Introduction
Gastric cancer (GC) results in a high mortality rate
in China (1). At present, the
diagnosis of GC is largely based on clinical symptoms in
combination with physical signs and endoscopy, B-mode ultrasound,
CT examination and exfoliative cytology (2), which, however, all have certain
defects. In addition, symptoms are not always obvious at the early
stages of GC (3). Numerous GC
patients are already in the advanced stages of the disease by the
time of diagnosis, and thus face a greater risk of developing
recurrence and metastasis following surgical resection (4). Therefore, improving the
understanding of the molecular mechanisms of GC proliferation and
metastasis will facilitate the discovery of novel biomarkers and
therapeutic targets for the treatment of GC.
circRNAs have stable structures and have recently
attracted ample research attention (5). Compared with linear RNAs, circRNAs
are not easily degraded by exonucleases, they are highly conserved
and remain stable in vivo, which suggests that circRNAs have
the potential to function as diagnostic markers (6). circRNAs primarily function as miRNA
'sponges', containing numbers and types of miRNA response elements
at varying degrees (5,7). circRNAs can bind to specific miRNAs
and competitively inhibit the binding of the miRNA to the
corresponding site, and subsequently regulate the expression of
downstream target genes (7).
Zhong et al demonstrated that circRNA MYLK functions a
competitive endogenous RNA in bladder cancer (8). The overexpression of circRNA MYLK in
bladder cancer cell lines can significantly reduce the expression
of miR-29a (8).
circRNAs play crucial roles in a variety of cancers
by functioning as tumor suppressor genes or oncogenes, suggesting
their potential to serve as tumor biomarkers (9). Han et alanalyzed the
expression profiles of circRNAs in hepatocellular carcinoma (HCC)
tissues, and found that patients with a low expression of circMTO1
had a shorter survival time, suggesting that it may be a tumor
suppressor (10). Furthermore,
miR-9, which is a circMTO1-related miRNA, was found to downregulate
p21 expression, and promote the proliferation and invasion of HCC
cells (10). A previous study
found that miRNAs can regulate the expression of almost 2/3 of
human proteins, and >50% of miRNAs are located in
tumor-associated genomic regions (11). Chromosomal abnormalities directly
lead to alterations in gene copy numbers, resulting in an abnormal
expression in various tumors. The indispensable roles of miRNAs and
circRNAs in the development of GC have been previously observed
(6,12).
The high expression of hsa_circ_0001982 (circRNA
RNF111) has been shown to promote the proliferation and migration
of breast cancer cells (13). In
addition, lncRNA UCA1 can enhance the multidrug resistance of GC by
downregulating miR-27b expression (14). However, whether circRNA RNF111 is
also regulated by miR-27b expression and whether it is involved in
GC remains to be determined. Therefore, the present study examined
the association between circRNA RNF111 and miR-27b by investigating
their potential molecular mechanisms and their biological effects
on GC.
Materials and methods
Ethics statement
The GC tissues were obtained from 48 patients who
were diagnosed with GC at the Nanjing First Hospital from November,
2018 to November, 2019. All the patients signed informed consent
forms, and agreed that their tissues would be used for clinical
research. The clinical trial program had been reviewed and approved
by the Ethics Committee of Nanjing First Hospital (W201810013).
Tissues, cell lines and culture
The GC tissues and their corresponding normal
adjacent tissues (at least 5 cm away from the tumor) were collected
from 48 patients with GC. Human normal gastric mucosa cells (GES-1,
CS0097, Beijing Dingguo Changsheng Biotechnology Co., Ltd.) and GC
cell lines [AGS (CRL-1739, ATCC), SNU-16 (CRL-5974, ATCC), MNK-45
(CS0096, Beijing Dingguo Changsheng Biotechnology Co., Ltd.) and
HGC27 (94042256, Sigma; Merck KGaA)] were used in the present
study.
The cells were cultured in DMEM (12491015, Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS, 30067334), 1% penicillin/streptomycin (15140163) at 37°C with
5% CO2 for 1 day. Following culture, the primary medium
was discarded, and the cells were washed with PBS 1-2 times; fresh
DMEM medium (5 ml) was then added and the cells were returned to
the CO2 incubator. The cells were sub-cultured after 80
to 90% of them had adhered to the wall.
Cell transfection
After 12 to 16 h, the cells reached 60 to 70%
confluency and were transfected with lipo-some. circRNA siRNF111
(5′-UAA GGA AAG CCU GAG GGA AC-3′), siNC (siN0000001-1-5) and
miR-27b-3p inhibitor (miR20000419-1-5), inhibitor negative control
(miR2N0000001-1-5) were purchased from Guangzhou RiboBio Co., Ltd.
Cell transfection was performed according to the protocol of the
manufacturer of Lipofectamine 3000 (L3000015, Thermo Fisher
Scientific, Inc.). In brief, after the seeded cells reached 70-90%
fusion and were ready for transfection, Lipofectamine 3000 reagent
and RNA (100 ng/well) were diluted in Opti-MEM medium,
respectively. The diluted Lipofectamine 3000 reagent was added with
the diluted RNA was added at 1:1. Following incubation of the
mixture for 10 min at room temperature, the RNA-lipid complex was
added to the cells and incubated for 2 days at 37°C. Finally, the
transfected cells were further analyzed.
Bioinformatics assay
The latent binding sites between circR-RNF111 and
miR-27b-3p were predicted using Starbase v2.0 (http://starbase.sysu.edu.cn/).
Dual-luciferase activity assay
WT-RNF111 and MUT-RNF111 DNA were cloned into
pmirGLO luciferase Vectors (E1330; Promega Corporation). To analyze
the latent binding sites between circRNA RNF111 and miR-27b-3p,
293T cells (CRL-11268, ATCC) were transfected with miR-27b-3p mimic
or inhibitor. The luciferase activity was determined using a
Double-Luciferase Reporter Assay kit (FR201-01, Beijing Transgen
Biotech Co., Ltd.). Briefly, the cell culture medium was removed,
and the cells were cautiously rinsed twice in PBS and then fully
lysed by 100 µl Cell Lysis Buffer for 10 min at room
temperature. The supernatant was obtained by centrifuging the cells
at 12,000 × g at 4°C for 10 min. Luciferase Reaction Reagent (100
µl) was added to the centrifugal tube at room temperature,
cell lysates (20 µl) were cautiously pipetted into the tube,
and the two were then gently mixed. The Firefly luciferase reporter
gene activity was measured using a luminescence meter (SpectraMaxL,
Molecular Devices, LLC). Finally, Luciferase Reaction Reagent II
(100 µl) was added to the above-mentioned reaction tube at
room temperature, vortexed, and the activity of the Renilla
luciferase reporter gene was determined using a luminescence
meter.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the GC tissues and
cells using the TRIzol method at 4°C. The reverse transcription of
the RNA was performed using the MicroRNA Reverse Transcription kit
(4366597, Thermo Fisher Scientific, Inc.) according to the
manufacturers' protocol. For regent preparation, briefly, 1.5
µl total RNA (10-200 ng), 2.0 µl One-Color Spike Mix
for Cyanine 3-labeling, 0.8 µl T7 Promoter Primer and 1.0
µl RNase free water were thoroughly mixed at 65°C for 10 min
and then reacted in ice bath for 5 min to form the Reaction 1. The
Reaction 2 was composed of 2.0 µl 5X First Strand Buffer,
1.0 µl 0.1 M DTT, 0.5 µl 10 m M d NTP mix, and 1.2
µl Affinity Script RNase Block Mix. The Reaction 1 was then
mixed with the Reaction 2 at 40°C for 120 min, at 70°C for 5 min,
and at 4°C for 5 min. qPCR was performed according to the
instructions of the detection system (ABI 7500, Life Technologies;
Thermo Fisher Scientific, Inc.). The relative expression levels
were calculated using the 2−ΔΔCq method (15). GAPDH was used as a housekeeping
gene for mRNA, and U6 as an internal control for miRNA. The primer
sequences were as follows: miR-27b-3p forward, 5′-AGT GGC TAA GTT
CTG CGT CG-3′ and reverse, 5′-GTA TCC AGT GCG TGT CGT GG-3′; RNF111
forward, 5′-TAG CAG TTC CCCAAT CCT TG-3′ and reverse, 5′-CAC AAA
TTC CCA TCA TTC CC-3′; GAPDH forward, 5′-TGT GGG CAT CAA TGG ATT
TGG-3′ and reverse, 5′-ACA CCA TGT ATT CCG GGT CAA T-3′; and U6
forward, 5′-AAA GCA AAT CAT CGG ACG ACC-3′ and reverse, 5′-GTA CAA
CAC ATT GTT TCC TCG GA-3′.
CCK-8 assay
The GC cells (1×104/well) were added to a
96-well plate and incubated for 24 h at 37°C. Following
transfection for 12, 24 and 48 h, 10 µl CCK-8 solution
(HY-K0301, MedChemExpress) was cultured with the cells for 2-4 h at
37°C. The absorbance (at 450 nm) was detected using a microplate
reader (24072800, Thermo Fisher Scientific, Inc.).
Colony formation assay
The monolayer cultured cells (250/ml) at logarithmic
growth phase were collected, digested with 0.25% trypsin at room
temperature for 3-4 min and pipetted into individual cells. The
cells were suspended in a culture solution containing 10% FBS, and
the cell suspension was diluted and then inoculated in DMEM. The
culture was terminated when macroscopic clones appeared in the
dish. The supernatant was discarded and the cells were carefully
rinsed twice with PBS. At room temperature, acetic acid/methanol at
1:3 was used to fix the cells for 15 min. After removing the
fixation solution, the cells were properly stained with Giemsa
(Beyotime Institute of Biotechnology, Inc.) at room temperature for
25 min. The cell clones were counted with the naked eye and the
colony formation rate was calculated as follows: Clonal formation
rate = (number of clone formation/number of cells inoculated)
×100%.
Flow cytometry
The AGS and SNU-16 cells (1×106) were
prepared as a single dispersed cell suspension. Before testing the
machine, 5 µl Annexin V-FITC and 5 µl PI staining
solution were added to the suspension for 15 min in the dark at
room temperature using the test kit (MA0220, Melone Pharmaceutical
Co., Ltd.). Cell apoptosis was determined using a FACSCalibur flow
cytometer (FACSCalibur, BD Biosciences) and FlowJo software
(version 10.0; FlowJo). The lower left quadrant
(AnnexinV−, PI−) represented healthy living
cells; the upper left quadrant (AnnexinV−,
PI+) represented dead cells; the lower right quadrant
(AnnexinV+, PI−) represented early apoptotic
cells; and the upper right quadrant (AnnexinV+,
PI+) represented late apoptotic cells.
Wound healing assay
The AGS and SNU-16 cells were seeded at a density of
1×104into a 96-well plate for 24 h at 37°C. The cell
culture layer was scratched using a sterile sampler tip, the
floating cells were washed away with PBS, and the cells were
cultured in serum-free DMEM medium for 24 h at 37°C. Cell migration
images were observed under a microscope (BZ-8100, Keyence) and the
migration distance was observed using Image-pro Plus 4.1 analysis
software (Media Cybernetics, Inc.).
Transwell assay
The invasive ability of the AGS and SNU-16 cells was
detected by 24-well Transwell plates (CLS3398, Sigma; Merck KGaA).
The cells (5×104 cells/well) were seeded into the upper
chamber pre-coated with 200 mg/ml Matrigel (354230, BD
Biosciences), and the upper chamber was supplemented with
serum-free medium. The lower chamber was supplemented with 10%
serum medium to form a nutrient concentration gradient, which
promoted the cell invasion into the lower chamber. Following
incubation for 24 h at 37°C, the cells were fixed with 4%
paraformaldehyde and stained with 0.1% crystal violet (Beyotime
Institute of Biotechnology, Inc.) for 20 min at room temperature.
Cell numbers in each group were counted from 4 fields of view under
an inverted microscope (CKX41, Olympus Corporation).
Western blot analysis
The GC cells was separated using RIPA lysate (89901,
Thermo Fisher Scientific, Inc.), and the protein concentration was
determined using a Protein Assay kit (A53227, Thermo Fisher
Scientific, Inc.). A volume of 30 µg proteins of each tissue
was separated on 10% SDS-polyacrylamide gels and transferred to a
PVDF membrane (HVLP04700, EMD Millipore), which the membrane was
then washed with TBS and blocked with 5% non-fat milk at 37°C for 1
h and shaken for 2 h at room temperature. The membrane was then
incubated with the following primary antibodies: Anti-Bcl2
(1:1,000; 26 kDa; rabbit; ab32124), anti-procaspase-3 (1:10,000; 35
kDa; rabbit; ab32499), anti-cleaved caspase-3 (1:500; 17 kDa;
rabbit; ab32042), Vimentin (1:1,000; 54 kDa; rabbit; ab92547),
N-cadherin (1 µg/ml; 130 kDa; rabbit; ab18203), E-cadherin
(1:10,000; 97 kDa; rabbit; ab40772) or GAPDH (1:10,000; 36 kDa;
rabbit; ab181602) (all from Abcam) overnight at 4°C. The target
band was incubated with a goat anti-rabbit IgG H&L (HRP)
(1:5,000; ab205718; Abcam) for 2 h at room temperature. Finally,
the signals were measured by ECL reagent (6883, Cell Signaling
Technology, Inc.), and the gray values of the bands were analyzed
and counted using ImageJ software (version 5.0, Bio-Rad
Laboratories, Inc.) (16).
Statistical analysis
The data are presented as the means ± SD.
Comparisons between groups were conducted using a paired Student's
t test or one-way analysis of variance (ANOVA), followed by
Dunnett's test or Tukey's test. The Chi-square test or rank sum
test were used to analyze the data presented in Table I. All the data were analyzed using
GraphPad Prism 7.0 software. The correlation between circRNA RNF111
and miR-27b-3p in GC was analyzed by Pearson's correlation
analysis. A P<0.05 was considered to indicate a statistically
significant difference.
 | Table IAssociation between circRNA RNF111
and miR-27b-3p and clinicopathological characteristics of patients
with gastric cancer. |
Table I
Association between circRNA RNF111
and miR-27b-3p and clinicopathological characteristics of patients
with gastric cancer.
| Characteristic | Total | circRNA RNF111
expression
| P-value | miR-27b-3p
expression
| P-value |
|---|
| Low n=24 | High n=24 | Low n=24 | High n=24 |
|---|
| Sex | | | | 0.763 | | | 0.365 |
| Male | 31 | 15 | 16 | | 17 | 14 | |
| Female | 17 | 9 | 8 | | 7 | 10 | |
| Age (years) | | | | 0.551 | | | 0.233 |
| ≤60 | 18 | 8 | 10 | | 7 | 11 | |
| >60 | 30 | 16 | 14 | | 17 | 13 | |
| Location | | | | 0.772 | | | 0.487 |
| Cardia | 9 | 4 | 5 | | 6 | 3 | |
| Body | 5 | 3 | 2 | | 2 | 3 | |
| Curvature | 6 | 2 | 4 | | 4 | 2 | |
| Antrum | 28 | 15 | 13 | | 12 | 16 | |
| Tumor size
(cm) | | | | 0.001 | | | <0.001 |
| ≥5 cm | 14 | 2 | 12 | | 13 | 1 | |
| <5 cm | 34 | 22 | 12 | | 11 | 23 | |
| Histological
differentiation | | | | 0.490 | | | 0.300 |
| Well | 5 | 3 | 2 | | 1 | 4 | |
| Moderate | 13 | 8 | 5 | | 6 | 7 | |
| Poor | 30 | 13 | 17 | | 17 | 13 | |
| TNM stage | | | | 0.002 | | | 0.014 |
| I | 11 | 9 | 2 | | 3 | 8 | |
| II | 10 | 8 | 2 | | 2 | 8 | |
| III | 22 | 5 | 17 | | 16 | 6 | |
| IV | 5 | 2 | 3 | | 3 | 2 | |
| Lymph node
metastasis | | | | 0.001 | | | 0.008 |
| Yes | 36 | 13 | 23 | | 22 | 14 | |
| No | 12 | 11 | 1 | | 2 | 10 | |
| Distant
metastasis | | | | 0.004 | | | 0.033 |
| Yes | 10 | 1 | 9 | | 8 | 2 | |
| No | 38 | 23 | 15 | | 16 | 22 | |
| Depth of
invasion | | | | <0.001 | | | 0.007 |
| m | 9 | 8 | 1 | | 2 | 7 | |
| sm | 10 | 9 | 1 | | 3 | 7 | |
| mp | 7 | 3 | 4 | | 2 | 5 | |
| se, sei | 22 | 4 | 18 | | 17 | 5 | |
Results
CircR-RNF111 and miR-27b were negatively
correlated and were abnormally expressed in GC
The associations between circRNA RNF111, miR-27b-3p
and the pathological characteristics of the clinical samples
obtained from the patients with GC were analyzed. The results
revealed that the high expression of circR-RNF111 and the low
expression of miR-27b-3p were significantly and positively
associated with tumor size, stage, lymphatic metastasis, distant
metastasis and invasion (Table
I). The expression levels of the 2 genes in GC and normal
samples were then compared, and it was found that the expression of
circRNA RNF111 in the GC tissues was significantly higher than that
in adjacent normal tissues (P<0.001; Fig. 1A), whereas the expression level of
miR-27b-3p was markedly lower in GC tissues than in adjacent normal
tissues (P<0.001; Fig. 1B).
Pearson's correlation curve analysis demonstrated that circRNA
RNF111 expression negatively correlated with miR-27b expression in
GC tissues (r=-0.352, P=0.014; Fig.
1C) and adjacent normal tissues (r=-0.379, P=0.008; Fig. 1D).
miR-27b-3p is targeted to bind to circRNA
RNF111
To explore the regulatory mechanisms of circR-RNF111
and miR-27b-3p in GC, starBase v2.0 was used to predict the
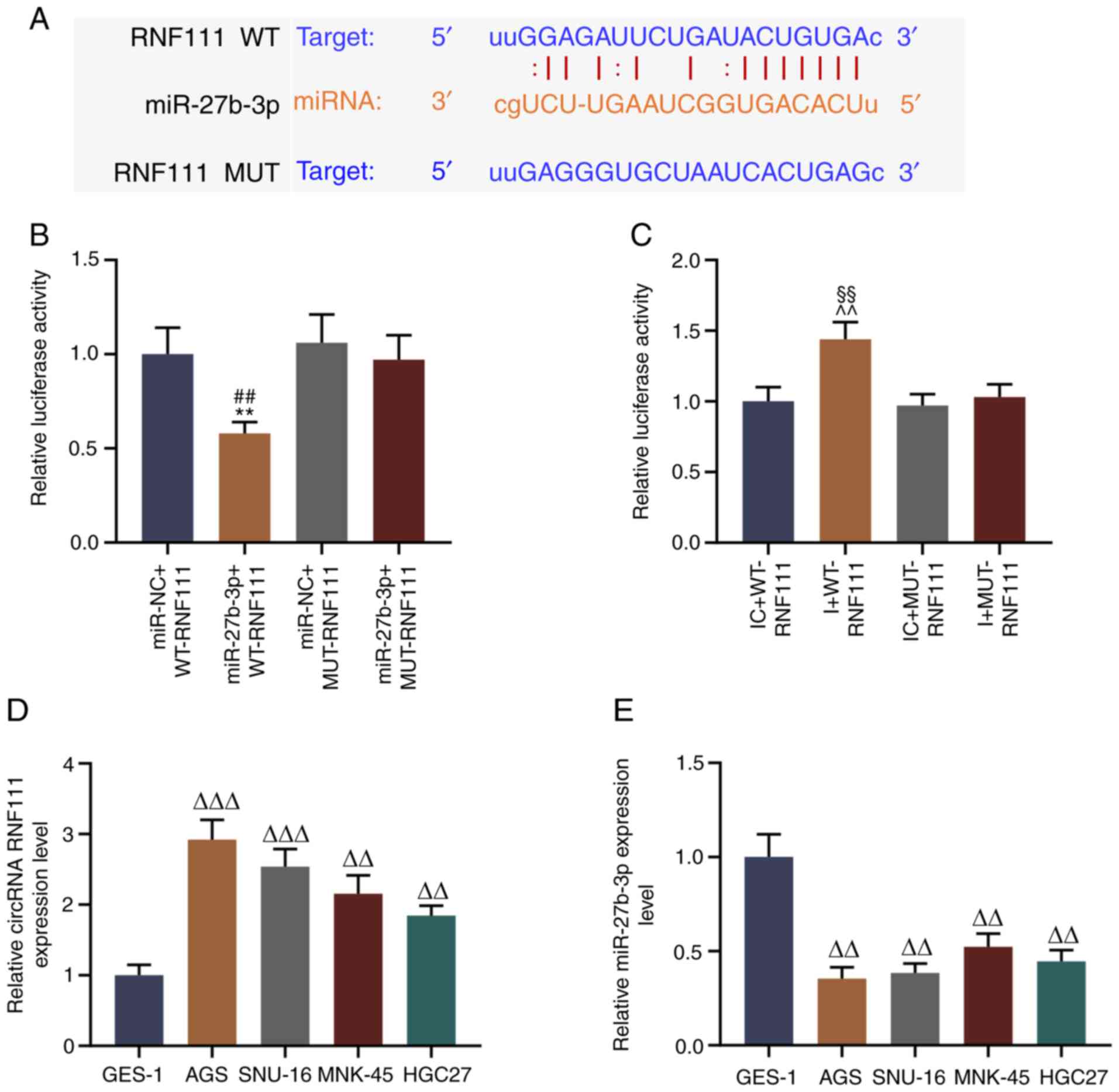
potential binding site between circR-RNF111 and miR-27b-3p. The
results revealed that circRNA RNF111 has a binding site specific
for miR-27b-3p (Fig. 2A). The
binding site between these 2 factors 293T cells was verified by
dual luciferase assay, and the data indicated that miR-27b-3p
effectively reduced the fluorescence activity of wild-type RNF111,
whereas it did not affect that of the mutant RNF111 fluorescent
plasmid (P<0.01; Fig. 2B);
miR-27b-3p inhibitor increased the fluorescence activity of
wild-type RNF111, whereas it did not affect that of the mutant
RNF111 fluorescent plasmid (P<0.01; Fig. 2C). Furthermore, the roles of
RNF111 and miR-27b-3p in GC were further examined by detecting the
expression levels of circR-RNF111 and miR-27b-3p in normal gastric
mucosal cells (GES-1) and GC cell lines (AGS, SNU-16, MNK-45 and
HGC27). It was also found that overall, circRNA RNF111 expression
was high-expressed in the GC cell lines (P<0.01; Fig. 2D), while miR-27b-3p expression was
low in GC cells (P<0.01; Fig.
2E). In addition, circRNA RNF111 expression was higher in the
AGS and SNU-16 cells and miR-27b-3p expression was lower in the AGS
and SNU-16 cells compared with the other GC cells; thus, these 2
cell lines were selected for use in subsequent experiments.
circRNA RNF111 binding to miR-27b-3p
regulates the growth and apoptosis of GC cells
To examine the effects of circRNA RNF111 and
miR-27b-3p on the biological functions of GC cells, circRNA
siRNF111 and miR-27b-3p inhibitor were successfully transfected
into the AGS cells and SNU-16 cells. As shown in Fig. 3A-D, in the AGS and SNU-16 cells,
the results revealed that the expression of circRNA RNF111 was
significantly lower in the siRNF111 group than in the siNC group
(P<0.001), and miR-27b-3p expression was decreased in the
inhibitor group (P<0.01). siRNA transfection significantly
inhibited circRNA RNF111 expression and increased that of
miR-27b-3p, while transfection with miR-27b-3p inhibitor partially
reversed the siRNA regulatory effects on circRNA RNF111 and
miR-27b-3p expression (P<0.01; Fig. 3E-H). The results of CCK-8 assay
revealed that circRNA RNF111 silencing markedly inhibited the
viability of the GC cells, and transfection with miR-27b-3p
inhibitor significantly increased the viability of the cells
(P<0.01; Fig. 3I and J).
Colony formation assay demonstrated that the silencing of
circR-RNF111 significantly inhibited the colony formation of the
cancer cells, and miR-27b-3p inhibitor noticeably reversed the
inhibitory effects of circRNA RNF111 silencing on cell
proliferation (P<0.001; Fig.
4). Flow cytometry was then performed to measure the apoptosis
of each group of cells; it was found that circRNA RNF111 silencing
markedly promoted apoptosis, which was markedly reversed by
miR-27b-3p inhibitor (P<0.05; Fig.
5).
 | Figure 3Effect of circRNA RNF111 silencing
and miR-27b-3p inhibitor on the proliferation of GC cells. (A-D)
RT-qPCR was used to detect the expression levels of circRNA RNF111
and miR-27b-3p after AGS or SNU-16 cells were transfected with
Control, siNC, siRNF111, inhibitor control, or inhibitor. (E-H)
RT-qPCR was used to detect the expression levels of circRNA RNF111
and miR-27b-3p after the AGS or SNU-16 cells were transfected with
Control, siNC + inhibitor control, siRNF111 + inhibitor control,
siNC + inhibitor, siRNF111 + inhibitor. (I and J) CCK-8 assay was
used to examine the cell viability of each group.
§§§P<0.001 vs. siNC, ΔΔP<0.01 vs. IC,
*P<0.05, **P<0.01,
***P<0.001 vs. siNC + IC, #P<0.05,
##P<0.01, ###P<0.001 vs. siRNF111 + IC,
^^P<0.01 vs. siNC + I. GC, gastric cancer; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; IC,
inhibitor control; I, inhibitor. |
 | Figure 4Effects of circRNA RNF111 and
miR-27b-3p on the growth of GC cells. (A) The numbers of colony
formation in AGS cells transfected with Control, siNC + inhibitor
control, siRNF111 + inhibitor control, siNC + inhibitor, siRNF111 +
inhibitor were determined by colony formation assay. (B) The
numbers of colony formation in SNU-16 cells transfected with
Control, siNC + inhibitor control, siRNF111 + inhibitor control,
siNC + inhibitor, siRNF111 + inhibitor were determined by colony
formation assay. **P<0.01, ***P<0.001
vs. siNC + IC, ###P<0.001 vs. siRNF111 + IC,
^^P<0.01 vs. siNC + I. GC, gastric cancer; IC,
inhibitor control; I, inhibitor. |
 | Figure 5Effects of circRNA RNF111 and
miR-27b-3p on the apoptosis of GC cells. (A) Flow cytometry was
performed to detect the apoptosis of AGS cells transfected with
Control, siNC + inhibitor control, siRNF111 + inhibitor control,
siNC + inhibitor, siRNF111 + inhibitor. (B) Flow cytometry was
performed to detect the apoptosis of SNU-16 cells transfected with
Control, siNC + inhibitor control, siRNF111 + inhibitor control,
siNC + inhibitor, siRNF111 + inhibitor. *P<0.05,
***P<0.001 vs. siNC + IC, ###P<0.001
vs. siRNF111 + IC, ^^P<0.01 vs. siNC + I. GC, gastric
cancer; IC, inhibitor control; I, inhibitor. |
circR-RNF111 binding to miR-27b-3p
regulates the metastasis of GC cells
To further examine the effects of circRNA RNF111 and
miR-27b-3p on GC, a wound healing assay and Transwell invasion
assay were performed. As shown in Fig. 6, circRNA RNF111 silencing
significantly inhibited the migration of the GC cells, while
miR-27b-3p inhibition promoted cell migration and effectively
reversed the inhibitory effects induced by circRNA RNF111 silencing
(P<0.05). In addition, circR-RNF111 silencing markedly decreased
cell invasion, whereas miR-27b-3p inhibition promoted cell invasion
and effectively reversed the inhibitory effects of circRNA RNF111
silencing (P<0.01; Fig.
7).
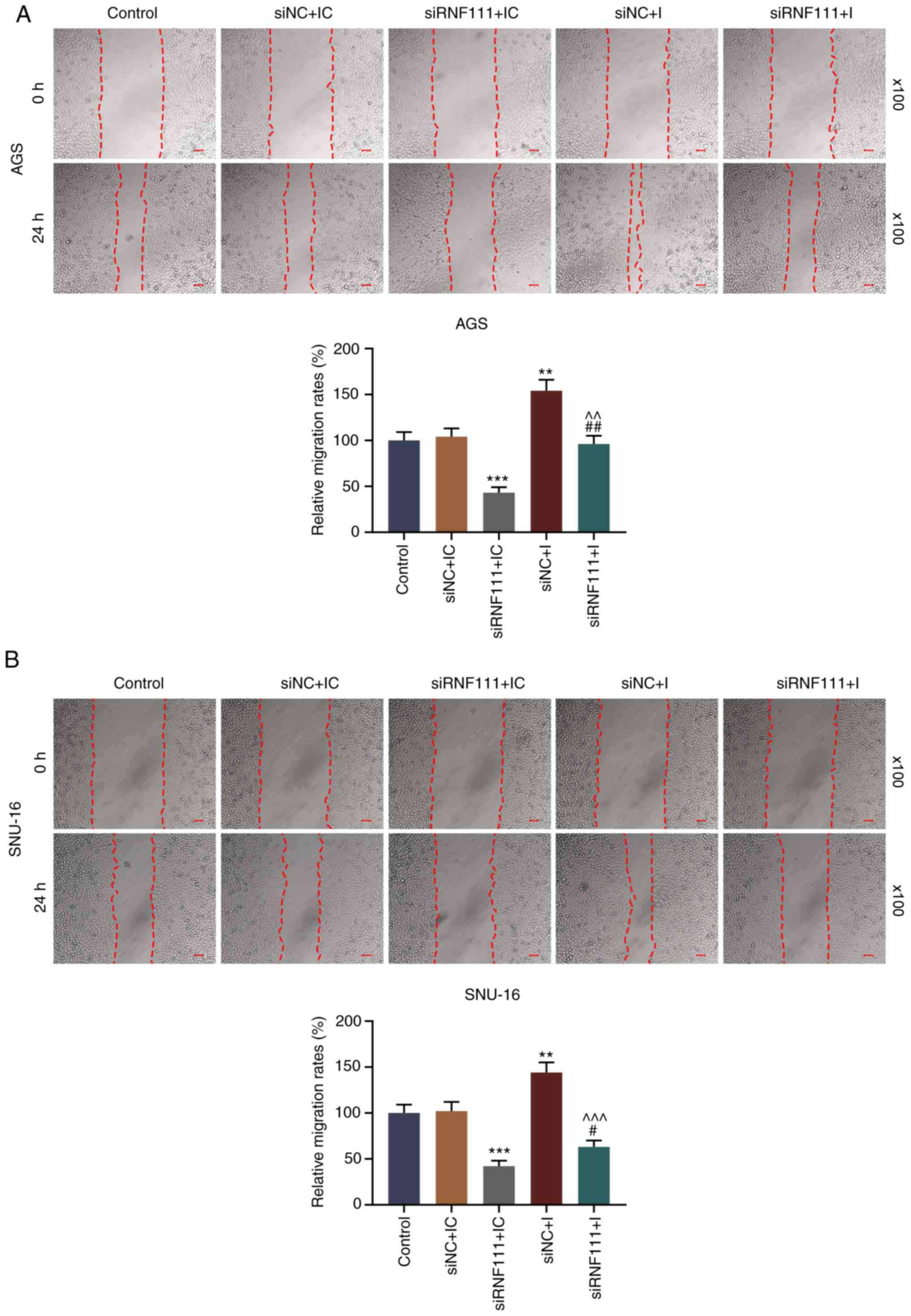 | Figure 6Effects of circRNA RNF111 and
miR-27b-3p on GC cell migration. (A) Migratory ability of AGS cells
following transfection with Control, siNC + inhibitor control,
siRNF111 + inhibitor control, siNC + inhibitor, siRNF111 +
inhibitor was determined by wound healing assay. (B) Migratory
ability of SNU-16 cells following transfection with Control, siNC +
inhibitor control, siRNF111 + inhibitor control, siNC + inhibitor,
siRNF111 + inhibitor was determined by wound healing assay. Scale
bars, 50 µm. **P<0.01,
***P<0.001 vs. siNC + IC, #P<0.05,
##P<0.01 vs. siRNF111 + IC, ^^P<0.01,
^^^P<0.001 vs. siNC + I. GC, gastric cancer; IC,
inhibitor control; I, inhibitor. |
 | Figure 7Effects of circR-RNF111 and
miR-27b-3p on GC cell invasion. (A) Detection of the invasive
ability of AGS cells by Transwell assay following transfection with
Control, siNC + inhibitor control, siRNF111 + inhibitor control,
siNC + inhibitor, siRNF111 + inhibitor. (B) Detection of the
invasion of SNU-16 cells by Transwell assay following transfection
with Control, siNC + inhibitor control, siRNF111 + inhibitor
control, siNC + inhibitor, siRNF111 + inhibitor. Scale bars, 50
µm. **P<0.01, ***P<0.001 vs.
siNC + IC, ##P<0.01, ###P<0.001 vs.
siRNF111 + IC, ^^P<0.01 vs. siNC + I. GC, gastric
cancer; IC, inhibitor control; I, inhibitor. |
To further explore the molecular mechanisms of
circR-RNF111 and miR-27b-3p in GC metastasis, the expression levels
of apoptosis-related and EMT-related proteins were detected by
western blot analysis. The results revealed that circRNA RNF111
silencing significantly decreased the expression levels of Bcl2,
Vimentin and N-cadherin, and increased and E-cadherin expression;
however, miR-27b-3p inhibition significantly reversed the
regulatory effects of circRNA RNF111 silencing (P<0.05; Fig. 8A and D). In addition, circRNA
RNF111 silencing significantly increased the expression of cleaved
caspase-3, whereas miR-27b-3p inhibition significantly decreased
cleaved caspase-3 expression and reversed the regulatory effects of
circRNA RNF111 silencing (P<0.05; Fig. 8B, C, E and F). However, no
significant differences were observed in the expression of
procaspase 3 in each group.
 | Figure 8Effects of circRNA RNF111 and
miR-27b-3p on the expression of apoptosis, EMT-related proteins in
GC cells. (A) Western blot analysis was used to detect the
expressions of Bcl2, Vimentin, N-cadherin and E-cadherin in AGS
cells following transfection with Control, siNC + inhibitor
control, siRNF111 + inhibitor control, siNC + inhibitor, siRNF111 +
inhibitor. (B) Western blot analysis was used to detect the
expression levels of procaspase-3 and cleaved caspase-3 in AGS
cells following transfection with Control, siNC + inhibitor
control, siRNF111 + inhibitor control, siNC + inhibitor, siRNF111 +
inhibitor. (C) Ratio of cleaved caspase-3/procaspase-3. (D) Western
blot analysis was used to detect the expression levels of Bcl2,
Vimentin, N-cadherin and E-cadherin in SNU-16 cells following
transfection with Control, siNC + inhibitor control, siRNF111 +
inhibitor control, siNC + inhibitor, siRNF111 + inhibitor. (E)
Western blot analysis was used to detect the expression level of
procaspase-3 and cleaved caspase-3 in SNU-16 cells after the
transfection with Control, siNC + inhibitor control, siRNF111 +
inhibitor control, siNC + inhibitor, siRNF111 + inhibitor. (F)
Ratio of cleaved caspase-3/procaspase-3. *P<0.05,
**P<0.01, ***P<0.001 vs. siNC + IC,
#P<0.05, ###P<0.001 vs. siRNF111 + IC,
^P<0.05, ^^^P<0.001 vs. siNC + I. GC,
gastric cancer; IC, inhibitor control; I, inhibitor. |
Discussion
GC is a malignant tumor with the fourth highest
incidence and the third highest mortality rate worldwide (1). The mechanisms of GC occurrence and
progression are highly complex, implicating several aspects of cell
biology and molecular biology, such as alterations in the adhesion
of tumor cells, detachment from primary tumors, and the degradation
of the extracellular matrix, local or vascular infiltration
(17). With the development of
high-throughput sequencing technology and bioinformatics analysis,
circRNAs have gradually become a main focus of tumor studies
(18).
The association between circRNAs and the
proliferation, invasion and metastasis of GC cells has been
investigated (19-21). In the present study, circRNA
RNF111 and miR-27b-3p were found closely related to tumor size,
stage, lymphatic metastasis, distant metastasis and invasion. Li
et al reported that in GC tissues, the expression of
circRNA002059 was significantly decreased compared to adjacent
normal tissues, and an association between the low expression of
circRNA and distant metastasis was observed (22). Chen et al detected the
expression of circPVT1 in GC tissues and adjacent normal tissues by
RT-qPCR (23), and found that the
expression of circPVT1 in GC tissues was significantly higher than
that in adjacent normal tissues (23). The expression levels of different
circRNAs in GC are not necessarily the same (24). In the present, it was found that
circRNA RNF111 was significantly highly expressed in GC tissues and
cell lines compared with normal tissues and cells. According to a
previous study, circRNAs can exert perform their biological
functions by interacting with miRNAs (24). For example, Zheng et al
found that circHIPK3 derived from HIPK3 gene Exon2 bound to 9
miRNAs with 18 potential binding sites; moreover, it was found to
specifically bind to miR-124 to inhibit its activity, and promote
tumor cell proliferation (25).
Differentially expressed genes, such as CD44, CXXC5, MYH9 and
MALAT1 can promote the occurrence of GC through various mechanisms,
such as the interactions between circRNAs and miRNAs, and between
miRNAs and mRNAs (26). Some
scholars have found that miR-27b-3p is involved in the development
of GC, and that it is also involved in other types of cancer, such
as lung cancer (27,28). miR-27b-3p is abnormally expressed
at low levels in GC tissues and negatively correlates with circRNA
RNF111. circRNAs function as a competitive endogenous RNAs of
miRNAs (6,7). The molecular mechanism underlying
the biological role of circRNA RNF111 in GC were further detected
in the present study. StarBase predicted that circRNA RNF111
contained a miR-27b-3p specific binding site, which was confirmed
by dual luciferase assay. The data indicated that circRNA RNF111
bound to miR-27b-3p to reduce the activity of miR-27b-3p and was
involved in the development of GC.
Tumor cells can secrete specific receptors and
chemokines to create a cell microenvironment facilitating their own
proliferation, invasion into surrounding normal cells and distant
metastasis (4,17). The role of RNF111 in lung cancer
has been investigated by scholars, and it has been found that
RNF111 promotes cancer growth and plays a key role in early cancer
cell metastasis (29); however,
its functions vary in different types of cancer (30). For example, RNF111 has been shown
to exert an inhibitory effect on colon cancer metastasis (31). circR-RNF111 is generated from the
RNF111 gene region. In 2013, some scholars identified a large
number of circular RNAs, including circRNA RNF111 (32,33), and circRNA RNF111 was then found
to be involved in physiological and pathological processes
(34,35). Tang et al found that
circRNA RNF111 was closely related to the occurrence of breast
cancer (13). However, to date,
circRNA RNF111 has rarely been reported in GC, at least to the
bests of our knowledge. According to previous studies, it was
hypothesized that circRNA RNF111 may indirectly increase the
expression levels of miR-27b-3p-related target genes by binding to
miR-27b-3p, and is involved in the regulation of GC. Unexpectedly,
the silencing circRNA RNF111 significantly inhibited the
proliferation and metastasis of GC cells and exerted a
pro-apoptotic effect. miR-27b-3p inhibition significantly promoted
the proliferation and metastasis of GC cells, and inhibited
apoptosis, indicating the biological functions of circRNA RNF111
and miR-27b-3p in GC. As an anti-apoptotic member of the Bcl
protein family, Bcl-2 plays a role in maintaining the
apoptotic-anti-apoptotic balance (36). The cascade reaction produced by
the caspase family is the central link of apoptosis. Activated
caspase-3 hydrolyzes the target substance in cells, thereby
degrading the intracellular protein and finally causing apoptosis
(36). E-cadherin and N-cadherin
are two proteins indicative of epithelial-mesenchymal transition
(EMT) (37,38). Tumor epithelial cells transform to
interstitial cells, resulting in an invasive phenotype and easy
distant metastasis (39). The
present study further examined the functions of circRNA RNF111 and
miR-27b-3p at the molecular level, and detected the expression
levels of Bcl2, cleaved caspase-3, Vimentin, N-cadherin and
E-cadherin in GC cells, and the findings revealed that circRNA
RNF111 may be involved in GC apoptosis and metastasis by targeting
miR-27b-3p to regulate apoptosis and EMT-related pathways.
It should be noted that the present study has some
limitations. The effects of circRNA RNF111/miR-27b-3p on the cell
cycle of GC remain to be analyzed. In addition, the effects of
silencing circRNA RNF111 on inhibiting the proliferation, migration
and invasion of GC cells should be confirmed by in vivo
experiments.
In conclusion, the present study demonstrated that
circRNA RNF111 was highly expressed in GC cells and tissues.
circRNA RNF111 was found to regulate GC cell growth and metastasis
possibly by targeting miR-27b-3p, suggesting that circRNA RNF111
and miR-27b-3p are potential targets for the treatment of GC.
Funding
The present was supported by the 2017 Medical
Science and Technology Development Program of Nanjing (grant no.
YKK17108).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
ZW and ZJ made substantial contributions to
conception and design of the study. ZW, ZJ, JZ and ZL were involved
in data acquisition, data analysis and interpretation. ZW and ZJ
drafted the article or critically revised it for important
intellectual content. All authors gave the final approval of the
final version of the manuscript to be published. All authors also
agree to be accountable for all aspects of the work in ensuring
that questions related to the accuracy or integrity of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All patients had signed informed consent forms, and
agreed that their tissues would be used for clinical research. The
clinical trial program had been reviewed and approved by the Ethics
Committee of Nanjing First Hospital (W201810013). No animals are
involved in this research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
RNF111
|
ring finger protein 111
|
|
GC
|
gastric cancer
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
HCC
|
hepatocellular carcinoma
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Correa P: Gastric cancer: Overview.
Gastroenterol Clin North Am. 42:211–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ang TL and Fock KM: Clinical epidemiology
of gastric cancer. Singapore Med J. 55:621–628. 2014. View Article : Google Scholar
|
|
4
|
Sano T: Gastric cancer: Asia and the
world. Gastric Cancer. 20(Suppl 1): S1–S2. 2017. View Article : Google Scholar
|
|
5
|
Greene J, Baird AM, Brady L, Lim M, Gray
SG, McDermott R and Finn SP: Circular RNAs: Biogenesis, function
and role in human diseases. Front Mol Biosci. 4:382017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patop IL and Kadener S: circRNAs in
cancer. Curr Opin Genet Dev. 48:121–127. 2018. View Article : Google Scholar :
|
|
7
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong Z, Huang M, Lv M, He Y, Duan C,
Zhang L and Chen J: Circular RNA MYLK as a competing endogenous RNA
promotes bladder cancer progression through modulating VEGFA/VEGFR2
signaling pathway. Cancer Lett. 403:305–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. . Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bernardo BC, Ooi JY, Lin RC and McMullen
JR: miRNA therapeutics: A new class of drugs with potential
therapeutic applications in the heart. Future Med Chem.
7:1771–1792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shin VY and Chu KM: MiRNA as potential
biomarkers and therapeutic targets for gastric cancer. World J
Gastroenterol. 20:10432–10439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R,
Yang SY, Yang DC and Wang XL: Circular RNA hsa_circ_0001982
promotes breast cancer cell carcinogenesis through decreasing
miR-143. DNA Cell Biol. 36:901–908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang Q, Chen X and Zhi X: Long non-coding
RNA (LncRNA) urothelial carcinoma associated 1 (UCA1) increases
multi-drug resistance of gastric cancer via downregulating miR-27b.
Med Sci Monit. 22:3506–3513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Singh C and Roy-Chowdhuri S: Quantitative
real-time PCR: Recent advances. Methods Mol Biol. 1392:161–176.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kurien BT and Scofield RH: Western
blotting: An introduction. Methods Mol Biol. 1312:17–30. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Röcken C: Molecular classification of
gastric cancer. Expert Rev Mol Diagn. 17:293–301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: CircRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017. View Article : Google Scholar
|
|
20
|
Tang W, Fu K, Sun H, Rong D, Wang H and
Cao H: CircRNA microarray profiling identifies a novel circulating
biomarker for detection of gastric cancer. Mol Cancer. 17:1372018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar
|
|
24
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol. 58:90–99.
2019. View Article : Google Scholar
|
|
25
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J,
Lin H, Liu F and Dai Y: Circular RNA and gene expression profiles
in gastric cancer based on microarray chip technology. Oncol Rep.
37:1804–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tao J, Zhi X, Zhang X, Fu M, Huang H, Fan
Y, Guan W and Zou C: miR-27b-3p suppresses cell proliferation
through targeting receptor tyrosine kinase like orphan receptor 1
in gastric cancer. J Exp Clin Cancer Res. 34:1392015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun Y, Xu T, Cao YW and Ding XQ: Antitumor
effect of miR-27b-3p on lung cancer cells via targeting Fzd7. Eur
Rev Med Pharmacol Sci. 21:4113–4123. 2017.PubMed/NCBI
|
|
29
|
Hedrick E, Mohankumar K and Safe S:
TGFβ-induced lung cancer cell migration is NR4A1-dependent. Mol
Cancer Res. 16:1991–2002. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen H, Yang T, Lei Z, Wang L, Yang H,
Tong X, Yang WT, Zhao J, Gu Y, Chen Y and Zhang HT: RNF111/Arkadia
is regulated by DNA methylation and affects TGF-β/Smad signaling
associated invasion in NSCLC cells. Lung Cancer. 90:32–40. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sharma V, Antonacopoulou AG, Tanaka S,
Panoutsopoulos AA, Bravou V, Kalofonos HP and Episkopou V:
Enhancement of TGF-β signaling responses by the E3 ubiquitin ligase
arkadia provides tumor suppression in colorectal cancer. Cancer
Res. 71:6438–6449. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
33
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maass PG, Glažar P, Memczak S, Dittmar G,
Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC and
Rajewsky N: A map of human circular RNAs in clinically relevant
tissues. J Mol Med (Berl). 95:1179–1189. 2017. View Article : Google Scholar
|
|
36
|
Li H, Lv B, Kong L, Xia J, Zhu M, Hu L,
Zhen D, Wu Y, Jia X, Zhu S and Cui H: Nova1 mediates resistance of
rat pheochromocytoma cells to hypoxia-induced apoptosis via the
Bax/Bcl-2/caspase-3 pathway. Int J Mol Med. 40:1125–1133. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Petrova YI, Schecterson L and Gumbiner BM:
Roles for E-cadherin cell surface regulation in cancer. Mol Biol
Cell. 27:3233–3244. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Derycke LD and Bracke ME: N-cadherin in
the spotlight of cell-cell adhesion, differentiation,
embryogenesis, invasion and signalling. Int J Dev Biol. 48:463–476.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|