974. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Storz P: KRas, ROS and the initiation of
pancreatic cancer. Small GTPases. 8:38–42. 2017. View Article : Google Scholar :
|
|
15
|
Storz P and Toker A: Protein kinase D
mediates a stress-induced NF-kappaB activation and survival
pathway. EMBO J. 22:109–120. 2003. View Article : Google Scholar :
|
|
16
|
Schroyer AL, Stimes NW, Abi Saab WF and
Chadee DN: MLK3 phosphorylation by ERK1/2 is required for oxidative
stress-induced invasion of colorectal cancer cells. Oncogene.
37:1031–1040. 2018. View Article : Google Scholar :
|
|
17
|
Carcoforo P, Ura B, Mischiati C,
Squerzanti M, Lanzara V, Cervellati C, Calza R, De Laureto PP,
Frare E, Portinari M, et al: Comparative proteomic analysis of
ductal breast carcinoma demonstrates an altered expression of
chaperonins and cytoskel-etal proteins. Mol Med Rep. 7:1700–1704.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ura B, Monasta L, Arrigoni G, Franchin C,
Radillo O, Peterlunger I, Ricci G and Scrimin F: A proteomic
approach for the identification of biomarkers in endometrial cancer
uterine aspirate. Oncotarget. 8:109536–109545. 2017. View Article : Google Scholar
|
|
19
|
Ura B, Scrimin F, Arrigoni G, Athanasakis
E, Aloisio M, Monasta L and Ricci G: Abnormal expression of
leiomyoma cytoskeletal proteins involved in cell migration. Oncol
Rep. 35:3094–3100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ura B, Scrimin F, Franchin C, Arrigoni G,
Licastro D, Monasta L and Ricci G: Identification of proteins with
different abundance associated with cell migration and
proliferation in leiomyoma interstitial fluid by proteomics. Oncol
Lett. 13:3912–3920. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lemmon MA and Schlessinger J: Cell
signaling by receptor tyrosine kinases. Cell. 141:1117–1134. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Basu A, Banerjee H, Rojas H, Martinez SR,
Roy S, Jia Z, Lilly MB, De León M and Casiano CA: Differential
expression of peroxiredoxins in prostate cancer: Consistent
upregulation of PRDX3 and PRDX4. Prostate. 71:755–765. 2011.
View Article : Google Scholar :
|
|
23
|
Jiang H, Wu L, Mishra M, Chawsheen HA and
Wei Q: Expression of peroxiredoxin 1 and 4 promotes human lung
cancer malignancy. Am J Cancer Res. 4:445–460. 2014.PubMed/NCBI
|
|
24
|
Yi N, Xiao MB, Ni WK, Jiang F, Lu CH and
Ni RZ: High expression of peroxiredoxin 4 affects the survival time
of colorectal cancer patients, but is not an independent
unfavorable prognostic factor. Mol Clin Oncol. 2:767–772. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rogalla T, Ehrnsperger M, Preville X,
Kotlyarov A, Lutsch G, Ducasse C, Paul C, Wieske M, Arrigo AP,
Buchner J and Gaestel M: Regulation of Hsp27 oligomerization,
chaperone function, and protective activity against oxidative
stress/tumor necrosis factor alpha by phosphorylation. J Biol Chem.
274:18947–18956. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garrido C, Brunet M, Didelot C, Zermati Y,
Schmitt E and Kroemer G: Heat shock proteins 27 and 70:
Anti-apoptotic proteins with tumorigenic properties. Cell Cycle.
5:2592–2601. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kamariah N, Sek MF, Eisenhaber B,
Eisenhaber F and Grüber G: Transition steps in peroxide reduction
and a molecular switch for peroxide robustness of prokaryotic
peroxiredoxins. Sci Rep. 6:376102016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kwon T, Rho JK, Lee JC, Park YH, Shin HJ,
Cho S, Kang YK, Kim BY, Yoon DY and Yu DY: An important role for
peroxiredoxin II in survival of A549 lung cancer cells resistant to
gefitinib. Exp Mol Med. 47:e1652015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abdullah M, Schultz H, Kähler D,
Branscheid D, Dalhoff K, Zabel P, Vollmer E and Goldmann T:
Expression of the acute phase protein haptoglobin in human lung
cancer and tumor-free lung tissues. Pathol Res Pract. 205:639–647.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fedorovych IP, Tymochko MF, Fedevych IuM,
Fetsich TG and Korobov VM: Serum haptoglobin in lung cancer
patients. Ukr Biokhim Zh 1978. 67:103–105. 1995.In Ukrainian.
|