Introduction
Non-typhoidal Salmonella (NTS) is a leading
cause of bacterial bloodstream infections in infants, the elderly
and immunocompromised people, particularly in sub-Saharan Africa,
where the case fatality rate is 20–25% (1,2).
In sub-Saharan Africa, invasive NTS (iNTS) has been reported to be
the second most common invasive bacterial disease type (29%),
followed by Streptococcus pneumoniae infection (3,4).
S. enterica serovar Typhimurium is the most common iNTS
isolate, followed by S. enterica serovar Enteritidis
(3,4). Although Salmonella may be
controlled using antibiotics, an increasing prevalence of
multidrug-resistant strains has been reported over previous decades
(5,6). Thus, vaccine development is a
potential prospect for controlling the epidemic prevalence of iNTS
in humans in addition to animals (7). At present, two types of licensed
Salmonella vaccines for typhoid fever are considered safe
and efficacious for people aged over 2 years (8); however, there is no vaccine directly
targeting iNTS. Vivotif®, a live oral vaccine containing
a mutated Salmonella (Ty21a), may effectively elicit
intestinal antibodies against O-antigens, which likely exhibits
cross-protective efficacy against NTS, including S.
Typhimurium and S. Enteritidis (9–11).
With the limitations of these existing vaccines,
numerous subunit and live-attenuated vaccines for iNTS diseases are
being developed, and a number of them are undergoing clinical
studies (12,13). Approaches to subunit vaccines
mainly focus on the application of the O-antigens of
lipopolysaccharides and surface virulence factors, including
flagellin and outer membrane proteins (14–16). These subunit vaccines potentially
have the advantage of cross-protection, safety and low production
cost (17). However, numerous
subunit vaccines against bacterial pathogens have not been
successful as the conformational changes in conserved epitopes
during preparation result in poor humoral and long-lasting
immunity, and a single structurally conserved protein, even if it
is a critical virulence factor, would not be sufficient to provide
effective immunity (18,19). Previously, numerous novel
strategies for developing live NTS vaccines have been introduced.
Most of the live attenuated Salmonella vaccine strains have
been constructed by deleting functional metabolic gene(s) and/or
virulence gene(s) (20–23). Among the candidate vaccine strains
for iNTS, only a few were tested in clinical studies. LH1160, a
phoPQ mutant strain that controls the transcription of
multiple genes necessary for intracellular survival, had been
tested in phase 1 clinical trials, but an unacceptable fever was
reported in two of six volunteers immunized with LH1160 (21,24). WT05 is another attenuated
S. Typhimurium vaccine in which the aroC gene,
involved in aromatic amino acid biosynthesis, and the ssaV
gene, a component of a Type 3 secretion system (T3SS) apparatus of
Salmonella pathogenicity island 2 (SPI-2), are deleted.
However, this vaccine also failed in phase 1 clinical trial due to
the prolonged stool shedding of the vaccine strain in healthy
volunteers immunized with WT05 (25,26). More recently, CVD1921, which is
mutated in the guaBA genes that are involved in the
biosynthesis of guanine nucleotides and the clpP gene
affecting flagella expression, was revealed to be notably
attenuated with decreased shedding, systemic spread and clinical
disease manifestations in the digestive tract of the non-human
primate model (rhesus macaque) used (27).
When Salmonella are internalized into the
intracellular compartment of host cells, they need to adapt to the
metabolism of available nutrients, mainly the carbon source in host
cells (28,29). Multiple carbon transport systems
are known to be involved in the intracellular life cycle and linked
to the virulence of Salmonella (30–32). For example, the carbohydrate
phosphotransferase system (PTS) is composed of two cytoplasmic
proteins, enzyme I (ptsI) and histidine protein
(ptsH), which are common to all PTS sugars, and
membrane-bound enzyme II (EII) complex (crr), which is
critical for sugar uptake (33).
In a previous study, the removal of two components genes,
ptsI and crr, in PTS was reported, and was revealed
to be adequate to construct Salmonella-attenuated vaccine
strains to provide protective humoral and mucosal immune responses,
but a less effective cell-mediated immune response (34).
In the present study, a new attenuated
Salmonella strain vaccine was developed by deleting the
ptsI (EI) gene in the carbohydrate phosphotransferase
(ptsH, ptsI and crr) operon. Furthermore, the
humoral, cellular and protective effects of the vaccine were
evaluated in a mouse model.
Methods and materials
Ethics statement
The present study was performed in strict accordance
with the recommendations in the Guide for the Care and Use of
Laboratory Animals of the National Institutes of Health. All animal
experiments were ethically approved by the Committee on The Use and
Care of Animals at the Korea Atomic Energy Research Institute
(KAERI; approval nos. IACUC-2016-028, IACUC-2017-010 and
IACUC-2018-007) and performed according to accepted veterinary
standards set by the KAERI animal care center. An overall
weight-loss of 25% was considered to be an indication for
euthanasia. All efforts were made to minimize the suffering of the
animals employed in the present study. To euthanize the mice, a
CO2 inhalation method as specified by KAERI
Institutional Animal Care and Use Committee guidelines was used.
Less than five mice were placed in a visible cage chamber
(L27×W21×H17) and injected with compressed grade B CO2
gas in cylinders. A fill rate of 10–30% of the cage chamber volume
per minute with CO2, which was 1–3 liter(s) per min, was
appropriate to achieve a balanced gas mixture to fulfill the
objective of rapid unconsciousness with minimal distress to the
animals. Expected time to unconsciousness was usually reached at
within 1–3 min and the flow was maintained for additional 1 min
once respiration had ceased. If both signs were observed, then the
mice were removed from the cage.
Reagents
All chemical reagents used in the present study were
purchased from Sigma-Aldrich (Merck KGaA).
Bacterial strains and growth
conditions
All bacteria strains used in the present study are
listed in Table I.
Salmonella strains were grown at 37°C in Luria-Bertani (LB)
broth (Difco; BD Biosciences), M9 minimum media (Sigma-Aldrich;
Merck KGaA) or phagosome-mimicking MES-buffered medium [5 mM KCl,
7.5 mM (NH4)2SO4, 0.5 mM
K2SO4, 1 mM KH2PO4, 8
mM MgCl2, 0.1% casamino acids, 0.4% carbon source and
170 mM 2-(N-morpholino) ethanesulphonic acid, pH=5.6]
(35). The media were
supplemented with kanamycin (50 μg/ml), chloramphenicol (15
μg/ml) or ampicillin (100 μg/ml), if required.
 | Table IStrains and plasmids used in the
present study. |
Table I
Strains and plasmids used in the
present study.
| A, Escherichia
coli |
|---|
|
|---|
| Author, year | Strain | Relevant gene | Source | (Refs.) |
|---|
| na | DH5α |
F−r−m+Ø80dlacZΔM15 | Gibco; Thermo
Fisher Scientific, Inc. | na |
|
| B,
Salmonella Typhimurium |
|
| Author, year | Strain | Relevant gene | Source | (Refs.) |
|
| na | UK1 | Clinical isolate
from horse ATCC68169 | American Type
Culture Collection | na |
| Abed et al,
2014 | BH129 | UK1,
ΔptsI(Tn:10) | na | (1) |
| KST0555 | UK1,
ΔptsI | The present
study | |
| Abed et al,
2014 | KST0650 | UK1,
ΔhilD | na | (1) |
| KST0600 | UK1
hilA::HA-KanaR | The present
study | |
| KST0601 | KST0555
hilA::HA-KanaR | The present
study | |
| KST0597 | UK1
hilD::HA-KanaR | The present
study | |
| KST0598 | KST0555
hilD::HA-KanaR | The present
study | |
| Febriani et
al, 2010 | KST0134 | SL1344,
sseA::lacZY-KanaR | na | (2) |
| KST0589 | UK1,
sseA::LacZY-KanaR | The present
study | |
| KST0590 | KST0555,
sseA::LacZY-KanaR | The present
study | |
| KST0583 | UK1, ΔhilD,
sseA::LacZY-KanaR | The present
study | |
|
| C, Plasmid |
|
| Author, year | Strain | Relevant gene | Source | (Refs.) |
|
| Ao et al,
2015 | pKD46 |
PBAD-gam-beta-exo oriR101
repA101LS; AmpR | na | (3) |
| Ao et al,
2015 | pKD13 | FRT
KanaR FRT PS1 PS4 oriR6K; AmpR | na | (3) |
| Ao et al,
2015 | pCP20 | cI857
PR flp oripSC101ts; AmpR
CmR | na | (3) |
Salmonella mutants construction
S. Typhimurium strains used in the present
study were derived from wild type (WT) strain UK1 (KST0134)
(36). The ptsI-deficient
strain (KST0555) was constructed using λ red-recombinase-mediated
replacement as described previously (37). In brief, a kanamycin resistance
(KmR) cassette was amplified from the pKD13 plasmid that
was annealed to the flanking regions of the deletion-target sites
by using the primers listed in Table
II. The resulting PCR product was transferred to UK1 harboring
the plasmid pKD46 by electrophoresis, and the mutant was isolated
on a kanamycin plate. The generated mutant was transformed with
pCP20 to excise the KmR cassette. Salmonella
strains containing HA or 3×FLAG epitope tag in HilA and HilD and
transcriptional lac fusion with sseA promoter
(sseA::lacZY) were constructed using P22 transduction as
described previously (38,39).
 | Table IIPrimers used in the present
study. |
Table II
Primers used in the present
study.
| Gene | Forward primer
sequence (5′-3′) | Reverse primer
sequence (5′-3′) |
|---|
| hilA |
ATTAAGGCGACAGAGCTGGA |
ATAGCAAACTCCGACGATG |
| invF |
TGTCGCACCAGTATCAGGAG |
AAATAGCGCGAAACTCAGGA |
| ssaG |
GATATGCTCTCCACATGGC |
AGGCAAATTGCGCTTTAATC |
| ssaB |
GGATTCATGCTGGCAGTTTT |
GCAGGATGCCATCAATAGT |
| sseA |
AGGAGGCCGAGAAGGATTTA |
TTCCTGACGGTATCTCCACC |
| ssrA |
CATGTTTGTGGCACTATCCG |
GCGAGCTAAGGTAGCCAATG |
| sipA |
AAACGTTGATACCTGCTG |
GACATGGCTTCGGCTATTGT |
| sipB |
TGTTTCGCTGTCTCAACTGG |
TCGCAGCGTCATAAACACTC |
| sipC |
AAGTCAGTGACCTGGGGTTG |
AACGGCACTGGAAGACATTC |
| sipD |
ACTGCCGTGGTACCGTCTAC |
ATAATGACACGCCGGACTTC |
| fthD |
CAACGAAGAGATGGCAAACA |
GACGCGTTGAAAGCATGATA |
| fliA |
CGCTGAAGGTGTAATGGAT |
CCGCATTTAATAACCGATG |
| fliZ |
AAACATTTCCACGATCTGC |
CGGTAAAGGGGGATTTCTGT |
| flgM |
CTTTGAAACCGTTAGCAC |
GCCGTTTTTAATGCTTCGAC |
| fliC |
AACGACGGTATCTCCATTGC |
TACACGGTCGATTTCGTTCA |
| pts
Imutant |
AAGTTTTTTTTCCGGGTTCTTTTAAAAATCA |
GCAGTTCCTGTTTGTAGATTTCTTTGCGCAG |
| Construction |
GTCACAAGTAAGGTAGGGTTCATATGAATATCCTCCTTA |
CGCGCGAACTTCTTGTCAATCTAGGCTGGAGCTGCTTC |
β-galactosidase assay
A β-galactosidase assay was performed as described
previously (40). Overnight
cultures of Salmonella KST0134 (WT), KST0555 (ΔptsI)
and KST0650 (ΔhilD) were sub-cultured into fresh LB broth or
MES-buffered minimum media. Salmonella bacteria were
harvested at 2, 4, 6, 8, 10 and 12 h post-inoculation, followed by
resuspension with 1 ml Z-buffer (0.06 M
Na2HPO4, 7H2O, 0.04 M
NaH2PO4, H2O, 0.01 M KCl, 0.01 M
MgSO4, 7H2O and 0.05 M β-mercaptoethanol;
pH=7.0), and permeabilized with 20 μl 0.1% sodium
dodecylsulfate and 40 μl chloroform. Following 10 min of
incubation at room temperature, 200 μl O-nitrophenyl
β-glactopyranoside (ONPG; 4 mg/ml) was added as the substrate, and
the mixture was incubated at 30°C for 30 min in a water bath until
the sample turned bright yellow. The reaction was stopped by adding
1 M Na2CO3 (500 μl), and the
incubation period, i.e., the time between the addition of the ONPG
substrate and the addition of stop solution, was recorded. The
optical density420 was recorded, and the specific
activity and protein concentration were determined. The
β-galactosidase activity was expressed in Miller units as described
previously (40). Miller units
were calculated as follows:
1,000×(Abs420)-1.75*Abs550(Abs600 of culture sample)*(volume of culture)*(reaction time)
Cell invasion and replication assays
Experimental procedures were performed as described
previously (30). The mouse
macrophage cell lines (RAW264.7; cat. no. ATCC® TIB-71)
and rat small intestinal epithelial cells (IEC-6; cat. no.
ATCC® CRL-1592™) were purchased from the Korean Cell
Line Bank. Cells were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
μg/ml streptomycin at 37°C and 5% CO2. The cells
were plated in 48-well cell culture plates (SPL Life Sciences) at
IEC-6 (1×105 cells) and RAW264.7 (5×104
cells) per well and incubated for 2 h at 37°C. Overnight-cultured
Salmonella were washed with phosphate buffered saline (PBS)
and suspended with DMEM, followed by addition to the prepared cell
monolayers at multiplicity of infection (MOI) of 10. Following 1 h,
the cells were washed three times with PBS, followed by an addition
of pre-warmed medium containing 100 μg/ml gentamicin to
remove extracellular bacteria. At 2 and 18 h post-infection (hpi),
the wells were washed with PBS three times and lysed with 0.5%
Triton-X100. Finally, the cell lysate was serially diluted with
PBS, and 10 μl of the diluents was spotted on LB/agar plates
to determine the number of colony forming units (CFUs). CFUs in the
range of 20 to 50 colonies per spot from each dilution were
counted. All data were calculated from at least three independent
experiments performed in triplicate.
Fluorescence microscopic analysis
Infected Salmonella cells were visualized by
plating RAW264.7 cells on 15 μm Chamber 12-well glass slides
(Ibidi GmbH), followed by infection with WT and KST0555 strains at
a MOI of 10 at 37°C for 1 h. DMEM containing gentamicin (100
μg/ml) was used to eliminate extracellular bacteria.
Following 2 or 18 h of infection, the cells were fixed with 4%
paraformaldehyde for 30 min at 4°C and permeabilized with 0.1%
Triton-X100 in PBS for 20 min. The cells were then washed three
times with PBS and blocked with 3% bovine serum albumin (BSA;
Sigma-Aldrich; Merck KGaA) in PBS for up to 2 h at room
temperature. Intracellular Salmonella were detected by
incubating the cells with fluorescein isothiocyanate
(FITC)-conjugated anti-rabbit Salmonella-specific antibodies
(cat. no. ab20320; 1:1,000; Abcam) at room temperature for 30 min.
The nuclei were stained with 150 ng/ml 4′,6-diamino-2-phenylindole
for 5 min at 37°C (Thermo Fisher Scientific, Inc.). All images were
captured using an Olympus CX41 fluorescence microscope (Olympus
Corporation).
Mice experiments
Animal housing conditions (specific pathogen-free)
and the animal experimental design were ethically approved by the
Committee on The Use and Care of Animals at the KAERI and performed
according to accepted National Health Institute standards. A total
of 90 six-week-old male BALB/c mice (weight, 19–21 g) were
purchased from OrientBio Inc. Five mice were randomly assigned per
individually ventilated housing cage (OrientBio Inc.) maintained in
an animal biological safety level 2 facility at 22–23°C on a 12
h:12 h light:dark cycle. Cages were covered with high efficiency
particulate air-filtered microisolation lids (OrientBio Inc.) in a
static airflow environment. Bedding (Aspen Shaving; OrientBio Inc.)
at an approximate depth of 1.0 cm was changed weekly. Irradiated
rodent diet food and sterile water were provided ad libitum,
supported by the wire cage top. Conscious mice were weighed
(PB602-S balance; Mettler Electronics Corp.) at regular intervals
using the 10 sec dynamic weighing method. For measuring the
virulence of Salmonella in the settings of a mice colitis
model, mice (n=5) were administered 20 mg streptomycin
(Sigma-Aldrich; Merck KGaA) in sterile water. Water and food were
withdrawn for 4 h prior to streptomycin administration.
Subsequently, the animals were supplied with water and food. At 20
h following streptomycin treatment, water and food were withdrawn
again for 4 h prior to the administration of Salmonella
orally (41). At the time of
mortality or 72 h post-infection (hpi), mouse spleens, mesenteric
lymph nodes and ceca were aseptically isolated, and the number of
viable bacteria in each organ were determined by plating serially
diluted homogenates or blood on LB agar plates. For further
verifying the protection capacity of the KST0555 vaccination, five
mice were immunized orally with 107 CFU KST0555 in 100
μl PBS three times at two-week intervals. At 7 days
following the last immunization, 107 CFU WT were
inoculated orally, and mortality and body weight changes were
observed and recorded for 2 weeks. In all experiments, mice were
considered to have succumbed to mortality and euthanized when they
reached 75% of their original bodyweight.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Log-phase cultures of WT (KST0134) and KST0555 were
diluted 100-fold into fresh LB broth or MES-buffered minimum media
(pH=5.6), followed by incubation for 4 h at 37°C overnight with
shaking (42). Total RNA was
isolated from bacterial cultures by using an RNeasy®
mini kit (Qiagen GmbH) according to manufacturer’s protocol. For
RT-qPCR analysis, cDNA was synthesized from 1 μg total
purified RNA using a Primescript 1st strand cDNA synthesis kit
(Takara Bio, Inc.) according to manufacturer’s protocol. Primers
for different genes were designed based on Primer Express software
(version 2.0; Applied Biosystems, Thermo Fisher Scientific, Inc.)
and are listed in Table II.
RT-qPCR amplification was performed using SYBR Premix EX Taq
(Takara Bio, Inc.) on a Bio-Rad CFX Real-Time System (Bio-Rad
Laboratories, Inc.). Thermocycling conditions were as follows: One
cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec,
60°C for 30 sec, 72°C for 30 sec, and a final extension at 72°C for
10 min. The relative gene expression was quantified using the
2−ΔΔCq method (43).
The 16S rRNA (rrsH) was selected as a control to normalize
the expression levels.
Agar plate motility assay
A motility assay was performed as described
previously (44). Briefly, 5
μl overnight cultures grown in LB broth were spotted onto
the surface of a swarm plate (LB; 0.5% Bacto agar and 0.5% glucose)
and incubated at 37°C for 24 h.
Western blot analysis
Whole Salmonella lysates (108 CFU)
were lysed in buffer (1% v/v Triton X-100 and 0.1% w/v SDS in PBS)
and loaded and separated on 12% Bis-Tris BOLT gels (Invitrogen;
Thermo Fisher Scientific, Inc.), followed by transfer onto
nitrocellulose membranes. The membranes were blocked with 5% dry
skimmed milk (Bio-Rad Laboratories, Inc.) in PBS with 0.05%
Tween-20 (PBS-T) for 30 min at room temperature and then incubated
with monoclonal anti-FLAG antibody (1:3,000; cat. no. F2555;
Sigma-Aldrich; Merck KGaA), monoclonal anti-HA antibody (1:4,000;
cat. no. ab137838; Abcam) or rabbit polyclonal anti-DnaK (1:3,000;
cat. no. ab80161; Abcam). Following primary antibody incubation,
the membranes were incubated with an anti-mouse total
immunoglobulin (Ig) or anti-rabbit IgG conjugated with horseradish
peroxidase-conjugated secondary antibody (1:5,000; cat. no. A9044;
Sigma-Aldrich; Merck KGaA; Merck KGaA) for 1 h at room temperature.
The protein bands were visualized using Enhanced Chemiluminescent
Western Blotting Substrate (Thermo Fisher Scientific, Inc.).
Bio-Rad ChemiDoc™ Touch imaging system and Bio-Rad CFX Manager
software (version 3.1; Bio-Rad Laboratories, Inc.) were used for
data acquisition and analysis.
Measurement of mice immunoglobulin
Mice blood samples were obtained 7 days subsequent
to the last immunization. The serum was isolated by centrifugation
at 500 × g for 10 min at 4°C and stored at −80°C for long-term use.
Salmonella were cultured in LB and harvested at mid-log
phase. The absorbance of the Salmonella pellet was adjusted
to 0.1 at 600 nm by dilution with PBS. Next, 96-well immunoplates
(SPL Life Sciences) were coated with 100 μl
Salmonella or 1 μg Salmonella
lipopolysaccharide (LPS; Sigma-Aldrich; Merck KGaA) and incubated
overnight at 4°C to allow the immobilization of bacterial cells
onto the wells. The plates were washed five times with PBS-T,
followed by blocking with 1% BSA in PBS for 1 h at room
temperature. Subsequent to blocking, diluted sera were added to
each well and incubated at room temperature for 1 h, followed by
the washing of unbound antibodies with PBS-T. Secondary antibodies
(all 1:4,000) were added to the wells by and incubated for 30 min
at room temperature. The secondary antibodies were as follows: Goat
anti-mouse Ig-HRP (cat. no. A0412; Sigma-Aldrich, Merck KGaA), goat
anti-mouse IgG-HRP (cat. no. 1030-05; SouthernBiotech), goat
anti-mouse IgM-HRP (cat. no. 1020-05; SouthernBiotech), goat
anti-mouse IgG1-HRP (cat. no. 1071-05; SouthernBiotech), goat
anti-mouse IgG2a-HRP (cat. no. 1080-05; SouthernBiotech), goat
anti-mouse IgG2b-HRP (cat. no. 1090-05; SouthernBiotech) and goat
anti-mouse IgG3-HRP (cat. no. 1100-05; SouthernBiotech). The plates
were then washed 5 times with PBS-T, and 100 μl of
3,3′,5,5′-tetramethylbenzidine substrate reagent (BD Biosciences)
was added. When the color developed, 50 μl 2 N
H2SO4 was added, and the absorbance was
measured at 450 nm by using a Victor X3 light plate reader
(PerkinElmer, Inc.). The assigned titer value was indicative of the
last dilution in which A450 is <0.1.
Cytokine measurement
Splenocytes (1×105 cells/well) were
plated onto a round bottom 96-well plates (SPL Life Sciences) and
incubated with PBS or S. Typhimurium WT lysate prepared by
sonication for 72 h at 37°C. Interleukin-4 (IL-4) and
interferon-γ(IFN-γ) levels in the supernatants were measured using
mouse IL-4 (cat. no. 555232; BD Biosciences) and mouse IFN-γ (cat.
no. 555138; BD Biosciences) ELISA kits, respectively.
Adoptive transfer of sera,
CD4+ and CD8+ cell protection
A total of 5 mice per group were immunized with
107 CFU of KST0555 orally three times in two-weeks
intervals. Mice spleens were isolated at 7 days following the last
vaccination, and lymphocytes were prepared from a cell suspension
by pressing organ segments through a cell strainer (BD
Biosciences). The cell suspension was depleted of red blood cells
by using the hypertonic pressure of distilled water and then washed
three times with PBS, as previously described (45). Splenic CD4+ and
CD8+ T cells were isolated using a CD4+ T
cell isolation kit (cat. no. 130-104-454; Miltenyi Biotec GmbH) and
a CD8+ T cell isolation kit (cat. no. 130-104-075;
Miltenyi Biotec GmbH) according to the manufacturer’s protocol.
Mice serum was isolated as described above and heated at 55°C for
30 min to inactivate the complements. Cell purity was verified to
be >90% using flow cytometry with staining with
phycoerythrin-conjugated anti-mouse CD4 (1:200; cat. no. 100511,
BioLegend, Inc.) or FITC-conjugated anti-mouse CD8a (1:200; cat.
no. 100705; BioLegend, Inc.) for 15 min at room temperature. Mouse
serum (300 μl per mice), enriched CD4+ T cells
(106 cells per mice) or CD8+ T cells
(105 cells per mice) were administered intraperitoneally
to each recipient mouse. Subsequent to 24 h, the mice were
administered a lethal dose of WT (107 CFU), and their
mortality and body weight changes were recorded.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Data in the bar graphs and bacterial numbers between groups were
compared using an unpaired Student’s t-test. The survival of mice
was determined using Kaplan-Meier survival analysis, and
significance of the difference was analyzed using a Log rank test
by using GraphPad Prism software (version 6.0; GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Characterization of a ptsI mutant strain
(KST0555)
Since a previous study indicated that glucose and
glycerol are important carbon sources in the systemic infection of
Salmonella (30), the
growth of KST0555 in minimal medium (M9) supplemented with these
carbon sources was examined compared with that of its parent
strain. Comparable growth was noted between KST0555 and WT
(KST0134) in LB, and each of the strains were unable to grow in M9
medium with no carbon source (data not shown). When KST0555 was
cultured in M9 supplemented with glucose, it exhibited
substantially lower growth compared with the WT and did not reach
the stationary phase in 12 h (Fig.
1A). Furthermore, its growth in M9 with glycerol was almost
abolished (Fig. 1B) indicating
that KST0555 was the auxotrophic mutant in the uptake of the carbon
source.
Reduction of KST0555 virulence in vitro
and in vivo
The present study then investigated the abilities of
KST0555 to invade into and replicate within RAW264.7 and IEC-6
cells. Cell monolayers were infected with Salmonella at a
MOI=10, and their relative invasion and replication ratio were
calculated at 2 and 18 hpi (Fig. 2A
and B). The invasion abilities (at 2 hpi) of KST0555 to
RAW264.7 and IEC-6 cells were reduced significantly compared with
that of the WT (P<0.005). In addition, the replication fold
increase of WT were 12.6±2.9 times and 11.7±3.2 times, whereas
those of KST0555 were only 1.3±0.2 and 1.5±0.3 times inside the
RAW264.7 and IEC-6 cells at 18 hpi, respectively (P<0.005).
To directly visualize the invasion and replication
of Salmonella within RAW264.7 cells, immunofluorescence was
performed using a FITC-conjugated Salmonella-specific
monoclonal antibody (Fig. 2C). A
higher density of FITC signals was present in the cell monolayers
infected with WT, compared with cells infected with KST0555 at 2
hpi, and the FITC signal was notably increased at 18 hpi,
indicating efficient invasion and replication in RAW264.7 cells.
However, KST0555 was barely detected at 2 and 18 hpi. The results
were similar with the pattern of intracellular invasion and
replication assays presented in Fig.
2A. All results suggest that the expression of ptsI is
necessary for invasion and replication in host cells; in other
words, its expression is likely required for the intracellular
growth of S. Typhimurium.
To assess the toxicity of KST0555 in vivo,
streptomycin-treated mice (n=5) were infected orally with the
indicated doses of WT or KST0555, followed by monitoring of the
mortality of mice for 21 days (Fig.
3A and B). When the mice (n=5) were inoculated with
1×107 CFU/mouse Salmonella WT strain, all mice
succumbed to mortality at 10 days subsequent the challenge and
their body weight were revealed to substantially decrease day by
day. When the mice were inoculated orally with 1×108
CFU/mouse KST0555, the mice exhibited a noticeable drop in their
body weight and 40% of the mice succumbed to mortality. The mice
inoculated orally with either 5×107 or 1×107
CFU/mouse doses did not succumb to mortality, but the mice
inoculated orally with 5×107 CFU/mouse exhibited a
noticeable body weight loss by day 3 and exhibited a slow recovery
in weight from day 11, indicating that the lethal dose 50
(LD50) of KST0555 was ~108 CFU/mouse, but the
optimal oral vaccination dose in mice was lower compared with
1×107 CFU/mouse.
Salmonella infections typically are
initiated by the ingestion of contaminated food and water, followed
by the successful colonization in the distal ileum (46,47). The present study initially
examined the colonized Salmonella in the mice organs at the
time of mortality or 72 h subsequent to the oral inoculation of
1×107 CFU bacteria (Fig.
3C). Mice infected with the WT strain revealed substantially
greater numbers of colonized Salmonella, estimated between
108 and 109 CFU/g, which was higher compared
with the number of infection, indicating infected Salmonella
colonized and replicated in the mice intestine. In contrast, it was
revealed that there was <106 CFU/g Salmonella
in the cecum of the mice infected with KST0555 at the time of
mortality or 72 hpi.
Colonized Salmonella are occasionally
translocated to systemic sites or to the peritoneal cavity to cause
iNTS (48). The present study
further examined the levels of Salmonella colonization in
mice mesenteric lymphnodes, spleen, and blood (Fig. 3D–F) at the time of mortality or 72
hpi. A total of >105–109 CFU/tissue or
CFU/ml Salmonella were detected in the mice organs and blood
infected with WT, but no or <104 CFU/tissue or CFU/ml
Salmonella were detected in mice organs and blood infected
with KST0555, indicating that the KST0555 strain is attenuated and
causes less pathological symptoms in vivo.
Downregulation of virulence-associated
genes in KST0555
Next, the present study investigated the effect of
the ptsI mutation on flagella expression and
Salmonella motility. Surprisingly, only the expression of
the fliC gene encoding a main flagellin subunit was
significantly reduced by 95±3.7% in KST0555 compared with that in
WT (P<0.005); however, flhD, fliA, fliZ and
flgM were not significantly reduced (Fig. 4A). To verify whether the
downregulation of fliC effectively influenced the motility
of Salmonella, stationary phase cultures of WT or KST0555
were inoculated on motility agar medium followed by monitoring
Salmonella migration diameters on agar plates following 18 h
of incubation. Even though only fliC gene expression was
impaired in KST0555, it exhibited significantly smaller halos
compared with those of WT (P<0.001; Fig. 4B).
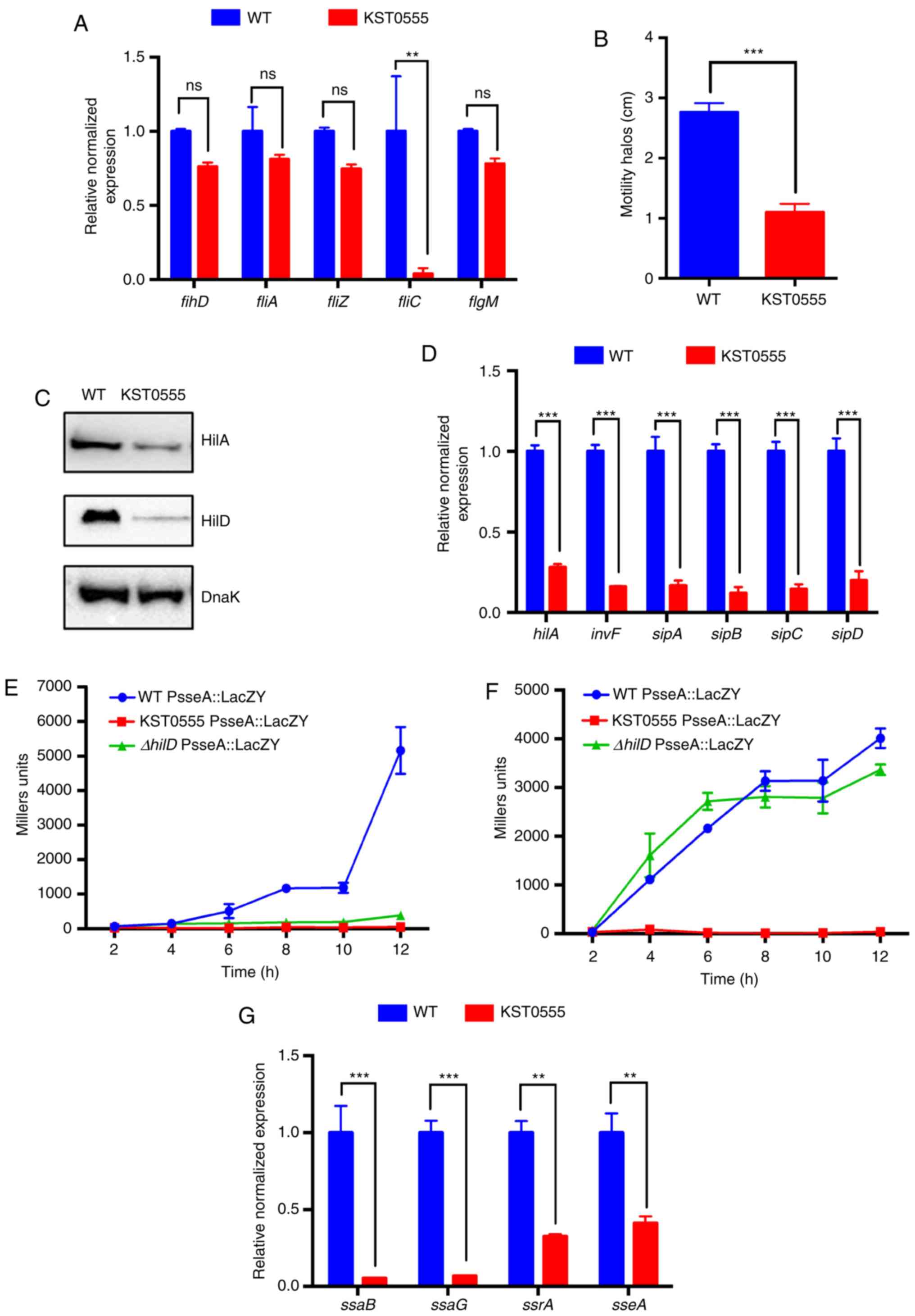 | Figure 4Downregulation of virulent genes and
key regulator proteins of Salmonella in T3SS. Lower mobility
caused by the deletion of the ptsI gene. (A) mRNA expression
levels of flagella biosynthesis genes, flhD, fliA,
fliZ, fliC and flgM in WT and KST0555 at
early-log phase in LB broth were measured using RT-qPCR. (B)
Salmonella motility was measured on an agar plate by
inoculating 10 μl WT or KST0555 culture broth. The zone size
of motility was measured at 18 h following inoculation. (C) Protein
expression of Salmonella T3SS key regulators (HilA and HiD)
in WT and KST0555 was examined using a HilA::FLAG and HilD::HA
chromosomal fusion system. (D) Reduced expression of
Salmonella SPI-1 genes and their regulators in KST0555. The
expression of SPI-1 encoded genes (hilA, invF,
sopA, sipB, sipC and sipD) in WT and
KST0555 at the early-log phase in LB broth was quantified using
RT-qPCR compared with that in WT. Reduced key virulence factor sseA
expression in KST0555 and ΔhilD was abolished, compared with
that in WT grown in (E) LB using the lacZ gene fused to the
promoter of sseA, but only KST0555 still exhibited no
expression of sseA, compared with ΔhilD and WT grown in (F)
LPM. (G) Downregulation of SPI-2 encoded genes in KST00555.
Transcription levels of SPI-2 encoded genes of KST0555 in LPM were
quantified using RT-qPCR compared with that in WT. Data are
representative of three independent experiments and presented as
the mean ± standard deviation. Asterisks indicate significant
differences between WT and KST0555. **P<0.05 and
***P<0.001 with comparisons shown by lines. WT, wild
type; RT-qPCR, reverse transcription-quantitative PCR; LB,
Luria-Bertani broth; LPM, MES-buffered minimum media; ns,
non-significant. |
To determine whether the substantial reduction of a
virulence phenotype in KST0555 was due to the downregulation of
major virulent genes in Salmonella, the expression of the
genes encoded in SPI-1 and -2 were examined, which have been
attributed to numerous virulence properties of Salmonella
(49,50). The majority of genes encoded
within SPI-1 are well known to serve an active function in
penetration into the intestinal epithelial barrier, and the
expression of SPI-1 genes are regulated by the fundamental
transcriptional regulators HilA, HilD and InvF (51). Thus, the present study firstly
compared the expression of HilA and HilD proteins, central
regulators of SPI-1 genes. As revealed in Fig. 4C, the expression of each of these
proteins was reduced in KST0555 compared with in WT. In addition,
invF mRNA levels, regulated by HilA and HilD, were
significantly downregulated in KST0555 compared with in WT, which
explains why invF-dependent sipBCDA mRNA expression
levels were reduced by ~80% in KST0555 compared with in WT
(P<0.001; Fig. 4D).
HilD is also known to activate SPI-2 genes, the key
virulence determinants for the survival/replication of
Salmonella inside the host cells, through inducing the
expression of the SsrA/B two-component system, the central positive
regulator of SPI-2 (52). Thus,
the expression of the key virulence factor sseA was compared
in WT, KST0555 and KST0583 (ΔhilD) grown in LB or LPM using
the lacZ gene fused to the promoter of sseA. As
expected, the sseA expression was almost abolished in
ΔhilD grown in only LB, whereas its expression was similar
with WT when cultured in LPM, SPI-2 gene expression media (Fig. 4E and F). However, no sseA
expression was revealed in KST0555 culture in LB and LPM. The
expression levels of the downstream genes of the SseA promoter
(SsaB, SsaG, SsrA and SseA) were examined using RT-qPCR. As shown
in Fig. 4G, these genes were
significantly downregulated in KST0555 culture in LB compared with
the WT [ssaB (P=0.001), ssaG (P=0.0003), ssrA
(P=0.0047) and sseA (P=0.0095)]. These results suggest that
the reduced SPI-2 expression in KST0555 was in part due to the
lower expression of HilD by a ptsI mutation, and additional
mechanism(s) may be responsible for the downregulation of SPI-2
expression other than HilD in KST0555.
Vaccination of KST0555 elicits protective
immune response against Salmonella
To determine whether KST0555 may be used as a live
oral vaccine, the mice were immunized orally (n=5) three times in
two-week intervals (0, 2 and 4 weeks) with 107 CFU of
KST0555 and the levels of Salmonella and Salmonella
LPS-specific antibodies in the serum were determined at 7 days
following the last immunization (5 weeks). As presented in Fig. 5A, KST0555 vaccination
significantly enhanced the serum levels of
Salmonella-specific IgG (mean titer=1024) and IgM (mean
titer=288) compared with those of PBS-immunized mice (mean
titer=36–72; P<0.05). LPS-specific IgG and IgM were also
elevated significantly by KST0555 vaccination compared with
PBS-treated mice (P<0.05; Fig.
5B). When the subclasses of IgG specific for Salmonella
(Fig. 5C) and LPS (Fig. 5D) were analyzed, Salmonella
and LPS specific-IgG1, IgG2a, IgG2b and IgG3 were significantly
increased compared with those of PBS groups (P<0.05). In
addition, significant increases in Th1 and Th2-spcific cytokines
(IFN-γ and IL-4) were detected in splenocytes from mice immunized
with KST0555 when compared with PBS-immunized control mice
(P<0.05) indicating that live KST0555 vaccine induces high
antigen-specific antibody responses and T-cell mediated immune
responses (Fig. 5E and F).
 | Figure 5Protective effect of KST0555
immunization against Salmonella infections in mice. BALB/c
mice (n=5) were orally immunized with KST0555 or PBS three times at
two-week intervals. Serum levels of (A) Salmonella- or (B)
LPS-specific total Ig, IgG and IgM were analyzed at 7 days
following the last immunization. Data are representative of three
independent experiments and are presented as the mean ± standard
deviation. Subclass levels of (C) Salmonellaand (D)
LPS-specific IgG1, IgG2a, IgG2b and IgG3 were analyzed at 7 days
subsequent to the last immunization. Data are representative of
three independent experiments and are presented as the mean ±
standard deviation. Splenocytes isolated from mice were incubated
with 108 CFU of S. Typhimurium WT lysate for 72
h, and concentrations of (E) IFN-γ and (F) IL-4 were measured from
culture supernatants using an enzyme-linked immunosorbent assay.
Bars indicate the mean of results of triplicate wells of a
representative experiment. Error bars indicate standard deviation.
Immunized mice were challenged orally with 107 CFU of WT
at 7 days following the last immunization and (G) mouse survival
and (H) body weight were monitored for 14 days.
*P<0.05, **P<0.005 and
***P<0.001 vs. PBS group. PBS, phosphate buffered
saline; Ig, immunoglobulin; LPS, lipopolysaccharide; CFU,
colony-forming units; WT, wild-type; IFN-γ, interferon-γ; IL-4,
interleukin-4. |
To directly determine the protective effect of a
live KST0555 vaccine, the immunized mice were challenged orally
with 107 CFU of WT at 7 days following the last
vaccination (week 5). All PBS-immunized mice succumbed to mortality
in 12 days post-injection along with a substantial reduction of
body weight daily, but 100% of KST0555-immunized mice were alive
for >14 days post-injection without an obvious change in body
weight (Fig. 5G and H). This
suggests that a live KST0555 vaccine may provide a protective
immune response.
Protection by adoptive transfer of serum,
CD4+ or CD8+ T cells
The present study then investigated whether a live
KST0555 vaccine may elicit functional humoral and cell-mediated
immunities by an adoptive transfer experiment. Five mice in each
group were administered intraperitoneally with 300 μl serum,
1×106 CD4+ T cells or 1×105
CD8+ T cells isolated from unvaccinated or live KST0555
(1×107 CFU) vaccinated mice as described above, followed
by challenging with 107 CFU of WT Salmonella.
Mice receiving the serum from live KST0555 immunized mice survived
significantly longer compared with mice receiving unimmunized serum
(P=0.0042; Fig. 6A), indicating
that the antibodies raised by KST0555 vaccination were functional.
The mice receiving splenic CD4+ T cells from
KST0555-immunized mice survived 2 or 3 days longer compared with
the naïve group, which was not significant, but modestly protected
(P=0.0547; Fig. 6B). The mice
receiving splenic CD8+ T cells from KST0555-immunized
mice exhibited significant and sufficient protection compared with
the transfer of those from the naïve group (P=0.0015; Fig. 6C), suggesting that a live oral
KST0555 vaccine might induce an efficient cell-mediated immune
response.
Discussion
NTS infection is the leading cause of mortality in
Africa and other developing countries (3). Previous comprehensive
epidemiological studies of iNTS indicate the urgent need of novel
therapeutic methods for these infections (3,4,53).
Live attenuated vaccines against NTS are a promising solution for
the spreading of NTS, because they have numerous advantages over
other vaccine types, including cost-effective production, induction
of mucosal and cell-mediated immunities and no hazardous waste
(21). In the present study, it
was hypothesized that the disruption of ptsI in S.
Typhimurium may create an attenuated strain, which is efficacious
for vaccine application, by interfering with the uptake of PTS
sugars, particularly glucose, the predominant carbohydrate source
during infection. This strain, deficient in the uptake of PTS
(glucose) and non-PTS (glycerol) sugars, exhibited a significantly
(P=0.0172) reduced virulence in vitro and in vivo and
elicited effective humoral and cell-mediated immune responses to
protect against Salmonella infection.
The vaccine strain attenuated in metabolic gene(s)
requires being metabolically active to reach immune inductive sites
and elicit a biologically relevant protective immunity in the
absence of overt disease. Furthermore, the hyperattenuation of
vaccine strains may require the administration of multiple oral
doses to achieve an effectively protective immune response
(54,55). The deletion of aroA, a gene
of the shikimate pathway associated with the synthesis of aromatic
amino acids, is most commonly used as a metabolic mutation to
attenuate Salmonella (56). The guaBA gene-deleted
vaccine candidates, which impair guanine biosynthesis, are based on
S. Typhimurium and S. Enteritidis isolates from
sub-Saharan Africa; preclinical studies were conducted using a
series of Center for Vaccine Development (University of Maryland
School of Medicine) by using relevant animal models for human
gastroenteritis, including streptomycin-pretreated mouse and rhesus
macaque models (20,21). Another approach for developing
live attenuated vaccines is the deletion of genes involved in the
uptake of essential elements or nutrients that are indispensable
for the growth of infectious microorganisms. In one example, the
mutant in the znuABC operon encoding the high-affinity zinc
transporter was revealed to be a safe and efficacious vaccine
candidate in the setting of mouse and pig vaccine models (57–60).
In a previous study, the extreme attenuation was
achieved by deleting two components genes, ptsI and
crr genes, in PTS, despite the fact that a high number
(>1×109 CFU) of ΔptsIcrr was administrated
without a noticeable body weight loss and its humoral immune
responses may protect against a lethal challenge of its parent
strain (34). However, the
hyperattenuation of the ΔptsIcrr strain also result in a
high dose of CFU being needed, along with the non-activation of
cell-mediated immune responses (34). In the present study, the KST0555
strain was defective in only one sugar transport gene
(ptsI), but also highly attenuated in the mouse model.
Although its targeting to immune sites, including spleen and
mesenteric lymph nodes, was substantially reduced compared with
that of the parent strain, its immune responses were sufficient to
provide the protection against Salmonella infection via both
humoral and cellular immunity.
In fact, numerous live attenuated Salmonella
vaccine strains under pre-clinical and clinical studies have been
revealed to have associations between the transcriptional and
translational expression for two notable groups of virulence genes,
SPI-1 and SPI-2 (21,24,61–63). Each are essential for
colonization, invasion and intracellular proliferation and thus
contribute to the successful infection of S. Typhimurium
into the host cells by assembling T3SS (64). PTS is the most well-known and
important carbon uptake system consisting of two general
components: Enzyme I (EI) encoded by ptsI and histidine
phosphocarrier protein (HPr) encoded by ptsH and the
membrane-associated EII complex. EII complexes, consisting of the
EIIA, EIIB and EIIC proteins, are usually carbon-specific; for
example, EIIAGlc encoded by crr stands for the
glucose-specific EIIA component (65). The first PTS component, PtsI,
autophosphorylates by using phosphoenolpyruvate as a phosphoryl
donor and transfers the phosphate group through HPr to the EII
complex to trigger the uptake of PTS sugars, including glucose,
mannose, mannitol, fructose and cellobiose (33). Further evidence suggests that
virulence functions and metabolism of pathogens are intertwined by
global regulatory networks, which serve an important function in
the regulation of virulence traits (66). The SirA/BarA two-component system
involved in various cellular responses, including carbohydrate
metabolism, motility, biofilm formation and stress survival, may
regulate the expression of SPI-1 and SPI-2 genes alone with the
carbon storage regulatory (csr) systems (67,68). Since the expression of sirA
and csrA is reduced by the mutation of the adenylate cyclase
gene, which synthesizes cyclic AMP (cAMP) from ATP, or the cAMP
receptor protein gene (crp) in S. Typhimurium
(67) and deletion of the
ptsI gene, prevents the formation of phosphorylated
EIIAGlc, the activator of adenylate cyclase, the
reduction of SPI-1 and SPI-2 gene expression in KST0555
(ΔptsI) is likely to be partially mediated via the
cAMP/crp regulation mechanism (69). In addition, it was revealed that
the reduction of virulence in KST0555 was not entirely due to the
downregulation of SPI genes. One novel result was that only the
fliC gene of the five flagella-associated genes tested was
significantly downregulated in KST0555, which impaired
Salmonella motility. Flagella-mediated motility is known to
be required for the efficient colonization and induction of colitis
in mice (70,71). Thus, further investigation on the
mechanisms by which ptsI mutation affects the flagella gene
expression of S. Typhimurium is warranted. The notable
downregulation of SPI-1 and SPI-2 genes in ptsI mutants
reveals the advantage of this mutant strain as an attenuated
vaccine candidate. However, the comparison of the immunological
efficacy of this strain with previously well-characterized vaccine
strains and the regulation axis between carbon uptake and virulence
in Salmonella should be investigated further at the
molecular level.
In summary, the present study provides further
strong evidence that the carbon transport system is an excellent
target to develop a live Salmonella vaccine. Since the
reduced virulence of KST0555 amy be attributed to not only the
impairment of the sugar uptake but also the downregulation of
virulence genes including SPI and flagellin, the molecular
mechanism of the association between virulence and PTS should be
studied further. However, these promising preclinical data provide
a strong rationale for the advancement of KST0555 as a vaccine
candidate. Furthermore, since PTS is widely conserved in
gram-negative and gram-positive bacteria, the present study offers
a novel insight for developing novel vaccine strains against
various pathogenic bacteria.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Research Foundation of Korea (grant nos. NRF-2017M2A2A6A020 20925
and NRF-2018K2A9A2A06023828 to HSS) and the Nuclear R&D Program
of the Ministry of Science, ICT & Future Planning (to SL).
Availability of data and materials
The data used and/or analyzed in this study are
available from the corresponding author upon reasonable
request.
Authors’ contributions
YZ and HSS conceived and designed the experiments.
YZ, HSS, SML, KBA and HJJ performed the experiments. YZ, HSS, KBA,
HCG, SR and SL analyzed the data. YZ, HSS, KBA and SL wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The animal experiments were ethically approved by
the Committee on The Use and Care of Animals at the Korea Atomic
Energy Research Institute and performed according to accepted
National Health Institute standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abed N, Grepinet O, Canepa S,
Hurtado-Escobar GA, Guichard N, Wiedemann A, Velge P and
Virlogeux-Payant I: Direct regulation of the pefI-srgC operon
encoding the Rck invasin by the quorum-sensing regulator SdiA in
Salmonella Typhimurium. Mol Microbiol. 94:254–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Febriani Y, Levallois P, Gingras S,
Gosselin P, Majowicz SE and Fleury MD: The association between
farming activities, precipitation, and the risk of acute
gastrointestinal illness in rural municipalities of Quebec, Canada:
A cross-sectional study. BMC Public Health. 10:482010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ao TT, Feasey NA, Gordon MA, Keddy KH,
Angulo FJ and Crump JA: Global burden of invasive nontyphoidal
Salmonella disease, 2010(1). Emerg Infect Dis. 21:2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feasey NA, Dougan G, Kingsley RA,
Heyderman RS and Gordon MA: Invasive non-typhoidal Salmonella
disease: An emerging and neglected tropical disease in Africa.
Lancet. 379:2489–2499. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Crump JA, Sjolund-Karlsson M, Gordon MA
and Parry CM: Epidemiology, clinical presentation, laboratory
diagnosis, anti-microbial resistance, and antimicrobial management
of invasive Salmonella infections. Clin Microbiol Rev. 28:901–937.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zansky S, Wallace B, Schoonmaker-Bopp D,
Smith P, Ramsey F, Painter J, Gupta A, Kalluri P and Noviello S:
From the centers for disease control and prevention. Outbreak of
multi-drug resistant Salmonella Newport-United States,
January–April 2002. JAMA. 288:951–953. 2002.PubMed/NCBI
|
|
7
|
Strugnell RA, Scott TA, Wang N, Yang C,
Peres N, Bedoui S and Kupz A: Salmonella vaccines: Lessons from the
mouse model or bad teaching? Curr Opin Microbiol. 17:99–105. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Typhoid vaccines: WHO position paper. Wkly
Epidemiol Rec. 83:49–59. 2008.(In English, Finnish). PubMed/NCBI
|
|
9
|
DeRoeck D, Ochiai RL, Yang J, Anh DD, Alag
V and Clemens JD: Typhoid vaccination: The Asian experience. Expert
Rev Vaccines. 7:547–560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
DeRoeck D, Clemens JD, Nyamete A and
Mahoney RT: Policymakers’ views regarding the introduction of
new-generation vaccines against typhoid fever, shigellosis and
cholera in Asia. Vaccine. 23:2762–2774. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kantele A, Pakkanen SH, Siitonen A,
Karttunen R and Kantele JM: Live oral typhoid vaccine Salmonella
Typhi Ty21a-a surrogate vaccine against non-typhoid salmonella?
Vaccine. 30:7238–7245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Watson DC, Robbins JB and Szu SC:
Protection of mice against Salmonella typhimurium with an
O-specific polysaccharide-protein conjugate vaccine. Infect Immun.
60:4679–4686. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gerke C, Colucci AM, Giannelli C, Sanzone
S, Vitali CG, Sollai L, Rossi O, Martin LB, Auerbach J, Di Cioccio
V and Saul A: Production of a shigella sonnei vaccine based on
generalized modules for membrane antigens (GMMA), 1790GAHB. PLoS
One. 10:e01344782015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Secundino I, Lopez-Macias C,
Cervantes-Barragan L, Gil-Cruz C, Ríos-Sarabia N, Pastelin-Palacios
R, Villasis-Keever MA, Becker I, Puente JL, Calva E and Isibasi A:
Salmonella porins induce a sustained, lifelong specific
bactericidal antibody memory response. Immunology. 117:59–70. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kodama C and Matsui H: Salmonella
flagellin is not a dominant protective antigen in oral immunization
with attenuated live vaccine strains. Infect Immun. 72:2449–2451.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gil-Cruz C, Bobat S, Marshall JL, Kingsley
RA, Ross EA, Henderson IR, Leyton DL, Coughlan RE, Khan M, Jensen
KT, et al: The porin OmpD from nontyphoidal Salmonella is a key
target for a protective B1b cell antibody response. Proc Natl Acad
Sci USA. 106:9803–9808. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sette A and Rappuoli R: Reverse
vaccinology: Developing vaccines in the era of genomics. Immunity.
33:530–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salazar-Gonzalez RM, Maldonado-Bernal C,
Ramirez-Cruz NE, Rios-Sarabia N, Beltrán-Nava J, Castañón-González
J, Castillo-Torres N, Palma-Aguirre JA, Carrera-Camargo M,
López-Macías C and Isibasi A: Induction of cellular immune response
and anti-Salmonella enterica serovar typhi bactericidal antibodies
in healthy volunteers by immunization with a vaccine candidate
against typhoid fever. Immunol Lett. 93:115–122. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hodak H and Galan JE: A Salmonella typhi
homologue of bacteriophage muramidases controls typhoid toxin
secretion. EMBO Rep. 14:95–102. 2013. View Article : Google Scholar :
|
|
20
|
MacLennan CA, Martin LB and Micoli F:
Vaccines against invasive Salmonella disease: Current status and
future directions. Hum Vaccin Immunother. 10:1478–1493. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tennant SM and Levine MM: Live attenuated
vaccines for invasive Salmonella infections. Vaccine. 33(Suppl 3):
C36–C41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tennant SM, Wang JY, Galen JE, Simon R,
Pasetti MF, Gat O and Levine MM: Engineering and preclinical
evaluation of attenuated nontyphoidal Salmonella strains serving as
live oral vaccines and as reagent strains. Infect Immun.
79:4175–4185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tacket CO, Sztein MB, Losonsky GA,
Wasserman SS, Nataro JP, Edelman R, Pickard D, Dougan G, Chatfield
SN and Levine MM: Safety of live oral Salmonella typhi vaccine
strains with deletions in htrA and aroC aroD and immune response in
humans. Infect Immun. 65:452–456. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Angelakopoulos H and Hohmann EL: Pilot
study of phoP/phoQ-deleted Salmonella enterica serovar typhimurium
expressing Helicobacter pylori urease in adult volunteers. Infect
Immun. 68:2135–2141. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tran TH, Nguyen TD, Nguyen TT, Ninh TT,
Tran NB, Nguyen VM, Tran TT, Cao TT, Pham VM, Nguyen TC, et al: A
randomised trial evaluating the safety and immunogenicity of the
novel single oral dose typhoid vaccine M01ZH09 in healthy
Vietnamese children. PLoS One. 5:e117782010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lyon CE, Sadigh KS, Carmolli MP, Harro C,
Sheldon E, Lindow JC, Larsson CJ, Martinez T, Feller A, Ventrone
CH, et al: In a randomized, double-blinded, placebo-controlled
trial, the single oral dose typhoid vaccine, M01ZH09, is safe and
immunogenic at doses up to 1.7 × 10(10) colony-forming units.
Vaccine. 28:3602–3608. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ault A, Tennant SM, Gorres JP, Eckhaus M,
Sandler NG, Roque A, Livio S, Bao S, Foulds KE, Kao SF, et al:
Safety and tolerability of a live oral Salmonella typhimurium
vaccine candidate in SIV-infected nonhuman primates. Vaccine.
31:5879–5888. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eisenreich W, Dandekar T, Heesemann J and
Goebel W: Carbon metabolism of intracellular bacterial pathogens
and possible links to virulence. Nat Rev Microbiol. 8:401–412.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abrahams GL and Hensel M: Manipulating
cellular transport and immune responses: Dynamic interactions
between intracellular Salmonella enterica and its host cells. Cell
Microbiol. 8:728–737. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bowden SD, Rowley G, Hinton JC and
Thompson A: Glucose and glycolysis are required for the successful
infection of macrophages and mice by Salmonella enterica serovar
typhimurium. Infect Immun. 77:3117–3126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Klose KE and Mekalanos JJ: Simultaneous
prevention of glutamine synthesis and high-affinity transport
attenuates Salmonella typhimurium virulence. Infect Immun.
65:587–596. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Postma PW, Lengeler JW and Jacobson GR:
Phosphoenolpy ruvate:Carbohydrate phosphotransferase systems of
bacteria. Microbiol Rev. 57:543–594. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Deutscher J, Francke C and Postma PW: How
phosphotransferase system-related protein phosphorylation regulates
carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev.
70:939–1031. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhi Y, Lin SM, Jang AY, Ahn KB, Ji HJ, Guo
HC, Lim S and Seo HS: Effective mucosal live attenuated Salmonella
vaccine by deleting phosphotransferase system component genes ptsI
and crr. J Microbiol. 57:64–73. 2019. View Article : Google Scholar
|
|
35
|
Maze A, Glatter T and Bumann D: The
central metabolism regulator EIIAGlc switches Salmonella from
growth arrest to acute virulence through activation of virulence
factor secretion. Cell Rep. 7:1426–1433. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luo Y, Kong Q, Yang J, Golden G, Wanda SY,
Jensen RV, Ernst PB and Curtiss R III: Complete genome sequence of
the universal killer Salmonella enterica serovar typhimurium UK-1
(ATCC 68169). J Bacteriol. 193:4035–4036. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Datsenko KA and Wanner BL: One-step
inactivation of chromosomal genes in Escherichia coli K-12 using
PCR products. Proc Natl Acad Sci USA. 97:6640–6645. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ebel-Tsipis J, Fox MS and Botstein D:
Generalized transduction by bacteriophage P22 in Salmonella
typhimurium. II. Mechanism of integration of transducing DNA. J Mol
Boil. 71:449–469. 1972. View Article : Google Scholar
|
|
39
|
Davis RW, Botstein D and Roth JR: Advanced
Bacterial Genetics. Cold Spring Harbor Laboratory; Cold Spring
Harbor, NY: 1980
|
|
40
|
Miller JH: Experiments in molecular
genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY:
1972
|
|
41
|
Coombes BK, Coburn BA, Potter AA, Gomis S,
Mirakhur K, Li Y and Finlay BB: Analysis of the contribution of
Salmonella pathogenicity islands 1 and 2 to enteric disease
progression using a novel bovine ileal loop model and a murine
model of infectious enterocolitis. Infect Immun. 73:7161–7169.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu XJ, McGourty K, Liu M, Unsworth KE and
Holden DW: pH sensing by intracellular Salmonella induces effector
translocation. Science. 328:1040–1043. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−D elta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
44
|
Kim W and Surette MG: Swarming populations
of Salmonella represent a unique physiological state coupled to
multiple mechanisms of antibiotic resistance. Biol Proced Online.
5:189–196. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nishikawa F, Kita E, Matsui N and Kashiba
S: Transfer of protection to murine typhoid conferred by L-form
Salmonella typhimurium in dependence of cooperation between L
form-adopted macrophages and L form-induced Lyt-2+ T
cells. Microbiol Immunol. 38:201–207. 1994. View Article : Google Scholar
|
|
46
|
Meyerholz DK and Stabel TJ: Comparison of
early ileal invasion by Salmonella enterica serovars choleraesuis
and typhimurium. Vet Pathol. 40:371–375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Haque A, Bowe F, Fitzhenry RJ, Frankel G,
Thomson M, Heuschkel R, Murch S, Stevens MP, Wallis TS, Phillips AD
and Dougan G: Early interactions of Salmonella enterica serovar
typhimurium with human small intestinal epithelial explants. Gut.
53:1424–1430. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tierrez A and Garcia-del Portillo F: New
concepts in Salmonella virulence: The importance of reducing the
intracellular growth rate in the host. Cell Microbiol. 7:901–909.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Marcus SL, Brumell JH, Pfeifer CG and
Finlay BB: Salmonella pathogenicity islands: Big virulence in small
packages. Microbes Infect. 2:145–156. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hensel M: Salmonella pathogenicity island
2. Mol Microbiol. 36:1015–1023. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lostroh CP and Lee CA: The HilA box and
sequences outside it determine the magnitude of HilA-dependent
activation of P(prgH) from Salmonella pathogenicity island 1. J
Bacteriol. 183:4876–4885. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bustamante VH, Martinez LC, Santana FJ,
Knodler LA, Steele-Mortimer O and Puente JL: HilD-mediated
transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad
Sci USA. 105:14591–14596. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Haselbeck AH, Panzner U, Im J, Baker S,
Meyer CG and Marks F: Current perspectives on invasive nontyphoidal
Salmonella disease. Curr Opin Infect Dis. 30:498–503. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Darji A, zur Lage S, Garbe AI, Chakraborty
T and Weiss S: Oral delivery of DNA vaccines using attenuated
Salmonella typhimurium as carrier. FEMS Immunol Med Microbiol.
27:341–349. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Levine MM, Ferreccio C, Abrego P, Martin
OS, Ortiz E and Cryz S: Duration of efficacy of Ty21a, attenuated
Salmonella typhi live oral vaccine. Vaccine. 17(Suppl 2): S22–S27.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hoiseth SK and Stocker BA:
Aromatic-dependent Salmonella typhimurium are non-virulent and
effective as live vaccines. Nature. 291:238–239. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pasquali P, Ammendola S, Pistoia C,
Petrucci P, Tarantino M, Valente C, Marenzoni ML, Rotilio G and
Battistoni A: Attenuated Salmonella enterica serovar Typhimurium
lacking the ZnuABC transporter confers immune-based protection
against challenge infections in mice. Vaccine. 26:3421–3426. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pesciaroli M, Aloisio F, Ammendola S,
Pistoia C, Petrucci P, Tarantino M, Francia M, Battistoni A and
Pasquali P: An attenuated Salmonella enterica serovar Typhimurium
strain lacking the ZnuABC transporter induces protection in a mouse
intestinal model of Salmonella infection. Vaccine. 29:1783–1790.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gradassi M, Pesciaroli M, Martinelli N,
Ruggeri J, Petrucci P, Hassan WH, Raffatellu M, Scaglione FE,
Ammendola S, Battistoni A, et al: Attenuated Salmonella enterica
serovar Typhimurium lacking the ZnuABC transporter: An efficacious
orally-administered mucosal vaccine against salmonellosis in pigs.
Vaccine. 31:3695–3701. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pesciaroli M, Gradassi M, Martinelli N,
Ruggeri J, Pistoia C, Raffatellu M, Magistrali CF, Battistoni A,
Pasquali P and Alborali GL: Salmonella Typhimurium lacking the
Znuabc transporter is attenuated and immunogenic in pigs. Vaccine.
31:2868–2873. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sittka A, Pfeiffer V, Tedin K and Vogel J:
The RNA chaperone Hfq is essential for the virulence of Salmonella
typhimurium. Mol Microbiol. 63:193–217. 2007. View Article : Google Scholar :
|
|
62
|
Lopez-Garrido J and Casadesus J:
Regulation of Salmonella enterica pathogenicity island 1 by DNA
adenine methylation. Genetics. 184:637–649. 2010. View Article : Google Scholar :
|
|
63
|
Hindle Z, Chatfield SN, Phillimore J,
Bentley M, Johnson J, Cosgrove CA, Ghaem-Maghami M, Sexton A, Khan
M, Brennan FR, et al: Characterization of Salmonella enterica
derivatives harboring defined aroC and Salmonella pathogenicity
island 2 type III secretion system (ssaV) mutations by immunization
of healthy volunteers. Infect Immun. 70:3457–3467. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hansen-Wester I and Hensel M: Salmonella
pathogenicity islands encoding type III secretion systems. Microbes
Infect. 3:549–559. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Le Bouguenec C and Schouler C: Sugar
metabolism, an additional virulence factor in enterobacteria. Int J
Med Microbiol. 301:1–6. 2011. View Article : Google Scholar
|
|
66
|
Wilharm G and Heider C: Interrelationship
between type three secretion system and metabolism in pathogenic
bacteria. Front Cell Infect Microbiol. 4:1502014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Teplitski M, Goodier RI and Ahmer BM:
Catabolite repression of the SirA regulatory cascade in Salmonella
enterica. Int J Med Microbiol. 296:449–466. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Martinez LC, Yakhnin H, Camacho MI,
Georgellis D, Babitzke P, Puente JL and Bustamante VH: Integration
of a complex regulatory cascade involving the SirA/BarA and Csr
global regulatory systems that controls expression of the
Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol
Microbiol. 80:1637–1656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Poncet S, Milohanic E, Maze A, Abdallah
JN, Aké F, Larribe M, Deghmane AE, Taha MK, Dozot M, De Bolle X, et
al: Correlations between carbon metabolism and virulence in
bacteria. Contrib Microbiol. 16:88–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Stecher B, Hapfelmeier S, Muller C, Kremer
M, Stallmach T and Hardt WD: Flagella and chemotaxis are required
for efficient induction of Salmonella enterica serovar Typhimurium
colitis in streptomycin-pretreated mice. Infect Immun.
72:4138–4150. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Stecher B, Barthel M, Schlumberger MC,
Haberli L, Rabsch W, Kremer M and Hardt WD: Motility allows S.
Typhimurium to benefit from the mucosal defence. Cell Microbiol.
10:1166–1180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lim S, Han A, Kim D and Seo HS:
Transcriptional profiling of an attenuated Salmonella Typhimurium
ptsI mutant strain under low-oxygen conditions using microarray
analysis. J Bacteriol Virol. 45:1–15. 2015. View Article : Google Scholar
|
|
73
|
Lim S, Kim M, Choi J and Ryu S: A mutation
in tdcA attenuates the virulence of Salmonella enterica serovar
Typhimurium. Mol Cells. 29:509–517. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Baba T, Ara T, Hasegawa M, Takai Y,
Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL and Mori H:
Construction of Escherichia coli K-12 in-frame, single-gene
knockout mutants: The Keio collection. Mol Syst Biol. 2:2006 0008.
2006. View Article : Google Scholar : PubMed/NCBI
|




















