Introduction
Osteonecrosis of the femoral head (ONFH) leads to
inestimable labor loss to the whole society with its high rate of
disability (1). Typically, the
disease becomes progressive and leads to severe pain and
eventually, total hip arthroplasty is necessary for curing
end-stage ONFH (2). However, the
mechanism underlying the incidence and progression of ONFH remains
largely unknown, limiting the advances in diagnosis and treatment
for early-stage ONFH. Therefore, the underlying mechanisms of ONFH
are valuable research topics that could guide the prevention,
diagnosis and treatment of ONFH. In recent years, ONFH has been
considered to be associated with the dysfunction of bone marrow
stem cells (BMSCs). Previous studies have demonstrated well that
decreased proliferation capacity, enhanced apoptosis, impaired
osteogenesis and enhanced adipogenesis were found in BMSCs of ONFH
patients (3,4). In addition, aberrant gene expression
was also found in BMSCs of ONFH patients and thus, is considered as
the major cause of the dysfunction of BMSCs, particularly in the
imbalance between osteogenesis and adipogenesis (5).
Increasing attention has been paid to detecting
novel factors regulating gene expression in ONFH patients.
Extensive efforts have been made on studying the function of
non-coding RNAs (ncRNAs), especially microRNAs (miRNAs/miRs)
(6). miR-199b-5p has been proved
to promote osteogenesis of BMSCs by suppressing glycogen synthase
kinase-3β/β-catenin signaling pathway (7). miR-23a has been reported to inhibit
osteogenic differentiation of BMSCs by targeting low-density
lipoprotein receptor-related protein (8). However, it is inadequate to
elucidate the pathogenesis of ONFH only by miRNAs. Previous studies
have revealed that miRNAs could be dynamically regulated by other
ncRNAs, such as circular RNAs (circRNAs) through a competing
endogenous RNA (ceRNA) mechanism (9-11).
Circular RNAs (circRNAs) are a large group of non-coding RNAs
produced by non-canonical splicing of pre-mRNAs. Due to their
continuous closed loop structures, circRNAs are resistant to RNA
exonucleases or RNase R, leading to its stable and abundant
presence in the cytoplasm, body fluids and blood (12). Nowadays, the advanced sequencing
technology and bioinformatics analysis have facilitated the
identification and classification of circRNAs. An increasing number
of studies have investigated the regulatory function of circRNAs in
diverse orthopedic diseases, including osteoarthritis (OA),
rheumatoid arthritis (RA) and osteoporosis (13-15). However, research on the altered
expression of circRNAs in BMSCs of ONFH patients is still
lacking.
Based on previous studies, it was speculated that
circRNAs were implicated in the pathogenesis of ONFH by regulating
the functions of BMSCs, especially osteogenic and adipogenic
differentiation (16,17). In the present study, BMSCs were
isolated from seven patients with steroid-induced ONFH and seven
healthy controls and then the alteration in biological behavior of
BMSCs in steroid-induced ONFH patients was investigated. In ONFH
patients, a significantly reduced proliferation and osteogenesis of
BMSCs was observed, along with enhanced apoptosis and adipogenesis.
The expression of miRNA and circRNA in BMSCs was further screened
using next-generation sequencing. The expression of three circRNAs
and two miRNAs was validated by reverse transcription-quantitative
(RT-q)PCR. Then, integrated bioinformatics methods were used to
analyze the function of differentially expressed circRNAs and
miRNAs. A circRNA-miRNA co-expression network was further
constructed to detect the potential interaction between
differentially expressed circRNAs and miRNAs. To the best of our
knowledge, this study is the first to report the aberrant
expression and function of circRNA in ONFH patients. These findings
offer new insight into understanding the pathogenesis, facilitating
early diagnosis and guiding treatment of this disease.
Materials and methods
Isolation, labeling and RNA extraction of
BMSCs
Bone marrow tissue was acquired from seven female
patients (age range, 26-54) with steroid-induced ONFH and seven
female controls (age range, 26-75) who underwent hip arthroplasty
in the Department of Orthopedics at Peking Union Medical College
Hospital (Beijing, China) from June 2016 to December 2016. The
inclusion criteria of the patient group included female ONFH
patients with corticosteroids usage due to systemic lupus
erythematosus (SLE). Patients with ONFH due to trauma, alcohol or
corticosteroids use for treating other autoimmune diseases were
excluded. Male patients were also excluded. Isolation and culturing
of BMSCs was done using a method previously described (18). The growth medium used was a
mixture of 440 ml Dulbecco's modified Eagle medium (DMEM), 50 ml
fetal bovine serum (FBS; both Gibco; Thermo Fisher Scientific,
Inc.), 5 ml glutamine and 5 ml penicillin-streptomycin solution.
According to the manufacturer's protocol, total RNA of BMSCs was
extracted using TRIzol agent (Invitrogen; Thermo Fisher Scientific,
Inc.). Detailed information of the donors is listed in Table SI. Notably, the same RNA samples
were simultaneously used for profiling and validating the long
non-coding RNA and mRNA expression (not published yet). Surface
markers including cluster of differentiation (CD)29, CD45 and CD90
were identified using mouse anti-human Alexa Fluor 647-CD29,
CD45-fluorescein isothiocyanate (FITC), and CD90-FITC antibodies
and analyzed by an Accuri C6 flow cytometer system.
This study was approved by the Ethics Committee at
Peking Union Medical College Hospital and written consent was
obtained from the all 14 tissue donors included in this study.
Proliferation and apoptosis assay
To investigate the proliferation ability, cells were
seeded and cultured at a density of 5,000 cells in 96-well plates.
Then, Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies,
Inc.) assay was performed after 24, 48 and 72 h of culturing. The
Cell-Light™ EdU cell proliferation detection (EdU) assay (Guangzhou
Ribobio Co., Ltd.) was performed 12 h after culturing, according to
the manufacturer's protocol. Briefly, the cells were labeled with
EdU. Subsequently, 100 µl Apollo reaction cocktail and 100
µl Hoechst 33342 solution were used in turn to stain the
cells for 30 min on ice. Then the cells were visualized under a
fluorescence microscope at a magnification of ×100.
Cell apoptosis was analyzed by Annexin V-PI double
staining after culturing the cells for 48 h, according to the
manufacturer's protocol. The cytographs were analyzed using FlowJo
V10 software (FlowJo LLC). For each sample, the apoptosis assay was
performed in triplicate.
Osteogenic and adipogenic differentiation
in vitro
BMSCs were seeded in growth medium. Osteogenic
medium (a mix of 175 ml basal medium, 20 ml FBS, 2 ml
b-glycerophosphate, 400 µl L-ascorbic acid, 2 ml glutamine,
2 ml penicillin-streptomycin solution and 20 µl
dexamethasone) and adipogenic medium (A solution: A mix of 175 ml
basal medium, 20 ml FBS, 2 ml glutamine, 2 ml
penicillin-streptomycin solution, 400 µl insulin, 200
µl IBMX, 200 µl rosiglitazone, and 200 µl
dexamethasone; B solution: A mix of 175 ml basal medium, 20 ml FBS,
2 ml glutamine, 2 ml penicillin-streptomycin solution and 400
µl insulin) were obtained from Cyagen Biosciences, Inc. For
osteogenic differentiation, the growth medium was replaced with
osteogenic differentiation medium when the cells reached 70-80%
confluence. For adipogenic differentiation, growth medium was
replaced with the adipogenic differentiation medium when cells
reached 90-100% confluence. Subsequent procedures were done
according to the protocol of Cyagen Biosciences, Inc.
Alizarin red staining and oil red O
staining
Alizarin red staining was conducted on day 12 after
the osteogenic induction. In brief, cells were washed with PBS,
fixed in 4% neutral buffered formalin for 1 h at room temperature,
washed with PBS again and stained using alizarin red solution at
37°C for 30 min.
Oil red O staining was performed on day 20 after
adipogenic induction. The stock solution of Oil red O was diluted
in deionized water and filtered prior to use. BMSCs were first
washed with PBS, then fixed in 4% neutral buffered formalin for 1 h
at room temperature, washed with deionized water and stained with
Oil red O solution for 30 min at room temperature. Excess Oil red O
stain was washed away with 60% isopropanol. The staining results
were observed and recorded under a light microscope at a
magnification of ×100.
Sequencing and differential expression
analysis of miRNA and circRNA
After evaluating the RNA integrity, concentration
and purity, sequencing procedures were performed by Annoroad Gene
Technology, Co., Ltd. Briefly, ribosomal RNA was first removed
using Epicentre Ribo-Zero™ Gold kits (Epicentre; Illumina, Inc.)
and generated sequencing libraries following manufacturer's
protocols with varied index label using NEBNext® Ultra™
Directional RNA Library Prep kit from Illumina, Inc. Then the
libraries were sequenced on an Illumina Hiseq 4000 platform and 150
bp paired-end reads were generated. Raw Data acquired was processed
with Perl scripts to ensure the quality of data. CIRI 1.2 software
was used to recognize circRNA (19). Spliced Reads per Billion Mapping
was then used to quantify the expression of each circRNA (20). MiRDeep2 (21) was used to identify miRNA and
Transcripts Per Million was used for estimation and normalization.
Then, DEGseq v1.18.0 was used for differential expression analysis
of miRNA and circRNA. The P-value was adjusted by the Benjamini and
Hochberg's approach for controlling the false discovery rate. Genes
with adj. P≤0.05 and |log2_foldchange|≥1 were identified as
differentially expressed (DE) circRNA and DE miRNA.
RT-qPCR
This analysis was conducted prior to the induction
of differentiation as well as on day 3, 6, and 12 of the induction.
PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio, Inc.) was
used for reverse transcription of RNA samples. The reaction was
performed at 37°C for 15 min and then at 85°C for 5 sec. qPCR was
performed using SYBR® Premix Ex Taq™ II (Tli RNaseH
Plus), ROX plus (Takara Bio, Inc.) for the analysis of
hsa_circ_0000219, hsa_circ_0004588, hsa_circ_0005936, alkaline
phosphatase (ALP), osteocalcin (OCN), peroxisome
proliferator-activated receptor γ (PPARγ) and lipoprotein lipase
(LPL). The thermocycling conditions were as follows: Firstly, 95°C
for 10 min; then 40 cycles of 95°C for 15 sec, 60°C for 45 sec and
70°C for 30 sec. GAPDH mRNA was set as internal controls for
circRNAs and mRNAs. U6 mRNA was set as internal controls for
miRNAs. The relative expression of each DEG was calculated using
the 2−ΔΔCq method (22). Reactions of each sample were run
in triplicate. miRNAs primers were purchased from Guangzhou
RiboBio, Co., Ltd., and gene primers are displayed in Table SII.
Functional analysis of DEGs
The Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; http://www.kegg.jp/) analysis of DE circRNAs and DE
miRNAs was implemented by the hypergeometric test. GO and KEGG
terms with adj. P<0.05 was considered to indicate significant
enrichment.
Prediction of miRNA target and
circRNA-miRNA interaction
MiRanda 2010 Release (http://www.microrna.org/microrna/home.do), PITA v6
(http://genie.weizmann.ac.il/pubs/mir07/mir07_dyn_data.html)
and TargetScan 7.0 (http://www.targetscan.org/) were used for predicting
target genes of known or novel miRNAs. To identify target miRNAs of
selected DE circRNAs, Arraystar's home-made software v1.0 (23), based on TargetScan and miRanda,
was used. The predicted miRNAs were further compared to the list of
DE miRNAs and then, a circRNA-miRNA co-expression network was
constructed based on competitive endogenous RNA principle.
Cytoscape 3.4.0 software (https://cytoscape.org) was used to visualize the
circRNA-miRNA network.
Statistical analysis
All data in this study were presented as the mean ±
standard deviation and each experiment was performed in triplicate
independently. Prism 7 software (GraphPad Software, Inc.) was
employed to perform the statistical analysis. A two-tailed t-test
was used to compare the difference between two groups and
comparisons between multiple groups were done by Kruskal-Wallis
test along with Dunnett's multiple comparisons test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Phenotype and altered cellular behaviors
of BMSCs in ONFH patients
The isolated BMSCs displayed a swirling arranged
phenotype microscopically (Fig.
S1). A total of three surface antigens, including CD29, CD45
and CD90 were further investigated by flow cytometry analysis
(Fig. S1B-D). BMSCs from passage
3 were positive for CD29 and CD90 (89.5 and 99.9%, respectively)
and negative for CD45 (0.16%).
The cellular behaviors between BMSCs of patient and
control groups were then compared. A significant decrease in
proliferation of BMSCs of ONFH patients by CCK-8 and EdU
proliferation assays were identified (P<0.05; Fig. 1A). It was also found that the
total apoptotic rate of BMSCs of ONFH patients was elevated
(Fig. 1B).
 | Figure 1Altered cellular behavior in BMSCs in
the ONFH group. (A) Decreased proliferation activity of BMSCs in
ONFH patients was observed compared with the control group. (a)
Decreased optical density at 450 nm was found in BMSCs in ONFH
patients during in vitro expansion. (b) Representative image
of EdU assay of BMSCs in ONFH patients (upper row) and controls
(lower row). (c) The percentages of proliferating cells were
quantified by EdU assay. (B) Cytographs of Annexin V-PI apoptosis
assay of BMSCs in (a) ONFH patients and (b) controls. (c)
Statistical analysis of total apoptotic cells. (C) Fewer calcium
nodules revealed by alizarin red staining in BMSCs in (a) ONFH
patients and (b) controls at day 12 of osteogenic differentiation
(magnification, ×40). Decreased expression of (c) ALP and (d) OCN
in ONFH patients before and during the induction of osteogenic
differentiation. (D) Accumulation of lipid granules in BMSCs in (a)
ONFH patients and (b) controls, revealed by Oil red O staining at
day 20 of the induction of adipogenic differentiation
(magnification, ×100). Increased expression of (c) LPL and (d)
PPARγ in ONFH group before and during the induction of adipogenic
differentiation. *P<0.05, **P<0.01 and
***P<0.001. BMSCs, bone marrow stem cells; ONFH,
osteonecrosis of the femoral head; PI, propidium iodide; PPAR,
peroxisome proliferator-activated receptor; ALP, alkaline
phosphatase; LPL, lipoprotein lipase. |
Alizarin red staining and oil red O staining was
performed to compare the differentiation capacity of the cells.
BMSCs from the ONFH group exhibited significantly decreased calcium
mineralization and increased lipid accumulation in the cytoplasm
(P<0.05; Fig. 1C-a, C-b, D-a and
D-b). Consistent with the results of staining, the lower
expression of osteogenic markers ALP and OCN as well as enhanced
expression of adipogenic markers PPARγ and LPL was found during the
process of osteogenic and adipogenic induction, respectively
(Fig. 1C-a, C-b, D-a and
D-b).
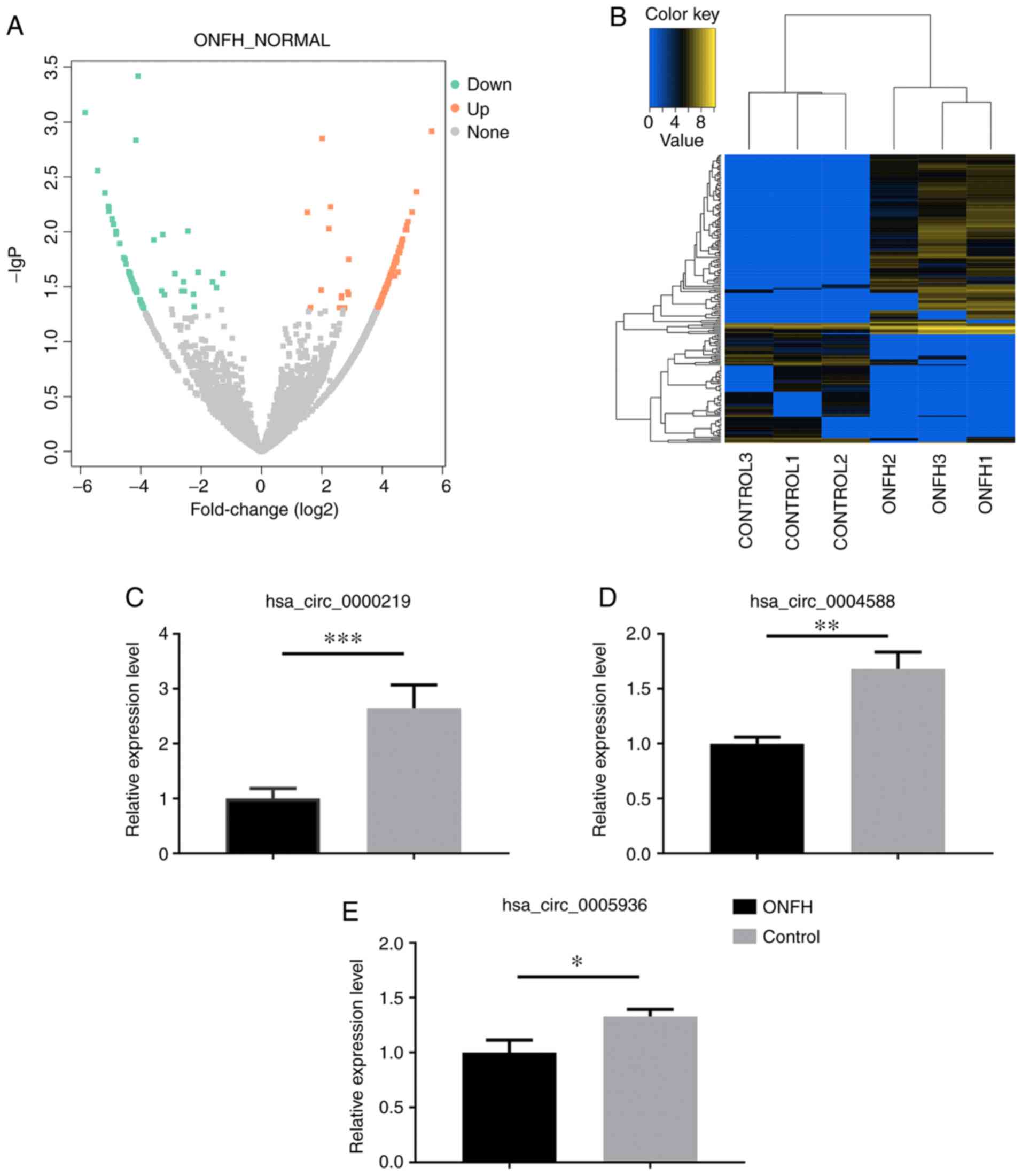
The miRNA and circRNA expression profile
in ONFH patients
Overall, 897 miRNAs and 14,131 circRNAs were
screened. miRNAs and circRNAs with |log2_foldchange|≥1 and adj.
P≤0.05 were considered as differentially expressed. A total of 129
DE miRNAs were detected, of which 47 miRNAs were downregulated and
82 miRNAs were upregulated in BMSCs of ONFH patients. In addition,
there were 231 DE circRNAs, including 141 upregulated circRNAs and
90 downregulated circRNAs. Among them, 215 circRNAs were
transcribed from the exonic region and only 16 from the intronic
region or the intergenic region. The expression profiles of miRNAs
and circRNAs are displayed in Figs.
2 and 3. Top 10 up- and
downregulated miRNAs and circRNAs are listed in Tables I and II.
 | Table ITop 10 up- and downregulated
miRNAs. |
Table I
Top 10 up- and downregulated
miRNAs.
A, Upregulated
miRNAs
|
|---|
| Gene |
Log2Fold-change | adj. P-value |
|---|
| hsa-miR-4451 | 14.043 |
1.16×10−39 |
|
hsa-miR-6757-5p | 13.25041 |
5.50×10−07 |
|
hsa-miR-7114-3p | 12.68606 |
5.64×10−07 |
| hsa-miR-124-3p | 12.00992 |
3.98×10−06 |
|
hsa-miR-4787-5p | 11.59704 |
7.07×10−19 |
| hsa-miR-1270 | 11.09448 |
4.84×10−22 |
|
hsa-miR-6750-5p | 10.97533 | 0.014683 |
|
hsa-miR-6800-3p | 10.97533 | 0.014683 |
| hsa-miR-6089 | 10.91785 |
2.69×10−21 |
| hsa-miR-9-5p | 10.83323 |
5.56×10−55 |
|
B, Downregulated
miRNAs
|
| hsa-miR-30a-5p | −1.53456 | 0.022986 |
| hsa-miR-582-5p | −1.60469 | 0.004317 |
| hsa-miR-496 | −1.67512 | 0.020004 |
|
hsa-miR-199b-5p | −1.69292 |
3.71×10−05 |
|
hsa-miR-302d-3p | −1.7246 | 0.041531 |
| hsa-miR-31-3p | −1.73719 | 0.000222 |
| hsa-miR-628-5p | −1.74575 | 0.003749 |
| hsa-miR-154-5p | −1.84565 | 0.001145 |
|
hsa-miR-30c-2-5p | −1.99482 | 0.001591 |
| hsa-miR-549a | −2.3367 | 0.014287 |
 | Table IITop 10 up- and downregulated circRNAs
and their source genes. |
Table II
Top 10 up- and downregulated circRNAs
and their source genes.
A, Downregulated
circRNAs
|
|---|
| Gene |
Log2Fold-change | Adj. P-value | Source_Gene |
|---|
|
hsa_circ_0013180 | −5.633964 | 0.001209 | Null |
|
hsa_circ_0011021 | −5.133924 | 0.004323 |
ENSG00000096060 |
|
hsa_circ_0000761 | −4.988706 | 0.006605 |
ENSG00000182985 |
|
hsa_circ_0010245 | −4.857244 | 0.008042 |
ENSG00000112972 |
|
hsa_circ_0006469 | −4.812017 | 0.00875 |
ENSG00000087586 |
|
hsa_circ_0003004 | −4.809939 | 0.009577 |
ENSG00000092439 |
|
hsa_circ_0000219 | −4.77287 | 0.009605 |
ENSG00000015171 |
|
hsa_circ_0000596 | −4.676935 | 0.011608 |
ENSG00000107771 |
|
hsa_circ_0005936 | −4.674644 | 0.011719 |
ENSG00000117419 |
|
hsa_circ_0012564 | −4.650421 | 0.012237 |
ENSG00000066697 |
|
B, Upregulated
circRNAs
|
|
hsa_circ_0010461 | 4.82842 | 0.008508 |
ENSG00000164176 |
|
hsa_circ_0001854 | 4.90173 | 0.007764 |
ENSG00000058272 |
|
hsa_circ_0002614 | 4.94959 | 0.007629 |
ENSG00000126773 |
|
hsa_circ_0003535 | 4.95424 | 0.006065 |
ENSG00000174939 |
|
hsa_circ_0004764 | 5.05521 | 0.006463 |
ENSG00000237440 |
|
hsa_circ_0011258 | 5.06074 | 0.005875 |
ENSG00000118412 |
|
hsa_circ_0005623 | 5.07195 | 0.005843 |
ENSG00000116991 |
|
hsa_circ_0011690 | 5.07232 | 0.004407 |
ENSG00000106571 |
|
hsa_circ_0003795 | 5.19475 | 0.002763 |
ENSG00000065559 |
|
hsa_circ_0002691 | 5.43192 | 0.000816 |
ENSG00000119707 |
|
hsa_circ_0010193 | 5.84601 | 0.008508 |
ENSG00000164190 |
Validation of the circRNA profile by
RT-qPCR
A total of 3 downregulated circRNAs
(hsa_circ_0000219, hsa_circ_0004588 and hsa_circ_0005936) were
selected for verification of the hypothesis that downregulated
circRNAs could potentially become genetic tools in early treatment
for steroid-induced ONFH by bone tissue engineering. The expression
of the selected circRNAs was examined in BMSCs from seven patients
with steroid-induced ONFH and seven controls using RT-qPCR.
Consistent with the authors' sequencing data, the expression levels
of hsa_circ_0000219, hsa_circ_0004588 and hsa_circ_0005936 were all
significantly decreased in BMSCs from ONFH patients compared with
the controls (P<0.05; Fig.
3C-E), indicating a potential positive correlation between the
expression of these circRNAs and proliferation and osteogenic
capacity of BMSCs, as well as a negative correlation between
circRNAs expression and apoptosis and adipogenic capacity of
BMSCs.
GO and KEGG analysis of DE miRNAs and DE
circRNAs
A total of 27,329 target genes were predicted by at
least two pieces of software for the validation of DE miRNAs and
the potential function of the DE miRNAs were was identified by GO
analysis and KEGG analysis of the target genes. Collectively, 1,886
biological process (BP) terms, 182 cellular component (CC) terms
and 237 molecular function (MF) terms were considered significant,
along with 129 KEGG terms. Table
SIII displays the 10 most significant BP, MF, and KEGG terms
for the DE miRNAs. The function of DE circRNAs was represented by
the functional analysis of their parental genes. Based on the
results of the present study, a total of 602 BP terms, 44 CC terms,
140 MF terms and 11 KEGG pathways were identified as significant.
Detailed information is shown in Table SIV.
Target miRNAs prediction and verification
for validated circRNAs
Based on TargetScan and miRanda, the target miRNAs
of the three validated downregulated circRNAs were predicted and
further compared with the results of miRNA sequencing. A number of
upregulated DE miRNAs were predicted as targets for
hsa_circ_0000219 and hsa_ circ_0005936 (Fig. 4A and B). However, no circRNA-miRNA
interactions were predicted for hsa_circ_0004588. Therefore,
miR-144-3p was selected, a predicted target miRNA for
hsa_circ_0000219 reported to inhibit the proliferation and
osteogenic differentiation of BMSCs, and miR-1270, a common target
for both hsa_circ_0000219 and hsa_circ_0005936 for further
verification (Fig. 4C and D).
Consistent with the sequencing data of the present study, these two
miRNAs were both significantly upregulated in BMSCs of ONFH
patients, indicating a potential ceRNA mechanism-mediated
regulation of progression of steroid-induced ONFH (P<0.001;
Fig. 4E).
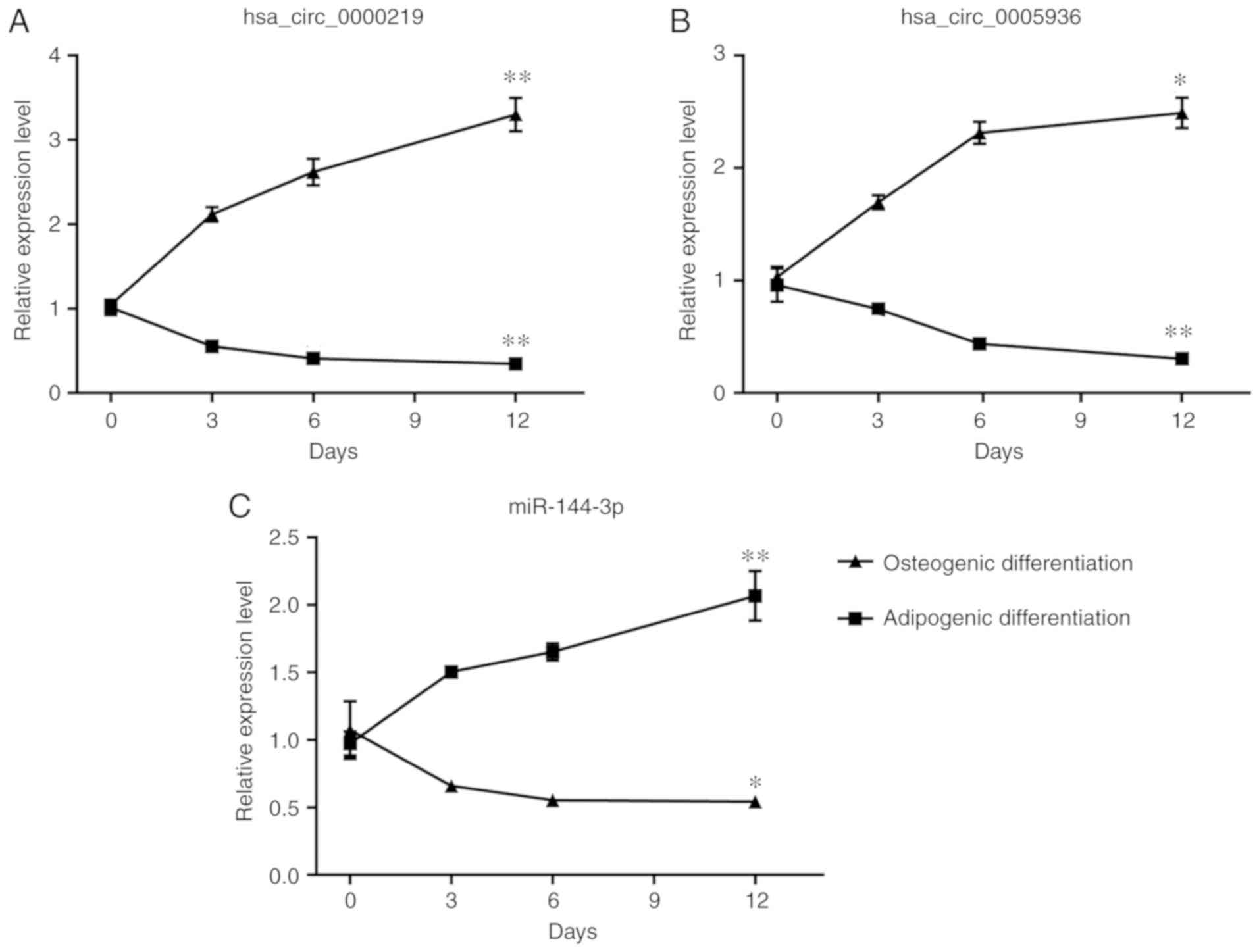
Time-dependent expression patterns of DE
circRNAs and miRNAs during differentiation of BMSCs
Considering that the imbalance of osteogenic and
adipogenic differentiation of BMSCs outweighs their proliferation
and apoptosis in the progression of ONFH, whether the selected
circRNAs and miRNAs could potentially regulate differentiation of
BMSCs was examined. BMSCs were induced with osteogenic medium and
adipogenic medium for 12 days and the expression of circRNAs and
miRNAs was detected on day 3, 6, and 12 of induction with RT-qPCR.
The expression of hsa_circ_0000219 and hsa_circ_0005936
consistently decreased during the osteogenic induction and reached
significance on day 12 (P<0.05; Fig. 5A and B). During the adipogenic
differentiation, significantly elevated expression of
hsa_circ_0000219 and hsa_circ_0005936 was also found on day 12
(P<0.05; Fig. 5A and B).
However, the relationship of hsa_circ_0004588 and differentiation
of BMSCs was not detected. The expression of miR-144-3p increased
during osteogenic differentiation and decreased during adipogenic
differentiation, displaying a negative relationship with
hsa_circ_0000219 and hsa_ circ_0005936 (Fig. 5C).
Discussion
Although the pathogenesis of ONFH remains elusive,
several studies have demonstrated the association of aberrant
behavior of BMSCs with the development of ONFH, as well as their
promising roles in treatment for ONFH via bone tissue engineering
(3,24,25). In the present study, BMSCs were
isolated from patients with ONFH and controls and the surface
markers were checked. CD29 is the marker of mesenchymal cells and
CD90 is the marker of stem cells. The absence of CD45 excluded
pan-hematopoietic cells (26).
The combination of the three markers in this study confirmed the
BMSCs were successfully isolated. Despite the limited number of
cases, it was confirmed that the proliferation capacity of BMSCs
decreased in ONFH patients, along with enhanced apoptosis. Reduced
osteogenic and increased adipogenic capacity of BMSCs in ONFH
patients was also verified. The results of the present study
substantiated previous studies regarding behaviors of BMSCs in
steroid-induced ONFH (3,4).
Aberrant gene expression has been identified in
BMSCs of ONFH patients and is considered to regulate the altered
cellular functions. Investigation of the regulation of gene
expression was emphasized and a large number of ncRNAs, especially
miRNAs were detected to be aberrantly expressed in ONFH patients.
These miRNAs were reportedly implicated in the progression of ONFH
mainly through the regulation of differentiation of BMSCs (27). Due to the advancement in
sequencing technology and bioinformatics analysis, circRNAs, which
have long been considered as a by-product produced during RNA
splicing, have been emerged as new hotspot in ONFH (28). Their stability makes them suitable
to be biomarkers and tools for gene therapy (12). circRNAs could bind miRNAs and
function through a ceRNA mechanism (29). Unfortunately, only a few studies
have focused on the roles of circRNAs in ONFH (17,30). In the present study, the
expression pattern of circRNAs and miRNAs were profiled
simultaneously by high-throughput RNA-sequencing. A total of 231 DE
circRNAs and 129 DE miRNAs were detected. Considering that
downregulated circRNAs could become potential tools used to rescue
the impaired proliferation and osteogenic capacity of BMSCs, while
promoting bone repair in necrotic area, three down-regulated
circRNAs, hsa_circ_0000219, hsa_circ_0004588, and hsa_circ_0005936,
were randomly selected to validate their expression levels by qPCR.
Compared with BMSCs of the control group, the expression of
hsa_circ_0000219, hsa_circ_0004588 and hsa_circ_0005936 was
decreased in BMSCs of the ONFH group, indicating the involvement of
these downregulated circRNAs in ONFH. Furthermore, target miRNAs
were predicted for validated circRNAs and a circRNA-miRNA network
was constructed based on the ceRNA principle (31). Although no circRNA-miRNA
interaction was predicted for hsa_circ_0004588, a total of 17
upregulated miRNAs were found to be targets for hsa_circ_0000219
and hsa_circ_0005936.
Since the functions of the selected circRNAs in ONFH
have not been reported yet, the authors speculated their functions
through the source genes and target miRNAs. Hsa_circ_0000219 is
located at chr10: 179994-221356 and its parental gene is zinc
finger MYND-type containing 11, whose mutation was found to be
associated with acute poorly differentiated myeloid leukemia
(32), indicating its potential
involvement in cell differentiation. However, the source genes of
hsa_circ_0004588 and hsa_circ_0005936 are phosphatidylinositol
glycan anchor biosynthesis class N and exoribonuclease family
member 3, respectively, whose functions have been rarely reported
previously. Although the functions of most target miRNAs of
hsa_circ_0000219 and hsa_circ_0005936 have been reported in cancer
instead of BMSCs, the present study speculated their functions in
BMSCs using previous literature. Only one study on miR-9a-5p
reported that the target gene of miR-9a-5p is Sirt1 (33). The expression of Sirt1 in BMSCs
promotes proliferation, inhibits apoptosis and pushes the direction
of differentiation towards the osteogenic lineage (34,35). miR-379-5p directly targets 3′-UTR
of focal adhesion kinase (FAK) and FAK loss has adverse effects on
proliferation and osteogenic differentiation of preosteoblasts by
repressing Wnt/β-catenin signaling (36,37). miR-214-3p overexpression
suppresses cell proliferation in gastric cancer by targeting
runt-related transcription factor (RUNX3). In mesenchymal stem cell
(MSCs), RUNX3 plays an important role in mediating the bone
morphogenic protein (BMP)9-induced osteogenic differentiation
(38,39). miR-144-3p inhibits the
proliferation and osteogenic differentiation in MSC lines
C3H10T1/2, by directly targeting mothers against decapentaplegic
homolog 4 (40). Forkhead box C1
has been reported to be a target gene of miR-204-5p and is a
crucial regulator of early osteogenic differentiation induced by
BMP4 (41,42). The target miRNA of
hsa_circ_0005936, miR-3074-5p, promotes cell apoptosis (43). Taken together, all the target
miRNAs were upregulated and impaired the proliferation and
osteogenic capacity while promoting cell apoptosis of BMSCs during
the progression of ONFH. Therefore, miR-144-3p was selected, whose
function has been well documented and miR-1270, a target miRNA for
both hsa_circ_0000219 and hsa_circ_0005936, to preliminarily
validate their expression by qPCR. The results showed that the
expression levels of these two miRNAs were elevated in BMSCs of
ONFH patients. The results of the present study indicated that the
downregulation of hsa_circ_0000219 and hsa_circ_0005936 may
contribute to ONFH progression by mainly affecting the
differentiation of BMSCs, through the hsa_circ_0000219-miR-144-3p
or hsa_circ_0005936-miR-1270 axis. However, more studies are needed
to elucidate the exact functions of the circRNAs and to evaluate
their potential in early interference in ONFH.
However, this study has certain limitations. First,
the conclusion may be compromised due to the limited number of
patients and control donors, and more samples are being collected
to further substantiate these conclusions. Second, whether there
are differentially expressed circRNAs between old trauma fracture
or DDH and trauma-related ONFH has not been proved. Thus, the
differentially expressed genes might result from loss of blood
supply instead of ONFH itself. Basically, loss of blood supply has
been proved to be one of the most important pathological changes in
ONFH of all causes. Thus, it was speculated that the DE circRNAs or
miRNAs associated with blood loss could also be correlated with
ONFH. Third, the DE circRNAs and miRNAs might also result from the
autoimmune diseases such as SLE instead of ONFH, which has been
demonstrated in previous studies (44-46). It is true that SLE itself could
lead to DE miRNAs and circRNAs. However, whether SLE leads to
functional alteration in BMSCs is still unclear. In the current
study, bone marrow tissue was collected from the proximal region of
the femur during hip replacement surgery, thus, BMSCs of SLE
patients without ONFH were not obtained, which made this problem
hard to answer. However, the time-dependent expression of two
circRNAs and a miRNA was detected in BMSCs to demonstrate that they
are associated with differentiation. Although it still cannot be
known whether the DE circRNAs and miRNAs related to SLE or steroid
treatment, it can be concluded that they are related to aberrant
differentiation of BMSCs, which has been proved in ONFH.
In the present study, BMSCs were isolated from seven
ONFH patients and seven controls, and profiled the expression
levels of circRNA in BMSCs of ONFH patients for the first time.
Then, the sequencing data of circRNA and miRNA were comprehensively
analyzed. Several signaling pathways regulating multiple cellular
functions were screened during functional analysis. circRNA-miRNA
co-expression networks were also constructed. A total of three
circRNAs and their predicted miRNAs were validated by qPCR in
BMSCs. The results of the present study could improve understanding
of the regulatory roles of circRNA-miRNA interaction in ONFH
development and provide new insights for early treatment of ONFH
using BMSCs and circRNAs as genetic tools.
Supplementary Data
Funding
The present study is supported by the National
Natural Science Foundation of China (grant no. 81572143 and
81630064).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SX performed the experiments and analyzed the
results. ZL was a major contributor in writing the manuscript, and
also helped in to perform several experiments and analyzing the
results. XW designed the research and was a major contributor in
recruiting the donors. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee at
Peking Union Medical College Hospital and written consent was
obtained from the all 14 tissue donors included in this study.
Patient consent for publication
All donors included in the current study provided
informed consent for publication at the point of recruitment.
Competing interests
The authors have declared that no conflict of
interest exists.
Acknowledgments
Not applicable.
References
|
1
|
Cui L, Zhuang Q, Lin J, Jin J, Zhang K,
Cao L, Lin J, Yan S, Guo W, He W, et al: Multicentric epidemiologic
study on six thousand three hundred and ninety five cases of
femoral head osteonecrosis in China. Int Orthop. 40:267–276. 2016.
View Article : Google Scholar
|
|
2
|
Wang C, Peng J and Lu S: Summary of the
various treatments for osteonecrosis of the femoral head by
mechanism: A review. Exp Ther Med. 8:700–706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Houdek MT, Wyles CC, Packard BD, Terzic A,
Behfar A and Sierra RJ: Decreased osteogenic activity of
mesenchymal stem cells in patients with corticosteroid-induced
osteonecrosis of the femoral head. J Arthroplasty. 31:893–898.
2016. View Article : Google Scholar
|
|
4
|
Yeh CH, Chang JK, Ho ML, Chen CH and Wang
GJ: Different differentiation of stroma cells from patients with
osteonecrosis: A pilot study. Clin Orthop Relat Res. 467:2159–2167.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Balla B, Pintér C, Kósa JP, Podani J,
Takács I, Nagy Z, Speer G, Horváth B, Korányi L and Lakatos P: Gene
expression changes in femoral head necrosis of human bone tissue.
Dis Markers. 31:25–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu X, Zhang Y, Guo X, Xu H, Xu Z, Duan D
and Wang K: Identification of differentially expressed microRNAs
involved in non-traumatic osteonecrosis through microRNA expression
profiling. Gene. 565:22–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao R, Li Y, Lin Z, Wan J, Xu C, Zeng Y
and Zhu Y: miR-199b-5p modulates BMSC osteogenesis via suppressing
GSK-3β/β-catenin signaling pathway. Biochem Biophys Res Commun.
477:749–754. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li T, Li H, Wang Y, Li T, Fan J, Xiao K,
Zhao RC and Weng X: microRNA-23a inhibits osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
by targeting LRP5. Int J Biochem Cell Biol. 72:55–62. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Q, Li Y and Zhang Y, Ma L, Lin L,
Meng J, Jiang L, Wang L, Zhou P and Zhang Y: LncRNA MEG3 inhibited
osteogenic differentiation of bone marrow mesenchymal stem cells
from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed
Pharmacother. 89:1178–1186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weng J, Peng W, Zhu S and Chen S: Long
noncoding RNA sponges miR-454 to promote osteogenic differentiation
in maxillary sinus membrane stem cells. Implant Dent. 26:178–186.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
13
|
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J
and Ao Y: Circular RNA related to the chondrocyte ECM regulates
MMP13 expression by functioning as a miR-136 'Sponge' in human
cartilage degradation. Sci Rep. 6:225722016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dou C, Cao Z, Yang B, Ding N, Hou T, Luo
F, Kang F, Li J, Yang X, Jiang H, et al: Changing expression
profiles of lncRNAs, mRNAs, circRNAs and miRNAs during
osteoclastogenesis. Sci Rep. 6:214992016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R,
Lou A, Zhu D, Shi GP and Yang M: Microarray expression profile of
circular RNAs in peripheral blood mononuclear cells from rheumatoid
arthritis patients. Cell Physiol Biochem. 42:651–659. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Peng W, Zhu S, Chen J, Wang J, Rong Q and
Chen S: Hsa_ circRNA_33287 promotes the osteogenic differentiation
of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed
Pharmacother. 109:1709–1717. 2019. View Article : Google Scholar
|
|
17
|
Qian DY, Yan GB, Bai B, Chen Y, Zhang SJ,
Yao YC and Xia H: Differential circRNA expression profiles during
the BMP2-induced osteogenic differentiation of MC3T3-E1 cells.
Biomed Pharmacother. 90:492–499. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Otsuru S, Hofmann TJ, Olson TS, Dominici M
and Horwitz EM: Improved isolation and expansion of bone marrow
mesenchymal stromal cells using a novel marrow filter device.
Cytotherapy. 15:146–153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Shi X, Chen C and Zhang L:
Improving RNA-Seq expression estimation by modeling isoform- and
exon-specific read sequencing rate. BMC Bioinformatics. 16:3322015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar :
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Gu W, Sun Y, Zheng X, Ma J, Hu XY, Gao T
and Hu MJ: Identification of gastric cancer-related circular RNA
through microarray analysis and bioinformatics analysis. Biomed Res
Int. 2018:23816802018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song H, Tao L, Wang F, Wang W, Wei Y, Shen
W and Zhou F: Effect of bone mesenchymal stem cells transplantation
on the micro-environment of early osteonecrosis of the femoral
head. Int J Clin Exp Pathol. 8:14528–14534. 2015.
|
|
25
|
Hernigou P, Beaujean F and Lambotte JC:
Decrease in the mesenchymal stem-cell pool in the proximal femur in
corticosteroid-induced osteonecrosis. J Bone Joint Surg Br.
81:349–355. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song K, Huang M, Shi Q, Du T and Cao Y:
Cultivation and identification of rat bone marrow-derived
mesenchymal stem cells. Mol Med Rep. 10:755–760. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yue J, Wan F, Zhang Q, Wen P, Cheng L, Li
P and Guo W: Effect of glucocorticoids on miRNA expression spectrum
of rat femoral head microcirculation endothelial cells. Gene.
651:126–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gu X, Li M, Jin Y, Liu D and Wei F:
Identification and integrated analysis of differentially expressed
lncRNAs and circRNAs reveal the potential ceRNA networks during
PDLSC osteogenic differentiation. BMC Genet. 18:1002017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
De Braekeleer E, Auffret R, Douet-Guilbert
N, Basinko A, Le Bris MJ, Morel F and De Braekeleer M: Recurrent
translocation (10;17)(p15;q21) in acute poorly differentiated
myeloid leukemia likely results in ZMYND11-MBTD1 fusion. Leuk
Lymphoma. 55:1189–1190. 2014. View Article : Google Scholar
|
|
33
|
Qi F, Hu JF, Liu BH, Wu CQ, Yu HY, Yao DK
and Zhu L: miR-9a-5p regulates proliferation and migration of
hepatic stellate cells under pressure through inhibition of Sirt1.
World J Gastroenterol. 21:9900–9915. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin CH, Li NT, Cheng HS and Yen ML:
Oxidative stress induces imbalance of adipogenic/osteoblastic
lineage commitment in mesenchymal stem cells through decreasing
SIRT1 functions. J Cell Mol Med. 22:786–796. 2018.
|
|
35
|
Yuan HF, Zhai C, Yan XL, Zhao DD, Wang JX,
Zeng Q, Chen L, Nan X, He LJ, Li ST, et al: SIRT1 is required for
long-term growth of human mesenchymal stem cells. J Mol Med (Berl).
90:389–400. 2012. View Article : Google Scholar
|
|
36
|
Chen JS, Li HS, Huang JQ, Dong SH, Huang
ZJ, Yi W, Zhan GF, Feng JT, Sun JC and Huang XH: MicroRNA-379-5p
inhibits tumor invasion and metastasis by targeting FAK/AKT
signaling in hepatocellular carcinoma. Cancer Lett. 375:73–83.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun C, Yuan H, Wang L, Wei X, Williams L,
Krebsbach PH, Guan JL and Liu F: FAK promotes osteoblast progenitor
cell proliferation and differentiation by enhancing Wnt signaling.
J Bone Miner Res. 31:2227–2238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu Y, Zhang G, Zou C, Zhang H, Gong Z,
Wang W, Ma G, Jiang P and Zhang W: LncRNA MT1JP suppresses gastric
cancer cell proliferation and migration through
MT1JP/miR-214-3p/RUNX3 axis. Cell Physiol Biochem. 46:2445–2459.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Feng Q, Ji C, Liu X, Li L and Luo
J: RUNX3 plays an important role in mediating the BMP9-induced
osteogenic differentiation of mesenchymal stem cells. Int J Mol
Med. 40:1991–1999. 2017.PubMed/NCBI
|
|
40
|
Huang C, Geng J, Wei X, Zhang R and Jiang
S: miR-144-3p regulates osteogenic differentiation and
proliferation of murine mesenchymal stem cells by specifically
targeting Smad4. FEBS Lett. 590:795–807. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao W, Wu Y, He X, Zhang C, Zhu M, Chen B,
Liu Q, Qu X, Li W, Wen S and Wang B: MicroRNA-204-5p inhibits
invasion and metastasis of laryngeal squamous cell carcinoma by
suppressing forkhead box C1. J Cancer. 8:2356–2368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hopkins A, Mirzayans F and Berry F: Foxc1
expression in early osteogenic differentiation is regulated by
BMP4-SMAD activity. J Cell Biochem. 117:1707–1717. 2016. View Article : Google Scholar
|
|
43
|
Gu Y, Shi Y, Yang Q, Gu WW, He YP, Zheng
HJ, Zhang X, Wang JM and Wang J: miR-3074-5p promotes the apoptosis
but inhibits the invasiveness of human extravillous
trophoblast-derived HTR8/SVneo cells in vitro. Reprod Sci.
25:690–699. 2018. View Article : Google Scholar
|
|
44
|
Li Z, Jiang C, Li X, Wu WKK, Chen X, Zhu
S, Ye C, Chan MTV and Qian W: Circulating microRNA signature of
steroid-induced osteonecrosis of the femoral head. Cell Prolif.
51:2018. View Article : Google Scholar
|
|
45
|
Zhang MY, Wang JB, Zhu ZW, Li LJ, Liu RS,
Yang XK, Leng RX, Li XM, Pan HF and Ye DQ: Differentially expressed
circular RNAs in systemic lupus erythematosus and their clinical
significance. Biomed Pharmacother. 107:1720–1727. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li LJ, Zhu ZW, Zhao W, Tao SS, Li BZ, Xu
SZ, Wang JB, Zhang MY, Wu J, Leng RX, et al: Circular RNA
expression profile and potential function of hsa_circ_0045272 in
systemic lupus erythematosus. Immunology. 155:137–149. 2018.
View Article : Google Scholar : PubMed/NCBI
|