Introduction
Breast cancer is a common type of cancer worldwide.
Although some progress has been made in the treatment of breast
cancer, the disease is still ranked as the second most prevalent
leading cause of cancer-related mortality among females (1-3).
The 5- and 10-year survival rates of patients with metastatic
breast cancer are relatively low and no marked improvements in
these rates have been noted over past 20-30 years; approximately
10% of breast cancer cases exhibit local recurrence, which severely
affects the prognosis of patients (4). Therefore, it is of utmost importance
to identify and develop effective measures with which to inhibit
the metastasis of breast cancer cells. It is also critical to
understand the pathogenesis of breast cancer and the expression
levels of related molecules so as to develop molecular therapies
which may be used to inhibit metastasis.
In recent years, an increasing number of studies
have found that the differential expression of genes is of great
significance to the occurrence and development of diseases, which
also helps to clarify the molecular mechanisms related to diseases
(5-7). Jensen et al in 2018 indicated
that tissue inhibitor of metal-loproteinases-2 (TIMP-2) can alter
the transformation of epithelial cells to mesenchymal cells, and
inhibit the growth, invasion and metastasis of breast cancer
(8). According to the study by
Raoof et al in 2019, in non-small lung cancer, the double
blocking of EGFR and FGFR may help to prevent and overcome the
occurrence of EMT-associated acquired drug resistance (9). In another study, growth
differentiation factor 15 (GDF15) was expected to be a novel
prognostic indicator for colorectal cancer, which may play a role
in promoting the metastasis of colorectal cancer by activating EMT
(10).
AT-rich interactive domain 1A (ARID1A) is located in
the chromosome 1p36 region (11),
and has been found to mutate in various types of cancer, such as
ovarian cancer (12), endometrial
cancer (13), gastric cancer
(14) and pancreatic cancer
(15). In addition, ARID1A
mutations are associated with an increased immune activity in
gastrointestinal cancer (16). It
has been reported that partial loss of ARID1A expression is related
to unfavorable outcome of patients with breast cancer (17). Epithelial-mesenchymal transition
(EMT) plays an important role in cancer development and progression
(18). Somsuan et al found
in 2019 that ARID1A knockout promoted EMT in cells, which was
characterized by an increased fusiform index and stromal markers,
decreased epithelial markers, and enhanced renal cell migration
activity and drug resistance (19). Wilson et al in 2019 also
indicated that the abnormal endometrial tissue diffusion was
closely related to the increase in EMT-related gene expression
induced by ARID1A deletion (20).
Therefore, the role of ARID1A in the inhibition of EMT is worthy of
attention. Moreover, whether ARID1A also plays a role in breast
cancer by intervening in the EMT process has attracted research
interests. Therefore, the present study conducted experiments to
exmaine the role of ARID1A in breast cancer, in order to provide
some basic referential strategies for the targeted treatment of
breast cancer in clinical practice.
Materials and methods
Patient samples and patient survival
rate
A total of 90 samples of breast cancer tissues and
their matched adjacent tissues were collected from 90 patients aged
between 30 and 50 years, who were diagnosed with breast cancer at
Dongzhimen Hospital, Beijing University of Chinese Medicine from
January, 2013 to January, 2014. The patients were divided into 2
groups according to the median expression level of ARID1A, and
their 5-year survival rates were calculated. Approval for the study
was obtained from the Dongzhimen Hospital Ethics Committee
(approval no. CH201309270). Written informed consent was obtained
from each patient. The demographics data of the patients are
presented in Table I.
 | Table IAssociation between ARID1A expression
and the clinicopathological characteristics of patients with breast
cancer. |
Table I
Association between ARID1A expression
and the clinicopathological characteristics of patients with breast
cancer.
| Variables | No. of
patients | ARID1A expression
| P-value |
|---|
| Negative | Positive |
|---|
| Total | 90 | 45 | 45 | |
| Age (years) | | 52.6±10.8 | 53.3±12.8 | 0.780 |
| TNM stage | | | | 0.003 |
| I | 27 | 9 | 18 | |
| II | 29 | 12 | 17 | |
| III | 34 | 24 | 10 | |
| Grade | | | | 0.660 |
| 1 or 2 | 58 | 28 | 30 | |
| 3 | 32 | 17 | 15 | |
| Tumor size | | | | 0.656 |
| ≤ cm | 27 | 14 | 13 | |
| >2, ≤5 cm | 30 | 13 | 17 | |
| >5 cm | 33 | 18 | 15 | |
| Lymph node
status | | | | 0.003 |
| Negative | 40 | 13 | 27 | |
| Positive | 50 | 32 | 18 | |
Cells and cell culture
Non-cancerous breast cells MCF-10A and breast cancer
cell lines, including BT20, BT474, T-47D, MCF7, MDA-MB-231,
MDA-MB-453 were acquired from the American Type Culture Collection
(ATCC). Dulbecco's modified Eagle's medium (DMEM) (GE Healthcare)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 µg/ml streptomycin and 100 U/ml penicillin was used to
culture the cells. The DMEM was maintained in a humidified
atmosphere at 37°C with 5% CO2.
Transfection, reagent treatment and
grouping
ARID1A siRNA (siARID1A; 5′-GGA GAU UGG UGG AUU GAC
UTT-3′) and scrambled siRNA (siNC; 5′-UUC UCC GAA CGU GUC ACG
UTT-3′) were purchased from Santa Cruz Biotechnology, Inc. The
pCMV6-XL4 and pCMV6-XL4-ARID1A (cat. no. SC303719) vectors were
obtained from OriGene Technologies, Inc., and pCMV6-XL4 was used as
a negative control (NC). For transfection, the MCF7 cells were
transfected with 50 nM siNC or 50 nM siARID1A, while the MDA-MB-231
cells were transfected with 100 ng pCMV6-XL4-ARID1A or 100 ng NC.
Cell transfection was performed using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer′s instructions. The cells were incubated in DMEM for
48 h at 37°C prior to all subsequent functional investigations.
Following transfection, the cells were treated with 5-fluorouracil
(5-FU; 5, 10, 20, 40 and 80 µg/ml) for 24 h, which was
purchased from Harbin Pharmaceutical Group.
In order to examine the effects of 5-FU on breast
cancer cell viability and ARID1A expression, the groups were set as
follows: The control group (untreated cells), siNC or NC group
(cells transfected with siNC or NC), siARID1A or ARID1A group
(cells transfected with siARID1A or pCMV6-XL4-ARID1A), 5-FU + siNC
or NC group (cells transfected with siNC or NC and treated with 5,
10, 20, 40 and 80 µg/ml 5-FU), 5-FU + siARID1A or ARID1A
group (cells transfected with siARID1A or pCMV6-XL4-ARID1A and
treated with 5, 10, 20, 40 and 80 µg/ml 5-FU). In order to
explore the function of ARID1A in breast cancer cells, the main
groups in the present study were set as follows: siNC or NC group,
siARID1A or ARID1A group, 5-FU + siNC or NC group (cells
transfected with siNC or NC and treated with 40 µg/ml 5-FU),
5-FU + siARID1A or ARID1A group (cells transfected with siARID1A or
pCMV6-XL4-ARID1A and treated with 40 µg/ml 5-FU).
Immunohistochemistry
A tissue microarray (TMA) slide set containing
duplicate or triplicate 0.6-mm cores from the 90 samples of breast
cancer tissues and their matched adjacent tissues from the same
surgical resection specimens was used. The immunohistochemical
staining of ARID1A was performed by TMA staining as follows: The
mouse monoclonal anti-ARID1A antibody (sc-81193; Santa Cruz
Biotechnology, Inc.) was diluted at 1/30 in blocking solution, and
the primary antibody was incubated with the samples for overnight
at 4°C. The bound antibody was detected with 2 µg/ml goat
anti-mouse biotin-conjugated secondary antibodies (cat. no. ab6789;
1:2,000; Abcam). Subsequently, the epithelial cells were evaluated
by 2 independent observers, who were selected to read the slides in
a blinded manner. The staining results were observed under a light
microscope (BX53; Olympus Corporation; magnification, x200). The
staining intensity was classified as 0 (negative), 1 (weak), 2
(moderate) and 3 (strong), the scores 0 and 1 were defined as low,
and the scores 2 and 3 were defined as high.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For RT-qPCR, 10 µg of total RNA were
extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and cDNA was then synthesized using oligodT and
SuperScript II reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.). The amplification of the RT-qPCR reaction was
conducted using a SYBR Premix Ex Taq kit (Takara Bio, Inc.) in the
7500 Real-time PCR system (Applied Biosystems). PCR commenced with
an initial DNA denaturation step (at 95°C for 3 min), followed by
30 cycles (denaturation at 95°C for 30 sec, annealing at 60°C for
30 sec, extension at 72°C for 30 sec). The result of mRNA levels
were normalized to GAPDH. PCR results were calculated using the
2−ΔΔCq method (21),
as previously described. The specific primers used are presented in
Table II.
 | Table IISequences of primers used for
RT-qPCR. |
Table II
Sequences of primers used for
RT-qPCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| ARID1A |
TCTTGCCCATCTGATCCATT |
CCAACAAAGGAGCCACCAC |
| E-Cadherin |
TGCCCAGAAAATGAAAAAGG |
GTGTATGTGGCAATGCGTTC |
| N-Cadherin CC |
ATCACTCGGCTTAATGGT |
ACCCACAATCCTGTCCACAT |
| Vimentin |
GACAATGCGTCTCTGGCACGTCTT |
TCCTCCGCCTCCTGCAGGTTCTT |
| VEGF |
TACCTCCACCATGCCAAGTG |
ATGATTCTGCCCTCCTCCTTC |
| Cyclin D1 |
GGATGCTGGAGGTCTGCGA |
AGAGGCCACGAACATGCAAG |
| Bcl-2 |
TTGTGGCCTTCTTTGAGTTCGGTG |
GGTGCCGGTTCAGGTACTCAGTCA |
| Bax |
CCTGTGCACCAAGGTGCCGGAACT CC |
ACCCTGGTCTTGGATCCAGCCC |
| GAPDH |
TGCCAAATATGATGACATCAAGAA |
GGAGTGGGTGTCGCTGTTG |
Cell proliferation assay
Following transfection, the breast cancer cells
(2×104/well) in each group were plated in 96-well plates
and incubated for 24 h at 37°C. Subsequently, 10 µl of CCK-8
solution (Dojindo Molecular Technologies, Inc.) were added to each
well to incubate the cells for 1 h at 37°C. The absorbance at a
wavelength of 450 nm was measured using a microtiter plate (Thermo
LabSystems; Thermo Fisher Scientific, Inc.).
Cell apoptosis assay
For the analysis of cell apoptosis, the breast
cancer cells in each group were collected and stained with 10
µl of Annexin V-fluorescein isothiocyanate (Annexin V;
Thermo Fisher Scientific, Inc.) and 5 µl of prop-idium
iodide (PI) according to the manufacturer's instructions, and the
solution was then incubated at room temperature for 15 min. The
percentage of Annexin V/PI-positive cells was quantified using a
FACScan flow cytometer (BD Biosciences).
Cell cycle assay
For the analysis of the cell cycle, the breast
cancer cells were collected, and further fixed using 70% ice-cold
ethanol at 4°C for 1 h. The cells were then washed once with PBS.
Subsequently, the breast cancer cells were incubated with RNase
(0.5 mg/ml) in PBS for 1 h at 37°C, followed by incubation with PI
for 30 min at 25°C in the dark. The cell cycle was analyzed by FACS
(BD Biosciences) at 488 nm.
Western blot (WB) analysis
The breast cancer cells were lysed with ice-cold
lysis buffer and the protein concentration was calculated with a
BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.).
Equal amounts of protein were separated by 10% SDS-PAGE and
transferred onto a polyvinylidene fluoride (PVDF) membrane. The
membrane was then blocked using 5% non-fat milk for 1 h and
incubated with the following primary antibodies overnight at 4°C:
ARID1A (1:2,000, 242 kDa, ab242377), E-cadherin (1:10,000, 97 kDa,
ab40772), N-cadherin (1:1,000, 130 kDa, ab18203), Vimentin
(1:1,000, 54 kDa, ab92547), vascular endothelial growth factor
(VEGF; 5-10 µg/ml, 21 kDa, ab1316), cyclin D1 (1:10,000, 34
kDa, ab134175), Bcl-2 (1:2,000, 26 kDa, ab59348), Bax (1:2,000, 21
kDa, ab32503) (all from Abcam). The secondary goat anti-mouse IgG
H&L (HRP) (1:2,000, ab205719) and goat anti-rabbit IgG H&L
(HRP) (1:2,000, ab205718) antibodies (both from Abcam) were then
used to incubate the membrane for 1 h at room temperature.
Image-Pro Plus 6.0 software (Media Cybernetics, Inc.) was used to
analyze protein expression. GAPDH (1:10,000, 36 kDa, ab181602;
Abcam) was used as an internal control.
Wound healing migration assay
For the determination of cell migration, the breast
cancer cells (1×105/well) were inoculated to 24-well
plates with DMEM. An equally wide single scratch was made using 10
µl pipette tip from the top to the bottom of the culture
plates when the cells of each group grew to reach 70% confluence.
The debris was removed with PBS and added serum-free DMEM. Images
of the cells were captured immediately after the scratch defect was
made and at 24 h following incubation at 37°C by using a light
microscope (magnification, x100; Nikon Corporation).
Cell invasion assay
Transwell chambers were used to assess cell
invasion. For cell invasion assay, the cells (3×105/ml)
were cultured in serum-free medium. The cells were then added to
the upper chamber precoated with Matrigel matrix (BD Biosciences),
and medium containing 10% FBS was added to the lower chamber.
Following incubation for 24 h at 37°C, cells remaining on the
surface of the upper chamber were removed with a cotton swab. The
invading cells were fixed with 4% paraformaldehyde for 15 min at
room temperature, and then stained with 0.1% crystal violet
solution for 20 min at room temperature. Images were obtained under
a light microscope (magnification, x100; Nikon Corporation) and the
number of migrated cells was calculated using ImageJ software 1.8.0
(National Institutes of Health).
Tube formation assay
The breast cancer cell culture medium was changed to
serum-free DMEM medium for 48 h, and the medium was then collected,
centrifuged (1,000 × g, 10 min, 4°C) and filtered to obtain
tumor-conditioned medium. Subsequently, 50 µl of ice-cold BD
Matrigel matrix (BD Biosciences) was added to a 24-well plate and
incubated for 30 min at 37°C. HUVECs (cat. no. PCS-100-010; ATCC)
in 100 µl of conditioned medium were then added to the
wells. Following incubation at 37°C for 4 h, the wells were
examined using an Olympus CKX41 microscope (Olympus Corporation).
Images were then captured with an Olympus DP20-5 digital camera
(Olympus Corporation) and the capillary tubes in the images were
counted using ImageJ software 1.8.0 (National Institutes of
Health).
Statistical analysis
The experimental data are expressed as the means ±
standard deviation and analyzed using SPSS 20.0 software (SPSS,
Inc.). The data in presented in Table
I were analyzed using the Chi-squared test and rank sum test.
The Student t-test was used to analyze differences between 2
groups, and one-way ANOVA was used to analyze differences between
>2 groups followed by the Bonferroni test. The Kaplan-Meier
method with the log rank test was adopted to compare the survival
rates in the 2 groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
ARID1A exhibits a low expression in
breast cancer
In order to examine the expression of ARID1A in
breast cancer, its expression in breast cancer tissues (n=3) and
their matched adjacent tissues (n=3) was analyzed by
immunohistochemistry, and the results revealed that ARID1A
exhibited a lower expression in the cancer tissues than in the
adjacent tissues (Fig. 1A).
Moreover, the staining intensity in adjacent tissue no. 1 was
higher than that in adjacent tissue no. 2, but lower than that in
adjacent tissue no. 3; among the 3 cancer tissues, the staining
intensity in cancer tissue no. 1 was lowest and that in cancer
tissue no. 3 was the highest (Fig.
1A). RT-qPCR was then performed to examine the mRNA expression
of ARID1A in these tissues, and the results obtained were similar
to those obtained by immunohistochemistry (Fig. 1B). Subsequently, the levels of
ARID1A in MCF-10A and breast cancer cell lines, including BT20,
BT474, T-47D, MCF7, MDA-MB-231, MDA-MB-453, were compared and the
results indicated that ARID1A was expressed at low levels in the
breast cancer cell lines compared with the normal MCF-10A cells
(Fig. 1C). Among the 6 breast
cancer cell lines, the expression level of ARID1A was the highest
in the MCF7 cells and the lowest in the MDA-MB-231 cells. Thus,
these 2 cells were selected for use in further experiments. The
data on the 5-year survival rate of the breast cancer cases were
then acquired, and the results revealed that the overall survival
rate of the breast cancer patients with a higher expression of
ARID1A was higher than those with a lower expression of ARID1A
(Fig. 1D). In addition, as shown
in Table I, ARID1A expression was
closely related to the TNM stage and the lymph node status of the
patients.
Silencing of ARID1A partially reverses
the inhibitory effect of 5-FU at various concentrations on cell
viability
The MCF7 and MDA-MB-231 cells were selected to
further conduct in vitro cell experiments. After verifying
the successful trans-fection efficiency of the cells (MCF7 cells
transfected with ARID1A siRNA and MDA-MB-231 cells transfected with
ARID1A overexpression vector; Fig.
2A-F), it was found that with the increase in the 5-FU
concentration (5, 10, 20, 40 and 80 µg/ml), the inhibitory
effects of 5-FU on cell viability were enhanced; however, the
silencing of ARID1A partially reversed the inhibitory effects of
5-FU, whereas the overexpression of ARIDIA further promoted the
inhibitory effects of 5-FU (Fig. 2G
and H). RT-qPCR was then performed to examine the effect of
5-FU on ARID1A expression. It was found that 5-FU increased the
mRNA level of ARID1A, and with the increase in the 5-FU
concentration, the promoting effect of 5-FU was enhanced. However,
transfection with ARID1A siRNA or pCMV6-XL4-ARID1A reversed or
promoted the effects of 5-FU, respectively (Fig. 2I and J).
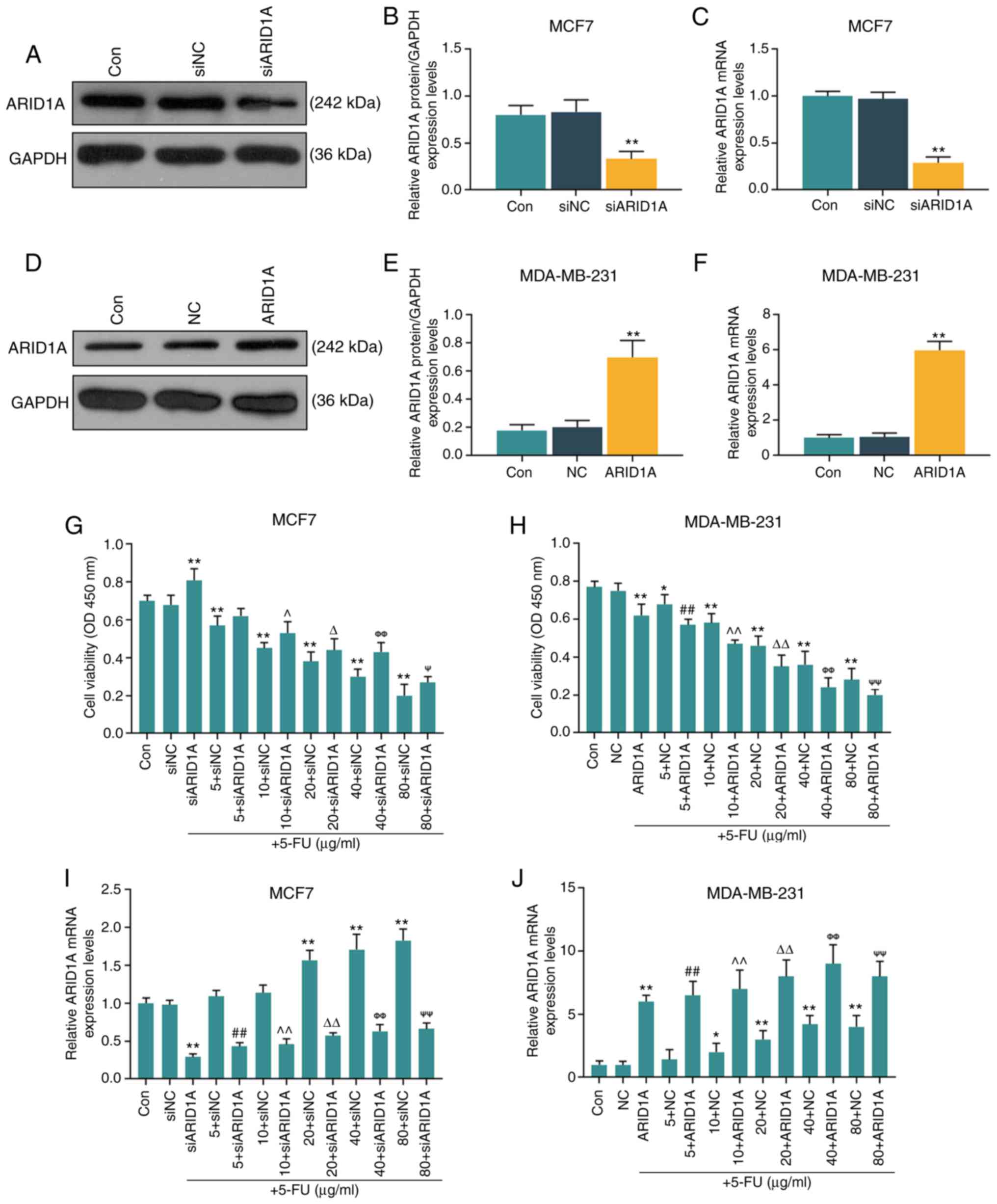 | Figure 2Effects of function of ARID1A on the
viability of breast cancer cells treated with 5-FU. (A and B) MCF7
cells were transfected with siARID1A and cultured for 48 h, and the
transfection efficiency of siARID1A was detected by WB analysis.
(C) The mRNA expression of ARID1A in MCF7 cells was detected by
RT-qPCR after the cells were transfected with siARID1A. (D and E)
MDA-MB-231 cells were transfected with ARID1A overexpression vector
and cultured for 48 h, and the transfection efficiency of ARID1A
was detected by WB analysis. (F) The mRNA expression of ARID1A in
MDA-MB-231 cells was detected by RT-qPCR after the cells were
transfected with ARID1A overexpression vector. (G) The effect of
ARID1A silencing on the viability of MCF-7 cells treated with
various concentrations of 5-FU (5, 10, 20, 40 and 80 µg/ml)
for 24 h was detected by proliferation assay. (H) Effect of ARID1A
overexpres-sion on the viability of MDA-MB-231 cells treated with
various concentrations of 5-FU (5, 10, 20, 40 and 80 µg/ml)
for 24 h was detected by proliferation assay. (I) The mRNA
expression of ARID1A in MCF-7 cells treated with various
concentrations of 5-FU (5, 10, 20, 40 and 80 µg/ml) and
siARID1A was detected by RT-qPCR. (J) The mRNA expression of ARID1A
in MDA-MB-231 cells treated with various concentrations of 5-FU (5,
10, 20, 40 and 80 µg/ml) and pCMV6-XL4-ARID1A was detected
by RT-qPCR. Data were obtained from 3 representative experiments
and are presented as the means ± standard deviation the experiment
was repeated 3 times (n=3). *P<0.05,
**P<0.001 vs. siNC or NC; ##P<0.001 vs.
5 + siNC or 5 + NC; ^P<0.05, ^^P<0.001
vs. 10 + siNC or 10 + NC; ΔP<0.05,
ΔΔP<0.001 vs. 20 + siNC or 20 + NC;
ΦΦP<0.001 vs. 40 + siNC or 40 + NC;
ΨP<0.05, ΨΨP<0.001 vs. 80 + siNC or 80
+ NC. ARID1A, AT-rich interactive domain 1A; 5-FU, 5-fluorouracil;
WB analysis, western blot analysis. |
Silencing of ARID1A suppresses the
effects of 5-FU on cells, whereas the overexpression of ARID1A
exerts an opposite effect
The concentration of 40 µg/ml 5-FU was then
used to treat the cells transfected with ARID1A siRNA or
pCMV6-XL4-ARID1A. First, the apoptotic rate of the cells was
calculated by flow cytometry. It was found that 5-FU promoted cell
apoptosis; however, the silencing of ARID1A partially reversed this
effect, whereas transfection with pCMV6-XL4-ARID1A enhanced the
effects of 5-FU (Fig. 3A and B).
Similar results were obtained with cell cycle analysis: 5-FU
exerted an inhibitory effect on the cell cycle at the G0 phase, and
the silencing of ARID1A partially reversed this effect. However,
the overexpression of ARID1A promoted the effects of 5-FU on the
cell cycle (Fig. 3C and D).
A scratch test was also conducted to examine the
migration rate of the cells. In this experiment, it was found that
the migration distance in the siARID1A group at 24 h was greater
than that in the siNC group, and the migration distance in the siNC
+ 5-FU group was less than that in the siNC group; siARID1A
partially reversed the effect of 5-FU on cell migration, whereas
the overexpression of ARID1A promoted the effect of 5-FU on cell
migration (Fig. 4A and B). In
addition, Transwell assay was conducted to detect the invasion rate
of the cells. It was found that the silencing of ARID1A promoted
cell invasion, whereas 5-FU exerted an opposite effect. The
silencing of ARID1A partially reversed the effect of 5-FU on cell
invasion, whereas ARID1A overexpression exerted an opposite effect
to ARID1A silencing (Fig. 4C and
D). Tube formation assay was performed to detect the
angiogenesis formation rate of the cells, and the results indicated
that cells transfected with ARID1A siRNA exhibited a strong tube
formation ability, whereas 5-FU inhibited this ability, and ARID1A
overexpression enhanced the effect of 5-FU (Fig. 4E and F).
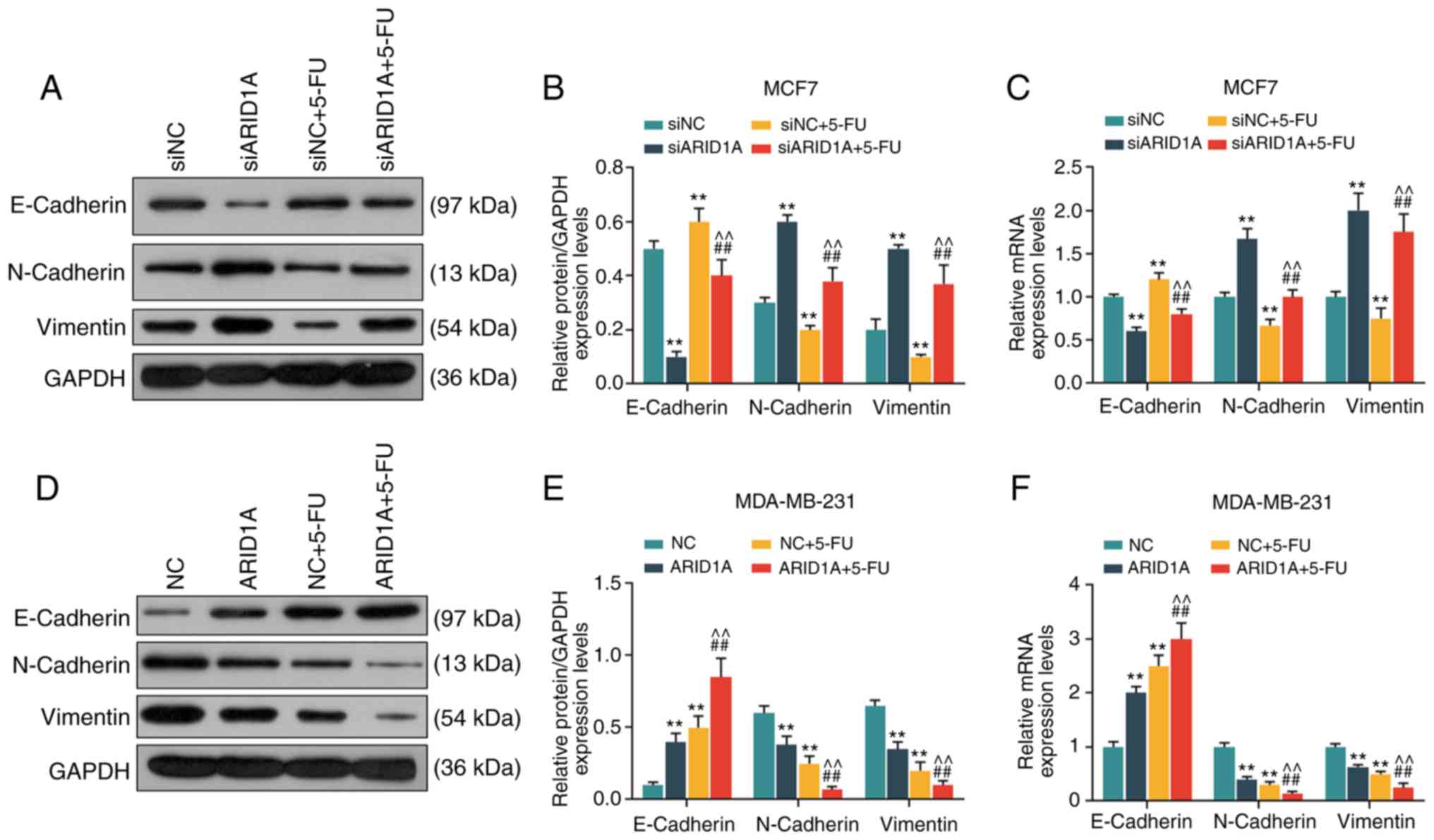
Silencing of ARID1A promotes the
expression of EMT-related proteins, whereas the overexpression of
ARID1A exerts an opposite effect
From the results of WB analysis and RT-qPCR, it was
revealed that the cells transfected with ARID1A siRNA exhibited a
lower expression of E-cadherin protein and mRNA, and a higher
expression of N-cadherin and Vimentin protein and mRNA. 5-FU
increased E-cadherin expression and decreased the expression of
N-cadherin and Vimentin, whereas the silencing of ARID1A partially
reversed the effects of 5-FU (Fig.
5A-C). On the contrary, the cells transfected with
pCMV6-XL4-ARID1A exhibited a higher expression of E-cadherin
protein and mRNA, and a lower expression of N-cadherin and Vimentin
protein and mRNA, and pCMV6-XL4-ARID1A promoted the effects of 5-FU
(Fig. 5D-F).
Silencing of ARID1A suppresses the
effects of 5-FU on the cell cycle, apoptosis and the expression of
angiogenesis-related proteins and mRNAs, whereas the overexpression
of ARID1A exerts an opposite effect
From the results of WB analysis and RT-qPCR, it was
revealed that the expression levels of ARID1A and Bax were
decreased, while those of VEGF, cyclin D1 and Bcl-2 were increased
in the siARID1A group; opposite results were observed in the siNC +
5-FU group, while the silencing of ARID1A partially reversed the
effects of 5-FU (Fig. 6A-C). On
the contrary, the expression levels of ARID1A and Bax were
increased, while those of VEGF, cyclin D1 and Bcl-2 were decreased
in the ARID1A over-expression group; similar results were observed
in the NC + 5-FU group, and ARID1A overexpression promoted the
effects of 5-FU (Fig. 6D-F).
 | Figure 6Effects of ARID1A on the cell cycle,
apoptosis, and the expression of angiogenesis-related proteins and
mRNAs. (A and B) Detection of the protein expression of ARID1A,
VEGF, cyclin D1, Bcl-2 and Bax in MCF7 cells by WB analysis. (C)
Detection of the mRNA expressions of ARID1A, VEGF, cyclin D1, Bcl-2
and Bax in MCF7 cells by RT-qPCR. (D and E) Detection of the
protein expression of ARID1A, VEGF, cyclin D1, Bcl-2 and Bax in
MDA-MB-231 cells by WB analysis. (F) Detection of the mRNA
expression of ARID1A, VEGF, cyclin D1, Bcl-2 and Bax in MDA-MB-231
cells by RT-qPCR. Data were obtained from 3 representative
experiments and are presented as the means ± standard deviation.
The experiment was repeated 3 times (n=3). **P<0.001
vs. siNC or NC; ##P<0.001 vs. siARID1A or ARID1A;
^P<0.05, ^^P<0.001 vs. siNC + 5-FU or
NC + 5-FU. ARID1A, AT-rich interactive domain 1A; 5-FU,
5-fluorouracil; WB analysis, western blot analysis. |
Discussion
EMT is a process through which epithelial cells are
transformed into mesenchymal cells, characterized by the loss of
E-cadherin and the increase in vimentin expression, and this
process has been found to be involved in the metastasis of several
types of cancer (22-24). The results of the present study
suggested that the upregulation of ARID1A expression enhanced the
sensitivity of breast cancer cells to 5-FU, inhibited cell growth,
migration and EMT, and promoted cell apoptosis.
5-FU has been widely used as a first-line drug in
human cancer chemotherapy. It can target thymidylate synthetase to
play an anticancer role by blocking DNA synthesis and interfering
in RNA processing. However, the clinical efficacy of 5-FU varies
greatly due to chemotherapeutic resistance (25,26). The study by Sagara et al in
2016 indicated that 5-FU was a good choice for patients with
triple-negative breast cancer with distant metastasis (27). However, drug resistance is an
issue worthy of attention in treatment, which was believed to be
related to the overexpression of drug efflux transporters or some
enzymes or EMT (27). Some
researchers have demonstrated that the downregulation of ADAM12-L
was helpful in reducing the resistance of breast cancer cells to
5-FU, which was related to the regulation of PI3K/Akt signaling,
thus proving that the regulation of ADAM12-L level may have a
positive effect on chemotherapy for breast cancer patients in
clinical practice (28). Another
study reported that the activation of FOXO3a can restore the
sensitivity of breast cancer cells to 5-FU (29).
In the present study, the role of ARID1A in
5-FU-treated breast cancer cells was investigated. It was found
that the expression of ARID1A was lower in the human cancer tissues
than in the normal tissues, and it was also expressed at low levels
in breast cancer cell lines. Furthermore, patients with a higher
level of ARID1A exhibited a longer survival time, which is
consistent with the antitumor effects of ARID1A observed in colon
cancer. Mathur et al in 2017 found that the loss of ARID1A
led to invasive colon adenocarcinoma in mice, and through the
detection of some genes, it was found that the regulation of
enhancer-mediated gene was probably one of the mechanisms through
which ARID1A exerts its anticancer effects on colon cancer
(30). Furthermore, Sasaki et
al in 2019 found that the change in ARID1A can be used as a
characteristic of the dual plate malformation type of small cell
lung cancer, and also a diagnostic immunohistochemical marker in
this disease (31).
Based on previous studies and the present
experimental results, the role of ARID1A in the sensitivity to
anticancer drugs was further explored. Consistent with findings
from previous research that 5-FU inhibits cancer growth (32-34), the results of the present study
revealed that 5-FU inhibited the activity of breast cancer cells,
by promoting the apoptosis and blocking the cell cycle in the G1
phase. The upregulation of the expression of ARID1A enhanced the
effects of 5-FU on the cells. In addition, it was found that 5-FU
inhibited the migration, invasion and tube formation of the cells,
and the upregulation of ARID1A expression enhanced the effects of
5-FU on the cells, while the downregulation of ARID1A expression
exerted an opposite effect to 5-FU. These data suggest that ARID1A
plays a role in enhancing the sensitivity of breast cancer cells to
5-FU. Furthermore, the possible mechanisms involved were
investigated, and it was found that 5-FU promoted the expression of
E-cadherin and inhibited the expression of N-cadherin and Vimentin,
whereas the silencing of ARID1A exerted an opposite effect.
Cadherin is an important factor in regulating homeostasis. It can
transfer adhesion signals into a network of signal effectors and
transcription programs, thus regulating cell functions (35). The decreased expression of
E-cadherin is a sign of EMT (36). The high expression of N-cadherin
is positively associated with EMT (37). Vimentin is an intermediate silk
protein expressed in stromal cells and some ectodermal cells. Its
abnormal expression level is related to changes in the cytoskeleton
protein structure, and it has the ability to promote cuboidal
epithelial cells to transform into fusiform fibroid cells, and
enhance their migratory ability. Therefore, the functions of
vimentin are mainly known as maintaining the morphology of cells
and organelles, signal transduction, transplantation immunity and
apoptosis (38). In the present
study, it was found that the upregulation of the expression of
ARID1A enhanced the sensitivity of breast cancer cells to 5-FU,
thus leading to the inhibition of cell migration and proliferation,
which is related to the regulation of the EMT process. Apart from
these factors, genes that are related to proliferation, migration,
invasion and tube formation, such as VEGF, cyclin D1, Bcl-2 and
Bax, were also influenced by 5-FU and the effect of 5-FU on these
genes were partially reversed by ARID1A silencing.
On the whole, the loss-of-function mutation of
ARID1A often occurs in breast cancer patients. The present study
demonstrated that the upregulation of the expression of ARID1A can
enhance the sensitivity of breast cancer cells to 5-FU, inhibit
cell growth, migration and EMT, and promote cell apoptosis.
However, there are also some limitations to the present study; for
example, in the future, animal models are required to further
verify the effects of ARID1A on breast cancer models to 5-FU, and
further experiments and test indicators are required to clarify the
mechanisms of action of ARID1A.
In conclusion, the present study demonstrates that
ARID1A plays an anticancer role in breast cancer and enhances the
sensitivity of breast cancer cells to 5-FU. These findings may
provide a novel treatment strategy for breast cancer.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW and XG made substantial contributions to the
conception and design of the study. KZ, TJ, SG, PL, XZ and XS were
involved in data acquisition, data analysis and interpretation. TW
and XG were involved in the drafting of the article or critically
revising it for important intellectual content. All authors gave
the final approval of the final version of the article to be
published and all authors agree to be accountable for all aspects
of the work in ensuring that questions related to the accuracy or
integrity of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
All procedures performed involving human
participants were in accordance with the ethical standards of the
institutional and/or national research committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards. Approval for the study was obtained from the Dongzhimen
Hospital Ethics Committee (approval no. CH201309270). Written
informed consent was obtained from each patient. No animals are
involved in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
ARID1A
|
AT-rich interactive domain 1A;
|
|
EMT
|
epithelial-mesenchymal transition;
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction;
|
|
WB
|
western blot;
|
|
CCK-8
|
Cell Counting kit 8;
|
|
5-FU
|
5-fluorouracil;
|
|
TIMP-2
|
tissue inhibitor of
metalloproteinases-2;
|
|
GDF15
|
growth differentiation factor 15;
|
|
DMEM
|
Dulbecco's modified Eagle's
medium;
|
|
PVDF
|
polyvinylidene fluoride
|
References
|
1
|
Liang HF, Zhang XZ, Liu BG, Jia GT and Li
WL: Circular RNA circ-ABCB10 promotes breast cancer proliferation
and progression through sponging miR-1271. Am J Cancer Res.
7:1566–1576. 2017.PubMed/NCBI
|
|
2
|
Balekouzou A, Yin P, Pamatika CM,
Bishwajit G, Nambei SW, Djeintote M, Ouansaba BE, Shu C, Yin M, Fu
Z, et al: Epidemiology of breast cancer: Retrospective study in the
Central African Republic. BMC Public Health. 16:12302016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calado A, Neves PM, Santos T and Ravasco
P: The effect of flax-seed in breast cancer: A literature review.
Front Nutr. 5:42018. View Article : Google Scholar
|
|
4
|
Yates LR, Knappskog S, Wedge D, Farmery
JHR, Gonzalez S, Martincorena I, Alexandrov LB, Van Loo P, Haugland
HK, Lilleng PK, et al: Genomic evolution of breast cancer
metastasis and relapse. Cancer Cell. 32:169–184.e7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goodarzi H, Nguyen HCB, Zhang S, Dill BD,
Molina H and Tavazoie SF: Modulated expression of specific tRNAs
drives gene expression and cancer progression. Cell. 165:1416–1427.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ketterer S, Gomez-Auli A, Hillebrand LE,
Petrera A, Ketscher A and Reinheckel T: Inherited diseases caused
by mutations in cathepsin protease genes. FEBS J. 284:1437–1454.
2017. View Article : Google Scholar
|
|
7
|
Butchbach ME: Copy number variations in
the survival motor neuron genes: Implications for spinal muscular
atrophy and other neurodegenerative diseases. Front Mol Biosci.
3:72016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jensen SM, Kumar S, Chowdhury A, Castro N,
Shih J, Salomon DS and Stetler-Stevenson WG: TIMP-2 inhibits triple
negative breast cancer growth and metastasis through EMT
suppression and promotion of vascular normalization. FASEB J.
32:678.6722018.
|
|
9
|
Raoof S, Mulford IJ, Frisco-Cabanos H,
Nangia V, Timonina D, Labrot E, Hafeez N, Bilton SJ, Drier Y, Ji F,
et al: Targeting FGFR overcomes EMT-mediated resistance in EGFR
mutant non-small cell lung cancer. Oncogene. 38:6399–6413. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li C, Wang J, Kong J, Tang J, Wu Y, Xu E,
Zhang H and Lai M: GDF15 promotes EMT and metastasis in colorectal
cancer. Oncotarget. 7:860–872. 2016. View Article : Google Scholar :
|
|
11
|
Chunder N, Mandal S, Basu D, Roy A,
Roychoudhury S and Panda CK: Deletion mapping of chromosome 1 in
early onset and late onset breast tumors-a comparative study in
eastern India. Pathol Res Pract. 199:313–321. 2003. View Article : Google Scholar
|
|
12
|
Bitler BG, Wu S, Park PH, Hai Y, Aird KM,
Wang Y, Zhai Y, Kossenkov AV, Vara-Ailor A, Rauscher FJ III, et al:
ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell
Biol. 19:962–973. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takeda T, Banno K, Okawa R, Yanokura M,
Iijima M, Irie-Kunitomi H, Nakamura K, Iida M, Adachi M, Umene K,
et al: ARID1A gene mutation in ovarian and endometrial cancers
(Review). Oncol Rep. 35:607–613. 2016. View Article : Google Scholar :
|
|
14
|
Wang K, Kan J, Yuen ST, Shi ST, Chu KM,
Law S, Chan TL, Kan Z, Chan AS, Tsui WY, et al: Exome sequencing
identifies frequent mutation of ARID1A in molecular subtypes of
gastric cancer. Nat Genet. 43:1219–1223. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shain AH, Giacomini CP, Matsukuma K,
Karikari CA, Bashyam MD, Hidalgo M, Maitra A and Pollack JR:
Convergent structural alterations define SWItch/Sucrose
NonFermentable (SWI/SNF) chromatin remodeler as a central tumor
suppressive complex in pancreatic cancer. Proc Natl Acad Sci USA.
109:E252–E259. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Li M, Jiang Z and Wang X: ARID1A
mutations are associated with increased immune activity in
gastrointestinal cancer. Cells. 8:6782019. View Article : Google Scholar :
|
|
17
|
Takao C, Morikawa A, Ohkubo H, Kito Y,
Saigo C, Sakuratani T, Futamura M, Takeuchi T and Yoshida K:
Downregulation of ARID1A, a component of the SWI/SNF chromatin
remodeling complex, in breast cancer. J Cancer. 8:1–8. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y and Weinberg RA:
Epithelial-to-mesenchymal transition in cancer: Complexity and
opportunities. Front Med. 12:361–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Somsuan K, Peerapen P, Boonmark W,
Plumworasawat S, Samol R, Sakulsak N and Thongboonkerd V: ARID1A
knockdown triggers epithelial-mesenchymal transition and
carcinogenesis features of renal cells: Role in renal cell
carcinoma. FASEB J. 33:12226–12239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wilson MR, Reske JJ, Holladay J, Wilber
GE, Rhodes M, Koeman J, Adams M, Johnson B, Su RW, Joshi NR, et al:
ARID1A and PI3-kinase pathway mutations in the endometrium drive
epithelial transdifferentiation and collective invasion. Nat
Commun. 10:35542019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Rokavec M, Kaller M, Horst D and Hermeking
H: Pan-cancer EMT-signature identifies RBM47 down-regulation during
colorectal cancer progression. Sci Rep. 7:46872017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Das V, Bhattacharya S, Chikkaputtaiah C,
Hazra S and Pal M: The basics of epithelial-mesenchymal transition
(EMT): A study from a structure, dynamics, and functional
perspective. J Cell Physiol. Feb 5–2019.Epub ahead of print.
View Article : Google Scholar
|
|
24
|
Koeck S, Amann A, Huber JM, Gamerith G,
Hilbe W and Zwierzina H: The impact of metformin and salinomycin on
transforming growth factor β-induced epithelial-to-mesenchymal
transition in non-small cell lung cancer cell lines. Oncol Lett.
11:2946–2952. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S,
Li M, You Q and Huang Z: miR-139-5p sensitizes colorectal cancer
cells to 5-fluoro-uracil by targeting NOTCH-1. Pathol Res Pract.
212:643–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He L, Zhu H, Zhou S, Wu T, Wu H, Yang H,
Mao H, SekharKathera C, Janardhan A, Edick AM, et al: Wnt pathway
is involved in 5-FU drug resistance of colorectal cancer cells. .
Exp Mol Med. 50:1012018. View Article : Google Scholar
|
|
27
|
Sagara A, Igarashi K, Otsuka M, Karasawa
T, Gotoh N, Narita M, Kuzumaki N, Narita M and Kato Y: Intrinsic
resistance to 5-fluo-rouracil in a brain metastatic variant of
human breast cancer cell line, MDA-MB-231BR. PLoS One.
11:e01642502016. View Article : Google Scholar
|
|
28
|
Wang X, Wang Y, Gu J, Zhou D, He Z, Wang X
and Ferrone S: ADAM12-L confers acquired 5-Fluorouracil resistance
in breast cancer cells. Sci Rep. 7:96872017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song Y, Lu M, Qiu H, Yin J, Luo K, Zhang
Z, Jia X, Zheng G, Liu H and He Z: Activation of FOXO3a reverses
5-Fluorouracil resistance in human breast cancer cells. Exp Mol
Pathol. 105:57–62. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mathur R, Alver BH, San Roman AK, Wilson
BG, Wang X, Agoston AT, Park PJ, Shivdasani RA and Roberts CW:
ARID1A loss impairs enhancer-mediated gene regulation and drives
colon cancer in mice. Nat Genet. 49:296–302. 2017. View Article : Google Scholar :
|
|
31
|
Sasaki M, Sato Y and Nakanuma Y:
Cholangiolocellular carcinoma with 'Ductal plate malformation'
pattern may be characterized by ARID1A genetic alterations. Am J
Surg Pathol. 43:352–360. 2019. View Article : Google Scholar
|
|
32
|
Wang X, Wang X, Gu J, Zhou M, He Z, Wang X
and Ferrone S: Overexpression of miR-489 enhances efficacy of
5-fluoro-uracil-based treatment in breast cancer stem cells by
targeting XIAP. Oncotarget. 8:113837–113846. 2017. View Article : Google Scholar
|
|
33
|
Hashemi-Moghaddam H, Kazemi-Bagsangani S,
Jamili M and Zavareh S: Evaluation of magnetic nanoparticles coated
by 5-fluorouracil imprinted polymer for controlled drug delivery in
mouse breast cancer model. Int J Pharm. 497:228–238. 2016.
View Article : Google Scholar
|
|
34
|
Gao F, Yu X, Meng R, Wang J and Jia L:
STARD13 is positively correlated with good prognosis and enhances
5-FU sensitivity via suppressing cancer stemness in hepatocellular
carcinoma cells. Onco Targets Ther. 11:5371–5381. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bruner HC and Derksen PW: Loss of
E-cadherin-dependent cell-cell adhesion and the development and
progression of cancer. Cold Spring Harb Perspect Biol.
10:a0293302018. View Article : Google Scholar
|
|
36
|
Ogasawara N, Kudo T, Sato M, Kawasaki Y,
Yonezawa S, Takahashi S, Miyagi Y, Natori Y and Sugiyama A:
Reduction of membrane protein CRIM1 decreases E-cadherin and
increases claudin-1 and MMPs, enhancing the migration and invasion
of renal carcinoma cells. Biol Pharm Bull. 41:604–611. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tanaka T, Goto K and Iino M: Sec8
modulates TGF-β induced EMT by controlling N-cadherin via
regulation of Smad3/4. Cell Signal. 29:115–126. 2017. View Article : Google Scholar
|
|
38
|
Liao S, Yu C, Liu H, Zhang C, Li Y and
Zhong X: Long non-coding RNA H19 promotes the proliferation and
invasion of lung cancer cells and regulates the expression of
E-cadherin, N-cadherin, and vimentin. Onco Targets Ther.
12:4099–4107. 2019. View Article : Google Scholar : PubMed/NCBI
|