Introduction
Common thyroid nodules are usually benign (1,2).
However, according to studies on fine-needle aspiration cytology,
among the nodules identified through clinical examinations, ~15% of
the nodules are malignant (3,4).
Correspondingly, the identification of a thyroid nodule with a
diameter of ≥1 cm frequently prompts the initialization of a more
thorough diagnostic evaluation (5). Fine-needle aspiration is the key
method for the evaluation of thyroid nodules, enabling the
evaluation of cellular morphology features that cannot be
identified through clinical imaging or assessment (6). Preoperative fine-needle
aspiration-guided ultrasonography has been indicated to accurately
identify ≤80% of all thyroid nodules, thus avoiding surgery for
diagnosis (7).
According to genome-wide sequencing results,
non-coding genes account for up to ≤98% of the whole genome
(8). Among these non-coding
genes, long non-coding RNAs (lncRNAs), which are >200 nt long,
exert crucial effects on different cellular processes and are
dysregulated under abnormal conditions, including rare diseases
such as cat eye, Klinefelter's and Down's syndromes (9,10).
In addition, lncRNAs modulate the expression of genes through
multiple mechanisms, such as miRNA degradation, epigenetic
modification, transcription control and gene imprinting (11,12). Other types of non-coding RNAs
include microRNAs (miRNAs), which are ~22 nt long and repress the
expression of genes under various biological conditions by binding
to the 3′-untranslated regions (UTRs) of target mRNAs and by
triggering the degradation or suppressing the translation of mRNAs
(13).
LncRNAs are involved in tumourigenesis. For example,
the expression of lncRNA nuclear enriched abundant transcript 1
(NEAT1) is significantly enhanced in cervical cancer cell lines and
tissues (14,15). In a molecular study, NEAT1 has
been demonstrated to function as a competing endogenous RNA (ceRNA)
by repressing miR-9-5p expression; in addition, high levels of
NEAT1 expression reduced the inhibitory effect of miR-9-5p on the
expression of POU class 2 transcription factor 1 (POU2F1) and PTEN
(16). Consistent with the
observation that NEAT1 negatively regulated miR-9-5p, the
upregulation of miR-9-5p suppressed NEAT1-induced cancer cell
proliferation (17,18). The expression of miR-9 was also
observed to be negatively associated with the expression of PTEN, a
tumour suppressor gene that was confirmed as a direct target gene
of miR-9, in laryngocarcinoma cells, and the introduction of miR-9
mimics into laryngocarcinoma cells facilitated cell metastatic
capability and proliferation (19). In nasopharyngeal carcinoma (NPC)
cells and tissues, NEAT1 was also upregulated, whereas miR-124 was
downregulated; knockdown of NEAT1 suppressed NPC progression and
facilitated apoptosis, whereas overexpression of NEAT1 resulted in
the opposite outcome (20).
Furthermore, NEAT1 was confirmed to inhibit the expression of
miR-124 through direct interaction in NPC cells (20). In addition, the anti-apoptotic and
pro-proliferative effects mediated by NEAT1 were reversed by
miR-124 (20).
Using bioinformatics methods and in vitro
experiments, programmed cell death protein 6 (PDCD6) was identified
as a protein directly targeted by miR-124-3p (21). The restoration of the expression
of PDCD6 impaired the suppressive effect of miR-124-3p on
metastasis by facilitating the infiltration of cancer cells
(21). In addition, miR-124-3p
expression levels were inversely related to the levels of PDCD6
mRNA in clinical samples of breast tumours (21). Therefore, these results suggested
that miR-124-3p suppressed the metastasis of tumours by repressing
the expression of PDCD6, and that the miR-124-3p/PDCD6 signalling
axis may be a target for new treatments for patients with advanced
breast cancer (BC) (21).
Deregulated miR-9 and miR-124 have been used as
biomarkers to differentiate benign and malignant thyroid nodules
(22). Previous studies have
reported that NEAT1 may function as a sponge of miR-9 and miR-124
(16,20). In addition, PTEN and PDCD6 have
also been identified as possible targets of miR-9 and miR-124,
respectively. PTEN and PDCD6 have been implicated in tumourigenesis
(23-25). In the present study, peripheral
blood and thyroid tissue samples were collected to investigate the
roles of NEAT1/miR-9/PTEN and NEAT1/miR-124/PDCD6 signalling in the
differentiation of benign and malignant thyroid nodules, and to
establish a practical method to determine the malignancy of thyroid
nodules.
Materials and methods
Human subjects and sample collection
A total of 98 patients diagnosed with thyroid
nodules between August 2014 and March 2017 at Shangluo Central
Hospital (Shangluo, China) were enrolled in the present study. The
subjects included 52 patients with malignant thyroid nodules (MTN
or the malignant group; nodule size range, 2.03-3.63 cm) and 46
patients with benign thyroid nodules (BTN or the benign group;
nodule size range, 2.06-3.46 cm). Peripheral blood and thyroid
tissue samples were collected from all participants. The
demographic and clinicopathological characteristics of the
participants, such as age, sex, nodule size, solitary nodule, free
triiodothyronine (FT3), free thyroxine (FT4) and
thyroid-stimulating hormone (TSH) levels, were recorded and
analysed. The inclusion criteria of the participants were as
follows: i) 18-70 years old without gender restrictions; ii)
confirmed diagnosis of thyroid nodules by imaging and histological
examinations; iii) not received medication that may have influenced
the metabolism of the thyroid in the previous three months; and iv)
no diseases that influence the metabolism of the thyroid, including
chronic thyroid dysfunction. The protocol of the present study was
approved by the Ethics Committee of Shangluo Central Hospital, and
all subjects signed informed consent forms.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
To establish the accuracy of the microRNA assays,
RT-qPCR was used to confirm the differential miRNA expression in
the samples. Total RNA was isolated from the collected samples and
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and the purification was performed using the
Qiagen RNeasy mini kit (Qiagen GmbH). RNA concentration was
measured by a spectrophotometer. Reverse transcription with the
cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.) and
qPCR (PowerUp SYBR®-Green; Thermo Fisher Scientific,
Inc.) were performed using an ABI real-time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The cDNA synthesis was
performed according to the manufacturer's instructions. The
thermocycling conditions of the qPCR were as follows: 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec.
U6 (for miRNA) and GAPDH (for mRNA) were used as the internal
control to calculate the normalized relative expression levels of
NEAT1, miR-124, miR-9, PTEN and PDCD6 mRNA using the
2-∆∆Cq method (26).
Primer Premier 5.0 (Premier Biosoft) was used to design primers for
qPCR. The primers used were as follows: NEAT1 forward,
5′-GCTGGACCTTTCATGTAACGGG-3′ and reverse, 5′-TGAACTCTGCCGGTACAGGGA
A-3′; miR-124 forward, 5′-TTCACAGCGGACCTTGA-3′ and reverse,
5′-GAACATGTCTGCGTATCTC-3′; miR-9 forward, 5′-TCTTTGGTTATCTAGCTGT-3′
and reverse, 5′-GAACATGTCTGCGTATCTC-3′; PTEN forward,
5′-TGAGTTCCCTCAGCCGTTACCT-3′ and reverse,
5′-GAGGTTTCCTCTGGTCCTGGTA-3′; PDCD6 forward,
5′-ACATCACGGACTGGCAGAACGT-3′ and reverse,
5′-AGGATGTCGTGGAACTGGTCAG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH forward,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′.
Cell culture and transfection
BCPaP and SW579 cells were obtained from the
Shanghai Cell Bank of the Chinese Academy of Sciences. All cells
were routinely cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS in an incubator with 5% CO2 at
37°C. When the cells were 80% confluent, they were transfected with
30 nM sh-NEAT1 (Guangzhou RiboBio Co., Ltd.), scramble control
(Guangzhou RiboBio Co., Ltd.), pcDNA3.1-NEAT1 overexpression vector
(p-lncRNA-NEAT1; Shanghai GenePharma Co., Ltd.) or pcDNA3.1 empty
vector (p-vector; Shanghai GenePharma Co., Ltd.) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions for
48 h at room temperature. At 48 h post-transfection, the expression
levels of NEAT1, miR-9 and miR-124 in the transfected cells were
measured by RT-qPCR.
Western blot analysis
Protein was extracted from the whole-cell lysate
using RIPA buffer (Cell Signaling Technology, Inc.). Subsequently,
western blotting was performed using a routine method. Briefly, the
protein concentration was measured with a BCA assay kit (Bio-Rad
Laboratories, Inc.), and 50 µg/lane of protein was resolved
by a 10% Mini-PROTEAN TG gel (Bio-Rad Laboratories, Inc.) and
loaded onto a PVDF membrane. The membranes were blocked using a
Tris buffer containing 5% skim milk and 0.1% Tween-20 for 60 min at
room temperature. Subsequently, the membrane was incubated
overnight at room temperature with anti-PDCD6 (1:1,000; cat no.
ab109181; Abcam) and anti-PETN (1:1,000; cat no. ab267787; Abcam)
primary antibodies, washed with a Tris buffer containing 0.1%
Tween-20 three times and incubated with an HRP-conjugated secondary
antibody (1:2,000; cat no. ab6721; Abcam) at room temperature for 1
h. The gel was visualized using Pierce™ ECL Western Blotting
Substrate (cat. no. 32109; Thermo Fisher Scientific, Inc.) and
analysed by ImageJ 1.44e software (National Institutes of Health).
Anti-β-actin (1:1,000; cat. no. MA5-15739; Invitrogen; Thermo
Fisher Scientific, Inc.) was used as the internal control to
calculate the relative expression levels of PDCD6 and PTEN
proteins.
Immunohistochemistry
The expression of PDCD6 and PTEN in
paraffin-embedded thyroid nodule tissues was determined using
immunohistochemistry. Briefly, 4-µm sections were prepared
from representative paraffin-embedded specimens of thyroid nodule
tissues. Surrounding normal thyroid nodule tissues were used as the
control. Xylene was used to deparaffinize the sections twice, and
five gradient ethanol baths (100, 95, 85, 75 and 60%) were used to
rehydrate the resultant sections. Antigen retrieval was performed
by heating the sections for 15 min in a pressure cooker, and EDTA
buffer (Ph 8.0) was added prior to incubation with 3%
H2O2 for 10 min to block endogenous
peroxidase activity. Subsequently, 1X PBS (Gibco; Thermo Fisher
Scientific, Inc.) was used to wash the sections, and the sections
were incubated with primary anti-PDCD6 (cat. no. ab9181; 1:1,000
dilution) and anti-PTEN (cat. no. ab267787; 1:1,000 dilution; both
from Abcam) antibodies was performed overnight at 4°C. Horseradish
peroxidase-labelled anti-rabbit IgG secondary antibodies (cat. no.
ab6721; 1:2,000 dilution; Abcam) were used to treat samples for 30
min at room temperature. 3,3′-Diaminobenzidine was used to develop
the sections, which were counterstained with haematoxylin and
observed under an Olympus light microscope (magnification, x200;
Olympus Corporation).
Statistical analysis
Data are presented as the mean ± SD. P-values and
fold-changes were used to evaluate the differences in mRNA, miRNA
and lncRNA expression between different groups. Receiver operating
characteristic (ROC) curves were generated to evaluate the
diagnostic values of NEAT1, miR-124 and miR-9 for malignant thyroid
nodules by analysing the area under the curve (AUC) of the
candidate genes. The intergroup differences were analysed with
unpaired Student's t-test using SPSS version 16.0 software (SPSS,
Inc.). The χ2 test was used for the analysis of
characteristic distribution of the patients enrolled in this study.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Demographic, clinicopathological and
genotypic parameters of the study participants
The demographic and clinicopathological
characteristics of the 98 participants diagnosed with thyroid
nodules are summarized in Table
I. The majority of the MTN or BTN subjects were female, aged
between 42 and 60 years. In addition, parameters including nodule
size, solitary nodule, FT3, FT4 and TSH were also compared between
the two groups; no significant differences in these parameters were
observed between the MTN and BTN groups.
 | Table ICharacteristics of the participants
of the present study. |
Table I
Characteristics of the participants
of the present study.
| Characteristics of
patients | BTN (N=46) | MTN (N=52) | P-value |
|---|
| Sex, male (%) | 9 (19.6) | 11 (21.2) | 0.685 |
| Age, years, mean ±
SD | 51.3±8.9 | 50.6±7.4 | 0.788 |
| Nodule size, cm,
mean ± SD | 2.83±0.8 | 2.76±0.7 | 0.453 |
| Solitary nodule, n
(%) | 18 (39.1) | 21 (40.4) | 0.963 |
| FT3, pmol/l, median
(IQR) | 4.6 (3.7-5.3) | 4.4 (3.8-5.1) | 0.748 |
| FT4, pmol/l, median
(IQR) | 15.3
(13.3-17.6) | 15.5
(14.3-16.9) | 0.957 |
| TSH, mIU/ml, median
(IQR) | 1.2 (0.6-2.1) | 1.2 (0.5-2.1) | 0.994 |
Differential expression of NEAT1, miR-9
and miR-124
RT-qPCR was performed to compare the levels of
NEAT1, miR-9 and miR-124 in the peripheral blood and thyroid tissue
samples between the MTN and BTN groups. The NEAT1 levels in the
peripheral blood (Fig. 1A) and
thyroid tissue (Fig. 2A) samples
of the BTN group were higher compared with those in the MTN group,
whereas lower levels of miR-124 (Figs. 1B and 2B) and miR-9 (Figs. 1C and 2C) were detected in the BTN group,
suggesting that malignant thyroid nodules were associated with the
downregulation of NEAT1 and upregulation of miR-124/miR-9.
Differential expression of PTEN and
PDCD6
RT-qPCR and IHC were performed to compare the levels
of PTEN and PDCD6 in the peripheral blood and thyroid tissue
samples between the MTN and BTN groups. An evident increase in the
mRNA (Fig. 3A) and protein
(Fig. 3B) levels of PTEN was
observed in thyroid tissues collected from the BTN group compared
with those from the MTN group, along with higher mRNA (Fig. 4A) and protein (Fig. 4B) levels of PDCD6, suggesting that
malignant thyroid nodules were associated with downregulation of
PTEN and PDCD6 expression.
Diagnostic values of NEAT1, miR-124 and
miR-9 for malignant thyroid nodules
ROC analysis was performed to evaluate the
diagnostic values of NEAT1, miR-124 and miR-9 for malignant thyroid
nodules. In the peripheral blood samples, NEAT1, miR-124 and miR-9
exhibited AUC values of 0.8946, 0.7657 and 0.7019, respectively
(Fig. 5A). In thyroid tissue
samples, NEAT1, miR-124 and miR-9 had AUC values of 0.9304, 0.8221
and 0.7757, respectively (Fig.
5B).
The regulatory relationship among NEAT1,
miR-124, miR-9, PTEN and PDCD6
RT-qPCR and western blot analyses were performed to
explore the regulatory relationship among NEAT1, miR-124, miR-9,
PTEN and PDCD6. The levels of miR-124, miR-9, PTEN and PDCD6 in
BCPaP and SW579 cells were determined following transfection with
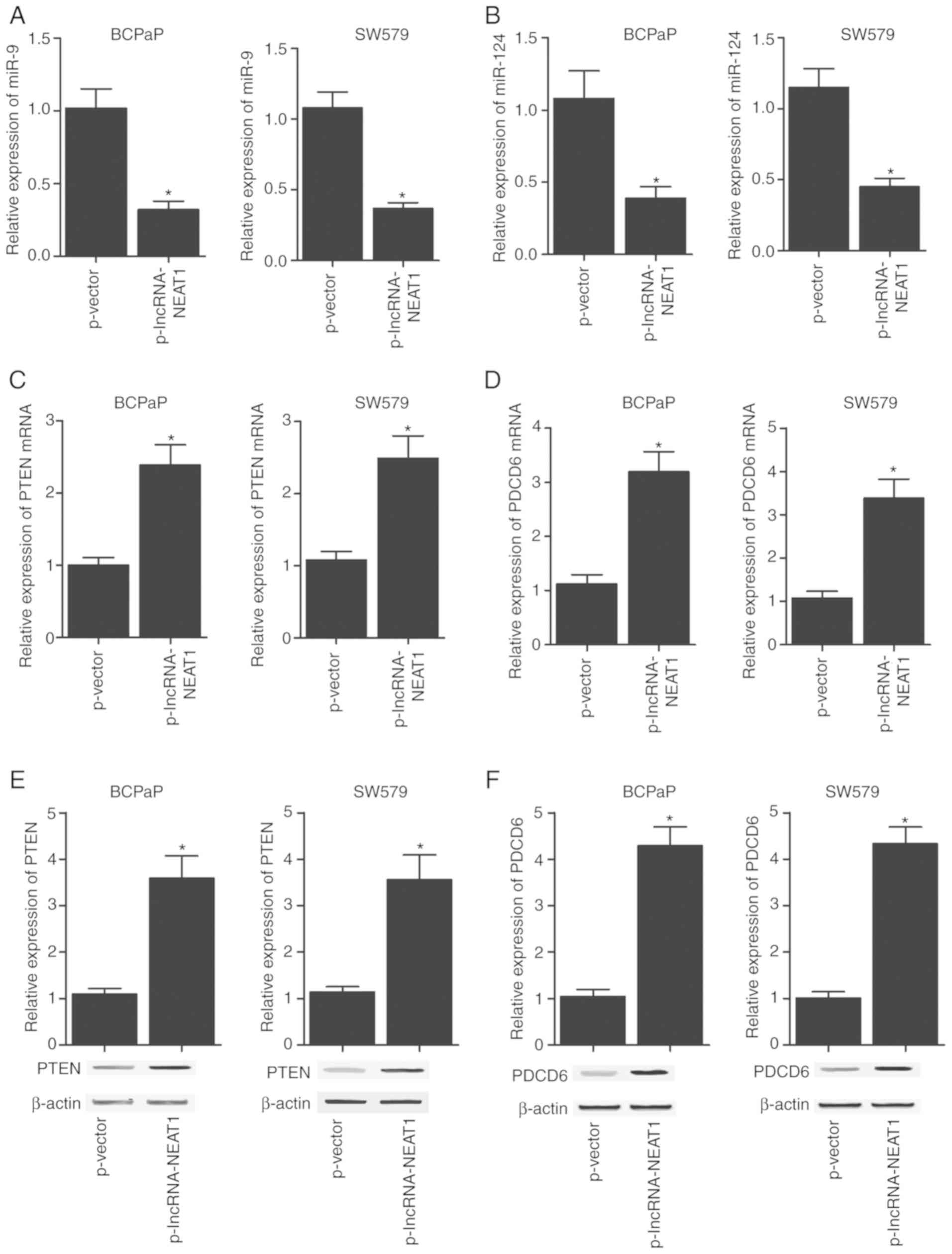
NEAT1 constructs. As presented in Fig. 6, miR-9 (Fig. 6A) and miR-124 (Fig. 6B) levels in BCPaP and SW579 cells
were markedly downregulated following transfection with the NEAT1
overexpression construct compared with those in the corresponding
negative control groups, along with markedly upregulated PTEN mRNA
(Fig. 6C) and protein (Fig. 6E) expression. In addition, BCPaP
and SW579 cells transfected with the NEAT1 overexpression construct
exhibited higher mRNA (Fig. 6D)
and protein (Fig. 6F) levels of
PDCD6 compared with those in cells transfected with the negative
control vector. The transfection of BCPaP and SW579 cells with
sh-NEAT1 inhibited NEAT1 expression (Fig. 7A), along with increased levels of
miR-9 (Fig. 7B) and miR-124
(Fig. 7C) compared with those in
the negative control groups. In addition, the transfection of BCPaP
and SW579 cells with sh-NEAT1 substantially downregulated the
expression of PTEN mRNA (Fig. 7D)
and protein (Fig. 7F), as well as
the expression of PDCD6 mRNA (Fig.
7E) and protein (Fig. 7G)
compared with those in the negative control groups. The successful
transfections of NEAT1 overexpression vector and sh-NEAT1 were
verified by the evidently upregulated (Fig. S1A) and downregulated (Fig. 1B) expression of NEAT1,
respectively.
Discussion
It is essential to identify an approach to
effectively differentiate benign nodules from thyroid carcinoma as
they are difficult to distinguish using fine-needle aspiration,
leading to unnecessary thyroidectomy (26). The present study compared the
levels of NEAT1, miR-9 and miR-124 in the peripheral blood and
thyroid tissue samples collected from subjects with malignant and
benign thyroid nodules. The results demonstrated that NEAT1
expression was significantly higher in the peripheral blood and
thyroid tissue samples of the BTN group compared with that in
samples from the MTN group, whereas the expression levels of
miR-124 and miR-9 were lower in the BTN group. The levels of PTEN
and PDCD6 expression in the peripheral blood and thyroid tissues of
the BTN group were higher compared with those in the tissues from
the MTN group. Previous studies have demonstrated that, compared
with patients with benign nodules, the expression of circulating
miR-5691, miR-9-3p and miR-124-3p was increased in patients with
papillary thyroid cancer (PTC), along with decreased levels of
miR-196b-5p and miR-4701 (22).
The abnormalities in the regulation of miR-196b-5p, miR-4701,
miR-9-3p and miR-124-3p were further confirmed using RT-qPCR; the
levels of miR-9-3p and miR-124-3p were increased in patients with
PTC compared with those of other candidate miRNAs (22). Another previous study demonstrated
that miR-124-3p and miR-9-2p could differentiate between benign and
malignant nodules with high specificity and sensitivity compared
with other candidate miRNAs (26), which was consistent with the
results of the present study. In addition, the present study
identified that the lncRNA NEAT1 regulated the expression of
multiple miRNAs by sponging them, and the results also demonstrated
that, compared with the other studied candidate genes, NEAT may be
a more accurate biomarker for the differentiation between malignant
and benign thyroid nodules with higher sensitivity and
specificity.
PTEN is involved in the regulation of metastasis,
apoptosis, proliferation and cell cycle progression, and low PTEN
expression has been reported to be associated with the progression
and development of human pharyngeal squamous cell cancer (27,28). A previous study reported that PTEN
acted as a downstream target gene of miR-9 and functioned as a
mediator of miR-9 in laryngocarcinoma cells, which suggested that
miR-9 could bind to the PTEN 3′-UTR to inhibit its transcription
(19). Another study demonstrated
that miR-9 modulated PTEN expression at the posttranscriptional
level (19). PTEN has been
reported to act as a cancer suppressor gene modulating the
PTEN/PI3K/AKT/mTOR signalling pathway, which is often dysregulated
in breast and prostate cancer (29). Therefore, by modulating the
miR-9/PTEN signalling axis, NEAT1 may participate in the
differentiation of benign and malignant thyroid nodules.
PDCD6 is a Ca2+-binding protein with a
molecular weight of 22 kDa and an open reading frame encoding a
sequence of 191 amino acids (31). In addition, PDCD6 has five
EF-hand-like motifs that are serially repetitive and form a region
comprising Ca2+-binding domains (32). The mRNA expression of PDCD6 is
ubiquitous among various tissues in mammals, and this pattern of
expression is associated with the process of apoptosis (31). In lower eukaryotes (such as
Dictyostelium discoideum), plants and insects, proteins
resembling PDCD6 have also been observed and identified to function
similarly to their homologs in mammals (33). The mRNA and protein levels of
PDCD6 are high in metastatic ovarian cancer as well as in
mesenchymal tumours (34,35). According to a previously published
study by Zhang et al (21), the role of miR-124-3p in BC
metastasis is regulated by the suppression of PDCD6. A functional
correlation between PDCD6 and miR-124 in primary tumours has been
indicated by the characterization of the miR-124-3p/PDCD6
signalling axis in patients with BC (21). Therefore, NEAT1 may interfere with
the differentiation of benign and malignant thyroid nodules,
possibly via the miR-124/PDCD6 signalling axis.
Previous studies have indicated that the expression
of NEAT1 is upregulated in various types of human cancer, such as
leukaemia, lung cancer and gastric carcinoma (17,18). NEAT1 upregulation facilitates the
malignancy of stomach tumours (17). In addition, the oncogenic effect
of NEAT1 has also been observed in BC (36,37). Han et al (38) also reported that NEAT1 enhanced
the resistance of cervical cancer to radiation by regulating
miR-193b-3p/cyclin D1 signalling. Overexpression of NEAT1 was
observed to promote cervical cancer cell proliferation, and NEAT1
acted as a ceRNA to negatively regulate miR-9-5p expression; the
levels of the downstream targets of miR-9-5p such as POU2F1 and
PTEN were accordingly elevated following NEAT1 overexpression
(16). Upregulation of miR-9-5p
could reverse the NEAT1-facilitated cancer cell proliferation,
suggesting that miR-9-5p may be a target molecule of NEAT1 in a
signalling cascade associated with cervical cancer progression
(16). It was also observed that
NEAT1 was located in the interchromatin space of the nucleus
(39). Previous studies have
reported that NEAT1 is an oncogene in a number of types of cancer
such as lung, oesophageal and colorectal cancer as well as
hepatocellular carcinoma (40).
Of note, radioimmunoprecipitation assays also confirmed that NEAT1
targeted miR-124 and was able to inhibit the expression of miR-124
(41). A previous study also
demonstrated a correlation between NEAT1 and miR-124-3p expression
levels in ovarian cancer cells (41). However, a contradictory statement
was published by Idogawa et al (42); in their study, lncRNA NEAT1 was
identified as a direct transcriptional target of p53. The
suppression of NEAT1 induced by p53 attenuated the inhibitory
effect of p53 on cancer cell proliferation and modulated gene
transactivation, indicating that NEAT1 may contribute to the
potential tumour-suppressor function of p53 (42). In the present study, the
diagnostic value of NEAT1, miR-124, and miR-9 was assessed using
ROC analysis. The results demonstrated that the AUC values of NEAT1
were higher compared with those of miR-124 and miR-9 in peripheral
blood samples and thyroid tissues. Therefore, the results of the
present study suggested that NEAT1 could be used as a biomarker to
differentiate between benign and malignant thyroid nodules.
In conclusion, the results of the present study
demonstrated the use of NEAT1 as a potential biomarker to
differentiate benign and malignant thyroid nodules. In addition,
NEAT1 may function as a sponge of miR-9 and miR-124. The present
study also identified PTEN and PDCD6 as possible targets of miR-9
and miR-124, respectively.
Supplementary Data
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ designed and supervised the study, participated
the experiments and drafted the manuscript. NZ participated the
experiments and analyzed the data. PZ performed literature
research, drafted and reviewed the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shangluo Central Hospital (Shangluo, China), and all
subjects provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
References
|
1
|
Shaheen Z, BenHamed NA and Elhamdani RM:
MON-436 cribriform-morular variant of papillary thyroid carcinoma:
A case report. J Endocr Soc. 4(Suppl 1): MON-4362020. View Article : Google Scholar :
|
|
2
|
Frates MC, Benson CB, Charboneau JW, Cibas
ES, Clark OH, Coleman BG, Cronan JJ, Doubilet PM, Evans DB,
Goellner JR, et al: Management of thyroid nodules detected at US:
Society of radiologists in ultrasound consensus conference
statement. Ultrasound Q. 22:231–240. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Papini E, Guglielmi R, Bianchini A,
Crescenzi A, Taccogna S, Nardi F, Panunzi C, Rinaldi R, Toscano V
and Pacella CM: Risk of malignancy in nonpalpable thyroid nodules:
Predictive value of ultrasound and color-Doppler features. J Clin
Endocrinol Metab. 87:1941–1946. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Frates MC, Benson CB, Doubilet PM,
Kunreuther E, Contreras M, Cibas ES, Orcutt J, Moore FD Jr, Larsen
PR, Marqusee E and Alexander EK: Prevalence and distribution of
carcinoma in patients with solitary and multiple thyroid nodules on
sonography. J Clin Endocrinol Metab. 91:3411–3417. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yassa L, Cibas ES, Benson CB, Frates MC,
Doubilet PM, Gawande AA, Moore FD Jr, Kim BW, Nosé V, Marqusee E,
et al: Long-term assessment of a multidisciplinary approach to
thyroid nodule diagnostic evaluation. Cancer. 111:508–516. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Haugen BR: 2015 American thyroid
association management guidelines for adult patients with thyroid
nodules and differentiated thyroid cancer: What is new and what has
changed? Cancer. 123:372–381. 2017. View Article : Google Scholar
|
|
7
|
Wang CC, Friedman L, Kennedy GC, Wang H,
Kebebew E, Steward DL, Zeiger MA, Westra WH, Wang Y, Khanafshar E,
et al: A large multicenter correlation study of thyroid nodule
cytopathology and histopathology. Thyroid. 21:243–251. 2011.
View Article : Google Scholar
|
|
8
|
Maher B: ENCODE: The human encyclopedia.
Nature. 489:46–48. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dianatpour A and Ghafouri-Fard S: Long
non-coding RNA expression intersecting cancer and spermatogenesis:
A systematic review. Asian Pac J Cancer Prev. 18:2601–2610.
2017.PubMed/NCBI
|
|
10
|
He JH, Han ZP and Li YG: Association
between long non-coding RNA and human rare diseases (Review).
Biomed Rep. 2:19–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maruyama R, Suzuki H, Yamamoto E, Imai K
and Shinomura Y: Emerging links between epigenetic alterations and
dysregulation of noncoding RNAs in cancer. Tumour Biol. 33:277–285.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu YC, Wang AM, Lu JK, Cen R and Liu LL:
Long noncoding RNA HOXD-AS1 regulates proliferation of cervical
cancer cells by activating Ras/ERK signaling pathway. Eur Rev Med
Pharmacol Sci. 21:5049–5055. 2017.PubMed/NCBI
|
|
14
|
Adriaens C and Marine JC: NEAT1-containing
paraspeckles: Central hubs in stress response and tumor formation.
Cell Cycle. 16:137–138. 2017. View Article : Google Scholar :
|
|
15
|
Choudhry H and Mole DR: Hypoxic regulation
of the noncoding genome and NEAT1. Brief Funct Genomics.
15:174–185. 2016. View Article : Google Scholar :
|
|
16
|
Xie Q, Lin S, Zheng M, Cai Q and Tu Y:
Long noncoding RNA NEAT1 promotes the growth of cervical cancer
cells via sponging miR-9-5p. Biochem Cell Biol. 97:100–108. 2019.
View Article : Google Scholar
|
|
17
|
Ma Y, Liu L, Yan F, Wei W, Deng J and Sun
J: Enhanced expression of long non-coding RNA NEAT1 is associated
with the progression of gastric adenocarcinomas. World J Surg
Oncol. 14:412016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao C, Zhang J, Wang Q and Ren C:
Overexpression of lncRNA NEAT1 mitigates multidrug resistance by
inhibiting ABCG2 in leukemia. Oncol Lett. 12:1051–1057. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu E, Su J, Zeng W and Zhang C: Enhanced
miR-9 promotes laryngocarcinoma cell survival via down-regulating
PTEN. Biomed Pharmacother. 84:608–613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cheng N and Guo Y: Long noncoding RNA
NEAT1 promotes nasopharyngeal carcinoma progression through
regulation of miR-124/NF-κB pathway. Onco Targets Ther.
10:5843–5853. 2017. View Article : Google Scholar :
|
|
21
|
Zhang L, Chen X, Liu B and Han J:
MicroRNA-124-3p directly targets PDCD6 to inhibit metastasis in
breast cancer. Oncol Lett. 15:984–990. 2018.PubMed/NCBI
|
|
22
|
Yu S, Liu X, Zhang Y, Li J, Chen S, Zheng
H, Reng R, Zhang C, Chen J and Chen L: Circulating microRNA124-3p,
microRNA9-3p and microRNA196b-5p may be potential signatures for
differential diagnosis of thyroid nodules. Oncotarget.
7:84165–84177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lei H, Gao Y and Xu X: LncRNA TUG1
influences papillary thyroid cancer cell proliferation, migration
and EMT formation through targeting miR-145. Acta Biochim Biophys
Sin (Shanghai). 49:588–597. 2017. View Article : Google Scholar
|
|
24
|
Bartolazzi A, Sciacchitano S and
D'Alessandria C: Galectin-3: The impact on the clinical management
of patients with thyroid nodules and future perspectives. Int J Mol
Sci. 19:4452018. View Article : Google Scholar :
|
|
25
|
Zhang YZ, Xu T, Gong HY, Li CY, Ye XH, Lin
HJ, Shen MP, Duan Y, Yang T and Wu XH: Application of
high-resolution ultrasound, real-time elastography, and
contrast-enhanced ultrasound in differentiating solid thyroid
nodules. Medicine (Baltimore). 95:e53292016. View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Morselli E, Galluzzi L, Kepp O, Vicencio
JM, Criollo A, Maiuri MC and Kroemer G: Anti- and pro-tumor
functions of autophagy. Biochim Biophys Acta. 1793:1524–1532. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Su CH, Chen LJ, Liao JF and Cheng JT:
Increase of phosphatase and tensin homolog by silymarin to inhibit
human pharynx squamous cancer. J Med Food. 16:778–784. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eng C: PTEN: One gene, many syndromes. Hum
Mutat. 22:183–198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hobert JA, Embacher R, Mester JL, Frazier
TW II and Eng C: Biochemical screening and PTEN mutation analysis
in individuals with autism spectrum disorders and macrocephaly. Eur
J Hum Genet. 22:273–276. 2014. View Article : Google Scholar :
|
|
31
|
Vito P, Lacanà E and D'Adamio L:
Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and
Alzheimer's disease gene ALG-3. Science. 271:521–525. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maki M, Narayana SV and Hitomi K: A
growing family of the Ca2+-binding proteins with five
EF-hand motifs. Biochem J. 328:718–720. 1997.
|
|
33
|
Ohkouchi S, Nishio K, Maeda M, Hitomi K,
Adachi H and Maki M: Identification and characterization of two
penta-EF-hand Ca(2+)-binding proteins in Dictyostelium discoideum.
J Biochem. 130:207–215. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Su D, Xu H, Feng J, Gao Y, Gu L, Ying L,
Katsaros D, Yu H, Xu S and Qi M: PDCD6 is an independent predictor
of progression free survival in epithelial ovarian cancer. J Transl
Med. 10:312012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
la Cour JM, Høj BR, Mollerup J, Simon R,
Sauter G and Berchtold MW: The apoptosis linked gene ALG-2 is
dysregu-lated in tumors of various origin and contributes to cancer
cell viability. Mol Oncol. 1:431–439. 2008. View Article : Google Scholar
|
|
36
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao D, Zhang Y, Wang N and Yu N: NEAT1
negatively regulates miR-218 expression and promotes breast cancer
progression. Cancer Biomark. 20:247–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han D, Wang J and Cheng G: LncRNA NEAT1
enhances the radio-resistance of cervical cancer via
miR-193b-3p/CCND1 axis. Oncotarget. 9:2395–2409. 2017. View Article : Google Scholar
|
|
39
|
Souquere S, Beauclair G, Harper F, Fox A
and Pierron G: Highly ordered spatial organization of the
structural long noncoding NEAT1 RNAs within paraspeckle nuclear
bodies. Mol Biol Cell. 21:4020–4027. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu X, Li Z, Zheng H, Chan MT and Wu WK:
NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif.
50:e123292017. View Article : Google Scholar
|
|
41
|
Chai Y, Liu J, Zhang Z and Liu L:
HuR-regulated lncRNA NEAT1 stability in tumorigenesis and
progression of ovarian cancer. Cancer Med. 5:1588–1598. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Idogawa M, Ohashi T, Sasaki Y, Nakase H
and Tokino T: Long non-coding RNA NEAT1 is a transcriptional target
of p53 and modulates p53-induced transactivation and
tumor-suppressor function. Int J Cancer. 140:2785–2791. 2017.
View Article : Google Scholar : PubMed/NCBI
|