Introduction
Osteosarcoma (OS) is a type of malignant bone tumor
which predominantly occurs in adolescents (1,2).
As one of the aggressive types of cancer, OS is associated with a
high risk of mortality. Over the past years, OS has been
effectively treated by complete surgical resection (3). With the deep understanding of OS
progression in recent years, neoadjuvant and adjuvant chemotherapy
have been applied to the treatment of patients with OS (4,5).
Despite great advances being made in OS treatment, the prognosis of
patients with OS remains unsatisfactory due to distant metastasis
(6,7). Therefore, it is essential to further
explore the potential molecular mechanisms underlying OS
progression in order to improve the prognosis of patients with
OS.
Long non-coding RNAs (lncRNAs) are a group of RNAs
with >200 nucleotides in length. Due to lack of a complete open
reading frame, they are limited to encode proteins (8). It has been demonstrated that
lncRNAs, aberrantly expressed in cancers, can regulate various
biological processes (9). For
example, LINC00163 was found at a low level in lung cancer and a
high LINC00163 expression predicted a better prognosis (10). lncRNA CRNDE has been shown to
serve as a tumor promoter and to promote cell proliferation,
migration and invasion in non-small cell lung cancer (NSCLC) via
sponging miR-338-3p (11). LncRNA
XIST was abnormally expressed in esophageal cancer and promoted
cancer development via sponging miR-494 and targeting CDK6
(12). According to previous
studies, lncRNA KCNQ1OT1 was discovered to be an oncogene in
multiple types of cancer, such as colorectal cancer, NSCLC and
colon cancer (13-15). Nevertheless, the functional role
and mechanisms of action of KCNQ1OT1 in OS warrant further
investigation.
Wnt/β-catenin signaling has been reported in various
types of cancer and plays pivotal role in cancer development and
progression. For example, LINC00675 promotes cell proliferation,
migration and invasion via activating Wnt/β-catenin signaling in
cervical cancer (16). LINC01606
has been shown to be associated with Wnt/β-catenin signaling, and
to promoted metastasis and invasion in gastric cancer (17). However, research on the
association between KCNQ1OT1 and Wnt/β-catenin signaling has not
been conducted in OS to date, at least to the best of our
knowledge.
The present study focused on investigating the
function of KCNQ1OT1 in OS. The results validated that KCNQ1OT1
predicted a poor prognosis, facilitated OS cell proliferative,
migratory and invasive abilities, and activated Wnt/β-catenin
signaling by targeting the miR-3666/KLF7 axis, which suggests that
KCNQ1OT1 may serve as a prognostic biomarker in OS.
Materials and methods
Tissue samples
A total of 40 pairs of OS tissues and matched normal
adjacent tissues were collected from the Eighth Affiliated
Hospital, Sun Yat-Sen University from November, 2013 to December,
2018. All patients provided written informed consent and the
approval of this study was obtained from the Ethics Committee of
the Eighth Affiliated Hospital, Sun Yat-Sen University.
Cells and cell culture
Three human OS cell lines (U2OS, SaoS-2 and MG-63)
and an osteoblast cell line (hFOB) were purchased from Cell Bank of
the Chinese Academy of Science. The cell lines were cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) in a humidified 5%
CO2 atmosphere at 37°C supplemented with 10% fetal
bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.), 100
µg/ml penicillin and 100 µg/ml streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.).
Cell transfection
For transfection, short hairpin RNA (shRNA; 50 nM)
plasmids directly targeting KCNQ1OT1 (sh-KCNQ1OT1#1/2/3), the
negative control (sh-NC; 50 nM), miR-3666 mimics (30 nM) and the
negative control (NC mimics; 30 nM) were all respectively
synthesized by GenePharma. The sequences were as follows: 5′-CTT
CAA CCC TTA GGT ACA ACA CCA AAA C-3′ (sh-KCNQ1OT1#1), 5′-ACC AAG
ACT CAG TCC CGG GCT TAA TCC T-3′ (sh-KCN Q1OT1#2), 5′-TCC TAG CCC
TCA GAC TCA ACC CCT GG AC-3′ (sh-KCNQ1OT1#3), 5′-CAA GCT TAA CAG
AGA GAC CAA AAG AAC A-3′ (sh-NC), 5′-CAG TGC AAG TGT AGA TGC CGA
GTC ACG TTC ACA TCT ACG GCT-3′ (miR-3666 mimics), 5′-AGA CAA GGA
CAG ATC GAA AAG TCT GTT CCT GTC TAG CTT TTC-3′ (NC mimics). For
KLF7 overexpression, the full-length of KLF7 cDNA was amplified and
inserted into pcDNA3.1. An empty vector pcDNA3.1 (30 nM,
Invitrogen; Thermo Fisher Scientific, Inc.) was served as a
negative control. Lipofectamine 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was applied to conduct all transfections
for 48 h and cells were collected for subsequent experiments at 3-4
passages.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from tumor tissues and
transfected cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Total RNA was transcribed into cDNA using the
cDNA Reverse Transcription kit (Applied Biological Materials Inc.).
The miR-3666 level was examined using the TaqMan MicroRNA Assays
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was
performed using the 7300 Real Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The PCR primers were as
followings: GAPDH forward, 5′-GGGAGAAGCTGAGTCATGGG-3′ and reverse,
5′-TCC CGG TGA CAT TTA CAG CC-3′; U6 forward, 5′-CGC GAT ATG GTT
TTG GCA GG-3′ and reverse, 5′-TGG ACG TAT TCG ATC AGC CG-3′;
miR-3666 forward, 5′-TGG GAT GGA TTG CAG GTT GAA-3′ and reverse,
5′-GCA GAG GCT TCT TCC TCA TGT-3′; KCNQ1OT1 forward, 5′-GGG TCT AGG
GTC CAC ATC CT-3′ and reverse, 5′-AGA CTC CCG ATC CTC TGT CC-3′;
KLF7 forward, 5′-TGT AGG CAG AAC AAG CGG G-3′ and reverse, 5′-ATG
TGG CCA CTT GTG AGA GC-3′. The reaction conditions comprised of a
pre-denaturation at 95°C for 10 min, with a total of 40 cycles of
denaturation at 95°C for 15 sec, annealing at an appropriate
annealing temperature for 1 min, and extension at 72°C for 30 sec.
All values were normalized to GAPDH or U6, and all data were
calculated using the 2−ΔΔCq method (18).
Cell Counting kit-8 (CCK8) assay
Cells were plated in a 96-well plate at a density of
2×104 cells/well and 10 µl of CCK-8 reagent was
added to each well daily. Following incubation for 2 h at 37°C,
microplate reader (Bio-Rad Laboratories, Inc.) was employed to
examine the absorbance value of each well at a wavelength of 450
nm.
Colony formation assay
Cells (500 per well) transfected with sh-KCNQ1OT1
were seeded in a 6-well plate. Following incubation at 37°C in 5%
CO2, cell colonies were fixed with 4% paraformaldehyde
for 10 min (Sigma Aldrich) and then stained with 0.1% crystal
violet for 5 min at room temperature (Sigma-Aldrich; Merck KGaA). A
microscope was then used to count colonies (Leica MZ8, Leica
Microsystems GmbH).
Transwell assay
The assessment of cell migration or invasion was
conducted by the use of Transwell chambers (Corning Costar, Inc.)
without or with Matrigel. A total of 2×104 cells were
starved in serum-free DMEM medium and then placed into the upper
chamber. DMEM medium with 20% serum was placed into lower chamber.
Following incubation for 48 h at 37°C, the migrated or invaded
cells in the lower chamber were fixed with 4% paraformaldehyde
(Sigma Aldrich; Merck KGaA) for 15 min and stained with 0.5%
crystal violet for 15 min at room temperature (C0121, Beyotime
Institute of Biotechnology), respectively. Finally, the cells were
counted under a light microscope (×200 magnification, Olympus
Corporation).
Xenograft tumor model
A total of 6 male 4-week-old BALB/c nude mice were
available from the Eighth Affiliated Hospital, Sun Yat-Sen
University and housed in the animal laboratory with laminar flow
equipment under special pathogen-free (SPF) conditions and 12 h
light-dark cycles. The temperature was set at 18-22°C and the
relative humidity was 50-60%, and the animals had free access to
autoclaved food and water during the study. The animal-related
protocol was approved by the Animal Care and Experiment Committee
of the Eighth Affiliated Hospital, Sun Yat-Sen University. U2OS
cells transfected with sh-KCNQ1OT1#1 or sh-NC were injected
subcutaneously into the mice (n=3 per group) for 28-day inoculation
purposes, with the health of the mice and tumor growth monitored
every 4 days. When mice exhibited tumor metastasis, lethargy,
weight loss in excess of 20% body weight or other signs of distress
consistent with the IACUC standards, they were sacrificed by
cervical dislocation. Mice were verified dead by observing
respiration, heartbeat, pupils and nerve reflex. Tumors were then
excised carefully from the mice and weighed for analysis. Tumor
volume was estimated using digital calipers and calculated using
the following the formula: 0.5 × length × width2.
Luciferase reporter assay
The KCNQ1OT1-WT/KLF7-WT vector was constructed by
cloning KCNQ1OT1/3′untranslated region (3′-UTR) of KLF7 covering
the miR-3666 binding site into the pmirGLO reporter vector (Promega
Corp.). The vector KCNQ1OT1-Mut/KLF7-Mut was inserted by the mutant
KCNQ1OT1/KLF7. miR-3666 mimics or NC mimics were then respectively
co-transfected with KCNQ1OT1-WT/KLF7-WT or KCNQ1OT1-Mut/KLF7-Mut.
All transfections were performed with the use of Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h later, the
Dual Luciferase Reporter Assay System (Promega Corp.) was used to
measure the luciferase activity and normalized to the activity of
the Renilla luciferase gene.
Nuclear-cytoplasmic fractionation
The nuclear and cytoplasmic fractions were separated
with a PARIS kit (Invitrogen; Thermo Fisher Scientific, Inc.).
Briefly, cells were re-suspended in cell fraction buffer and
incubated on ice for 10 min. The cytoplasmic fraction and nuclear
pellet were collected following centrifugation at 500 × g for 5 min
at 4°C. Subsequently, KCNQ1OT1 expression was evaluated by RT-qPCR
with GAPDH as cytoplasmic control and U6 as nuclear control.
Bioinformatics analysis
The potential bindings between miRNAs and KCNQ1OT1
were predicted by starBase (http://star-base.sysu.edu.cn/). Online tools of RNA22
(https://cm.jefferson.edu/rna22/Interactive/), PicTar
(https://pictar.mdc-berlin.de/),
TargetScan (http://www.targetscan.org/vert_72/) and microT
(http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=
microT_CDS/index), were used to predict the target gene of
miR-3666.
Western blot analysis
RIPA lysis buffer was applied to lyse the cells and
thereby the proteins were collected. Protein concentrations were
measured using the BCA Protein Assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Proteins were loaded on 10% SDS-PAGE with 20
µg protein per lane and transferred onto PVDF membranes.
Subsequently, 5% skimmed milk powder was employed to block the PVDF
membranes for 2 h, and the membranes were incubated with primary
antibodies to β-catenin (1:8,000, ab32572, Abcam), c-Myc (1:1,000,
ab166837, Abcam), matrix metalloproteinase (MMP)7 (1:1,000,
ab207299, Abcam) and GAPDH (1:2,000, ab8245, Abcam) overnight at
4°C. The membranes were then incubated with goat anti-mouse IgG
H&L conjugated to HRP (1:8,000; ab97040, Abcam) for 2 h at room
temperature. Finally, the immunofluorescence of protein bands was
visualized with ECL detection system (Pierce; Thermo Fisher
Scientific, Inc.). The Bio-Rad image analysis system (Bio-Rad
Laboratories, Inc.) was used to obtain images, and Quantity One
version 4.6.2 software (Bio-Rad Laboratories, Inc.) was used for
analysis. The relative protein level was expressed by the gray
value of the corresponding protein band value of the GAPDH protein
band.
RIP assay
RIP assay was carried out using the EZ Magna RNA
immunoprecipitation kit (EMD Millipore). Briefly, RIP lysis buffer
was used to lyse the OS cells. The cell lysates were then
cultivated with magnetic beads conjugated with specific antibodies
to human Ago2 (1:2,000, ab186733, Abcam) or IgG (1:2,000, ab190475,
Abcam) at 4°C for 3 h until overnight. Finally, the
immunoprecipitates were purified and subjected to RT-qPCR.
Statistical analysis
Statistical analyses were conducted with SPSS 21.0
statistical software (SPSS Inc.). Data are expressed as the means ±
SD and each experiment was conducted in triplicate. The differences
among groups were detected by one-way or two-way ANOVA followed by
Tukey's post-hoc test or the Student's t-test. Pearson's
correlation analysis was conducted to determine the correlation
between the expression of miR-3666 and KCNQ1OT1 or KLF7 in OS
tissues. An overall survival curve was drawn to determine survival
analysis using the Kaplan-Meier method and log-rank test. Data were
also analyzed by Pearson's χ2 test. A P-value <0.05
was considered to indicate a statistically significant
difference.
Results
Overexpression of KCNQ1OT1 accelerates OS
progression and activates Wnt/β-catenin signaling
To determine the expression pattern of KCNQ1OT1 in
OS, KCNQ1OT1 expression was detected in OS tissues and adjacent
non-tumor tissues. The results of RT-qPCR revealed that KCNQ1OT1
exhibited a higher level in OS tissues than in adjacent normal
tissues. Consistently, a high KCNQ1OT1 expression was found in OS
cell lines, particularly in the U2OS and SaoS-2 cells (Fig. 1A). Through Kaplan-Meier survival
analysis, it was concluded that patients with OS with a high
KCNQ1OT1 expression exhibited a relatively low overall survival
compared to those with a low KCNQ1OT1 expression (Fig. S1). In addition, it was found that
KCNQ1OT1 expression was not associated with age or sex, but was
associated with tumor size and TNM stage (Table I). These results indicated the
potential oncogenic role of KCNQ1OT1 in OS. T
 | Table IAssociation between KCNQ1OT1
expression and clinical features of patients with OS (n=40). |
Table I
Association between KCNQ1OT1
expression and clinical features of patients with OS (n=40).
| Variable | KCNQ1OT1 expression
| P-value |
|---|
| Low | High |
|---|
| Age, years | | | |
| <60 | 12 | 14 | 0.7410 |
| ≥60 | 8 | 6 | |
| Sex | | | |
| Male | 13 | 11 | 0.7475 |
| Female | 7 | 9 | |
| Tumor size, cm | | | |
| ≤5 | 15 | 3 | 0.0003 |
| >5 | 5 | 17 | |
| TNM stage | | | |
| I-II | 12 | 3 | 0.0079 |
| III-IV | 8 | 17 | |
To further explore biological function of KCNQ1OT1
in OS, a loss-of-function assay was conducted. Firstly, the
expression of KCNQ1OT1 was stably silenced in the U2OS and SaoS-2
cells by transfection with sh-KCNQ1OT1#1/2/3 (Fig. 1B). Owing to the optimal
transfection efficiency, sh-KCNQ1OT1#1 and sh-KCNQ1OT1#2 were
selected for use in subsequent experiments. Through CCK-8 and
colony formation assays, it was observed that the deficiency of
KCNQ1OT1 overtly suppressed the proliferation and colony formation
ability of the U2OS and SaoS-2 cells (Fig. 1C and D). Transwell assay exhibited
that OS cell migration and invasion were considerably suppressed
upon KCNQ1OT1 silencing (Fig. 1E and
F). Of note, KCNQ1OT1 was previously reported to promote
osteogenic differentiation by activating Wnt/β-catenin signaling
(19). Therefore, the present
study investigated whether KCNQ1OT1 exerts an effect on
Wnt/β-catenin signaling in OS. Through western blot analysis, it
was found that the protein levels of Wnt/β-catenin signaling
downstream genes (β-catenin, MMP7 and c-Myc) were significantly
decreased in the sh-KCNQ1OT1#1/2-trans-fected OS cells (Fig. 1G), which suggested that KCNQ1OT1
activated Wnt/β-catenin signaling in OS. Subsequently, the role of
KCNQ1OT1 in tumor growth was further investigated by using an in
vivo assay. In this experiment, each nude mouse was burdened
with one tumor, and the size of the tumors in the sh-KCNQ1OT1#1
group was smaller than that of those in the sh-NC group (Fig. 1H). Moreover, it was revealed that
sh-KCNQ1OT1#1 transfection suppressed tumor growth and volume
(Fig. 1I and J). The maximum
diameter of tumors obtained was 17 mm and the maximum volume was
1,224 mm3. In addition, the weight of tumors was found
to be decreased in the sh-KCNQ1OT1#1 group compared with the sh-NC
group (Fig. 1K). All these data
indicated that KCNQ1OT1 was overexpressed, and promoted cellular
progression and activated Wnt/β-catenin signaling in OS.
KCNQ1OT1 serves as a molecular sponge of
miR-3666
Subsequently, the underlying mechanisms of KCNQ1OT1
in OS were investigated. It has been widely reported that lncRNAs
can function competing endogenous RNAs (ceRNAs) by competitively
interacting with miRNAs to release their target mRNAs in the
cytoplasm (20,21). Thus, the present study assessed
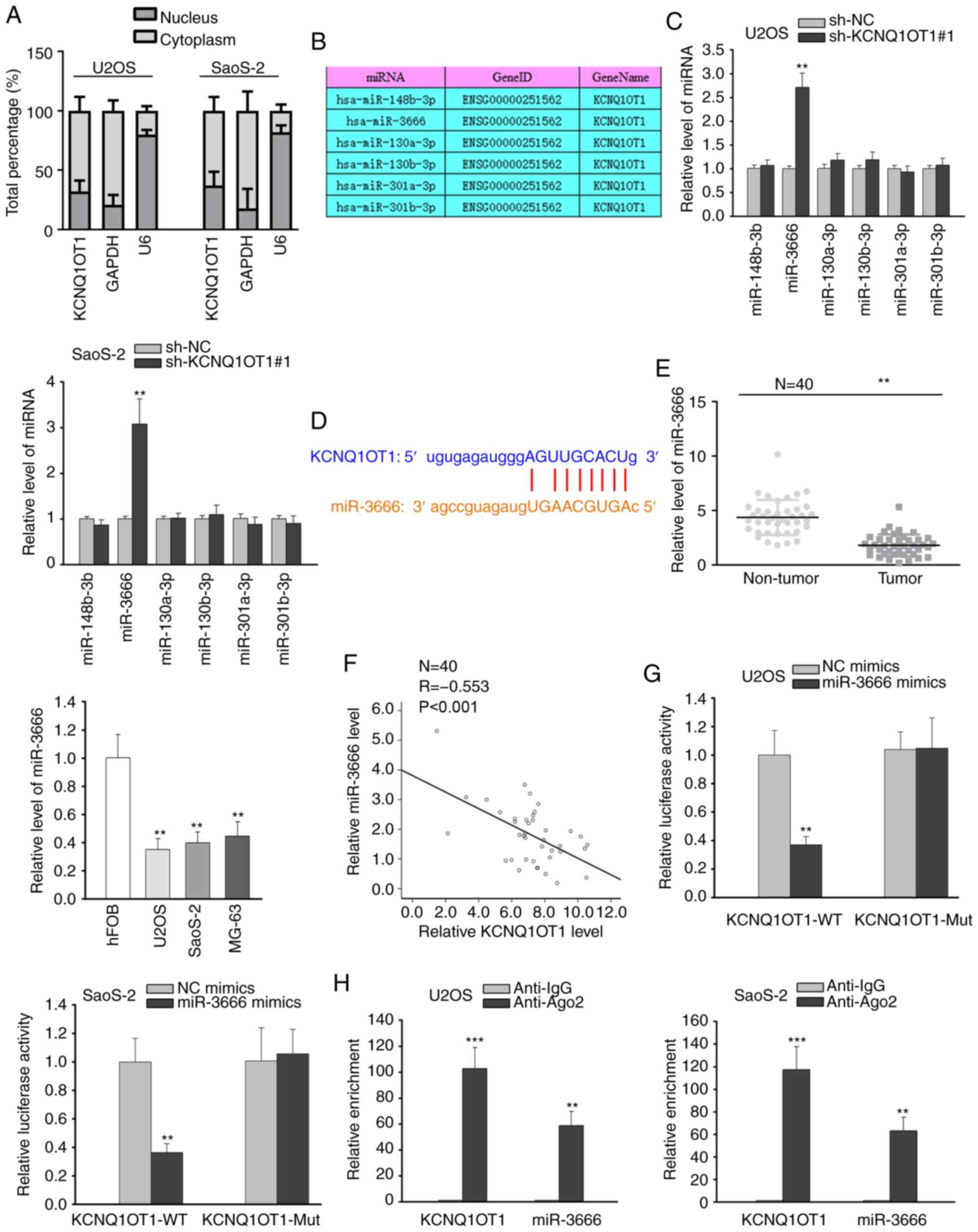
the distribution of KCNQ1OT1 in OS cells via nuclear-cytoplasmic
fractionation assay. The results revealed that KCNQ1OT1 was mainly
distributed in the cytoplasm of OS cells (Fig. 2A), indicating the ceRNA
hypothesis. Using starBase (http://starbase.sysu.edu.cn/), several miRNAs
possessing binding sites for KCNQ1OT1 were predicted (Fig. 2B). RT-qPCR analysis revealed that
KCNQ1OT1 silencing markedly increased the expression of miR-3666,
whereas it did not affect the expression of other miRNAs (Fig. 2C). Thus, miR-3666 was selected for
analysis. The binding site between miR-3666 and KCNQ1OT1 is
illustrated in Fig. 2D.
Subsequently, miR-3666 expression in OS tissues and cells was
evaluated, and the results demonstrated that miR-3666 expression
was downregulated in both OS tissues and cell lines (Fig. 2E). Pearson's correlation analysis
further validated the negative correlation between the expression
of KCNQ1OT1 and miR-3666 in OS tissues (Fig. 2F). Luciferase reporter assay
revealed that the overexpression of miR-3666 caused an overt
decrease in the luciferase activity of the KCNQ1OT1-WT reporter,
while that of the KCNQ1OT1-Mut reporter remained unaltered
(Fig. 2G). The results from RIP
assay also revealed the abundant enrichment of KCNQ1OT1 and
miR-3666 in the anti-Ago2 group compared with anti-IgG group
(Fig. 2H). From the
above-mentioned findings, it was thus concluded that KCNQ1OT1 can
sponge miR-3666 in OS cells.
KLF7 is a downstream target of
miR-3666
Subsequently, the downstream target of miR-3666 was
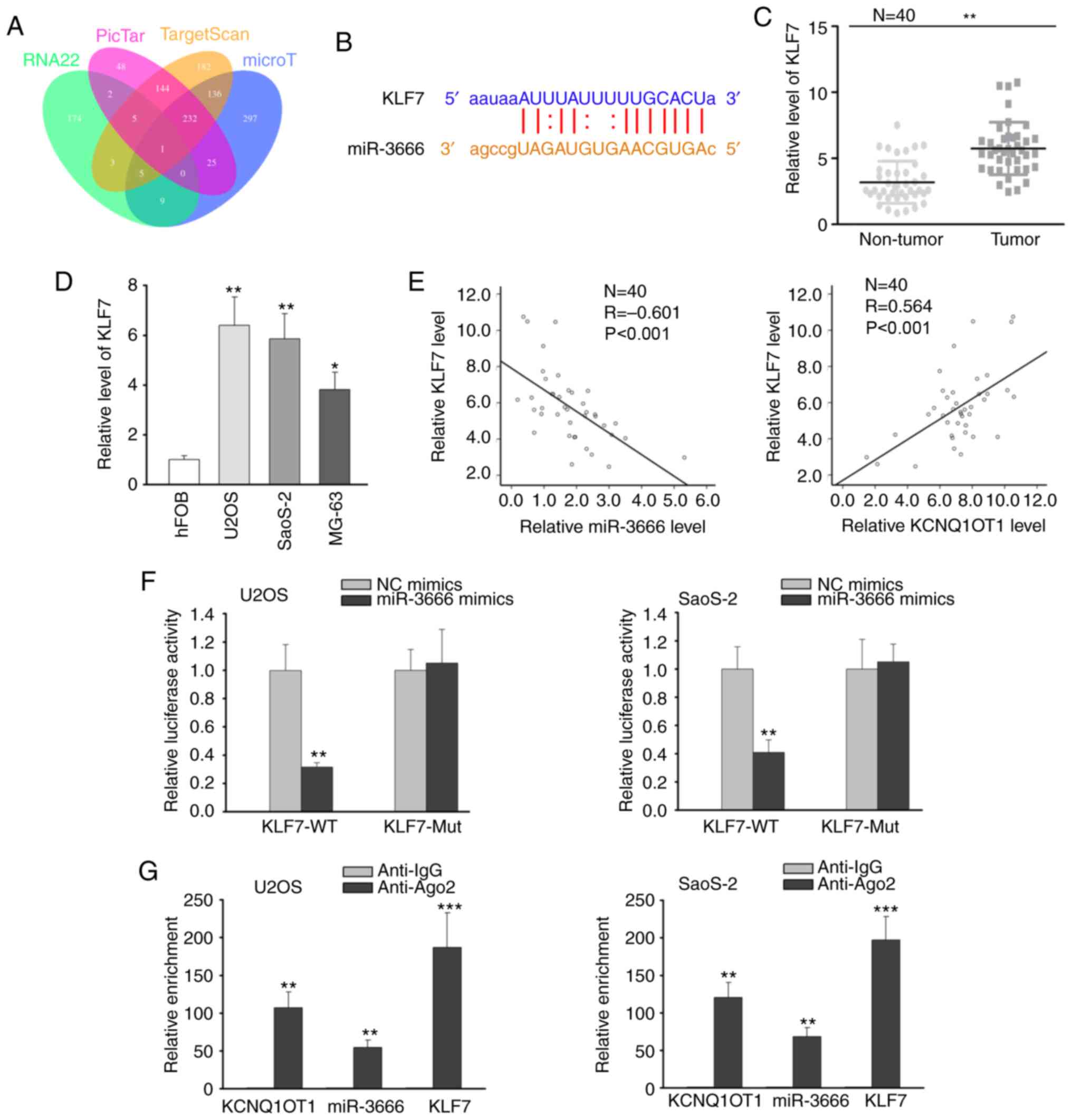
further investigated. Combining the online tools of RNA22, PicTar,
TargetScan and microT, KLF7 was predicted to be the target gene of
miR-3666 (Fig. 3A). As shown in
Fig. 3B, miR-3666 contained a
binding site on KLF7 3′UTR. Thereby, the expression of KLF7 in OS
tissues and cells was analyzed by RT-qPCR, and the results revealed
the high expression of KLF7 in OS tissues and cells (Fig. 3C and D). Furthermore, the
expression of KLF7 was found to negatively correlate with miR-3666
expression, and to positively correlate with KCNQ1OT1 expression by
Pearson's correlation analysis (Fig.
3E). Luciferase reporter assay disclosed that miR-3666
overexpression reduced the luciferase activity of the KLF7-WT
reporter, but exhibited no effect on the KLF7-Mut reporter
(Fig. 3F). On the basis of RIP
assay, the anti-Ago2 group rather than the anti-IgG group exhibited
a notable enrichment of KCNQ1OT1, miR-3666 and KLF7 (Fig. 3G). Taken together, these findings
indicate that miR-3666 can bind to KLF7 in OS cells.
KCNQ1OT1 promotes OS progression and
activates Wnt/β-catenin signaling by targeting the miR-3666/KLF7
axis
Finally, the effect of the KCNQ1OT1/miR-3666/KLF7
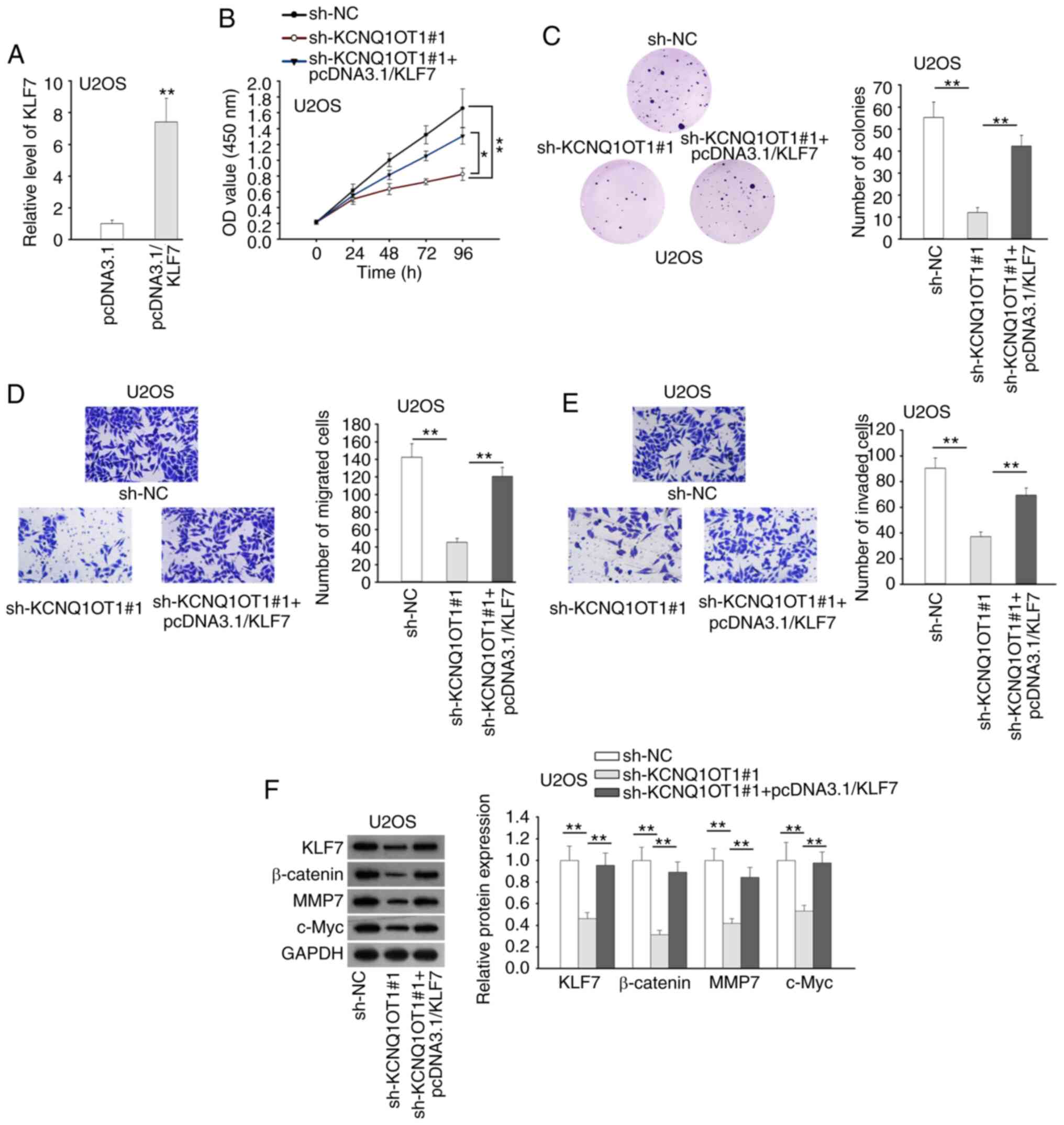
axis on OS progression was verified. KLF7 was firstly overexpressed
in U2OS cells, and the expression of KLF7 was markedly increased by
pcDNA3.1/KLF7 transfection (Fig.
4A). CCK-8 and colony formation assays revealed that the
inhibitory effects of KCNQ1OT1 suppression or miR-3666
overexpression on OS cell proliferation were reversed by the
upregulation of KLF7 (Figs. 4B and
C and S2A and B). Based on
Transwell assay, KLF7 overexpression recovered the impaired
migration and invasion induced by the silencing of KCNQ1OT1 or
overexpression of miR-3666 (Figs. 4D
and E and S2C and D).
Western blot analysis revealed that the upregulation of KLF4
counteracted the suppressive effects of KCNQ1OT1 silencing on
Wnt/β-catenin signaling in U2OS cells (Figs. 4F and S3). The above-mentioned data indicated
that KCNQ1OT1 promoted cell proliferation, migration, invasion and
Wnt/β-catenin signaling in OS by targeting the miR-3666/KLF7
axis.
Discussion
In recent years, mounting evidence has indicated
that lncRNAs play a role in the development of OS. For instance,
lncRNA B4GALT1-AS1 has been shown to be upregulated in OS, and to
promote OS cell stemness and migration (22). lncRNA SNHG4 has also been shown to
facilitate tumor growth by sponging miR-224-3p and a high SNHG4
expression is associated with poor prognosis in OS (23). In the present study, KCNQ1OT1 was
discovered to be highly expressed in OS tissues and cell lines.
Patients with OS with a high KCNQ1OT1 expression exhibited a worse
prognosis than those with a low KCNQ1OT1 expression. Moreover, the
inhibition of KCNQ1OT1 suppressed cell proliferation, migration and
invasion in OS. More importantly, KCNQ1OT1 silencing also
inactivated Wnt/β-catenin signaling. The results of the in
vivo assay indicated that KCNQ1OT1 knockdown suppressed tumor
growth in OS. All these data suggested that KCNQ1OT1 served as a
tumor promoter in OS.
miRNAs belong to the cluster of non-coding RNAs
which are 21-23 nucleotides in length and have been reported to
play a key role in cancer progression (24,25). It has been validated that lncRNAs
can serve as ceRNAs to increase target gene expression by
competitively combining with miRNAs (26). The ceRNA network has been reported
in diverse cancers, such as gastric cancer (27), bladder cancer (28), colorectal cancer (29) and OS (30). In the present study, miR-3666 was
predicted to be a downstream miRNA of KCNQ1OT1. Previously,
miR-3666 was proposed as a tumor suppressor in several types of
cancer. miR-3666 was shown to suppress cell growth in lung cancer
by targeting BPTF (31). miR-3666
was also shown to suppress the expression of SIRT7 to impair the
growth of NSCLC cells (32). In
the present study, miR-3666 expression was found to be markedly
downregulated in OS tissues and cell lines. Moreover, there was a
negative correlation between the expression of KCNQ1OT1 and that of
miR-3666 in OS tissues. Furthermore, KCNQ1OT1 was corroborated to
interact with miR-3666. Taken together, these findings indicate
that KCNQ1OT1 serves as a sponge of miR-3666 in OS.
KLF7 has been widely reported in cancers and plays a
critical role in cancer progression. For example, miR-185 has been
shown to block cell proliferation and invasion in NSCLC cells by
targeting KLF7 (33). LINC00668
facilitates cell motility in NSCLC cells by sponging miR-193a and
increases the expression of KLF7 (34). In the present study, KLF7
expression was conspicuously upregulated in OS tissues and cell
lines. Moreover, it was verified that KLF7 bound to miR-3666 in OS
cells. Notably, the overexpression of KLF7 abolished the miR-3666
amplification-mediated suppression of OS cell proliferation,
migration and invasion. Rescue assays manifested that the
suppressed OS progression and Wnt/β-catenin signaling induced by
KCNQ1OT1 silencing were abrogated by KLF7 overexpression.
In conclusion, the present study demonstrates that
KCNQ1OT1 predict an unfavorable prognosis, accelerates tumor growth
and activates Wnt/β-catenin signaling in OS by sponging miR-3666
and targeting KLF7, suggesting that KCNQ1OT1 is a promising
prognostic biomarker in OS. The findings of this study may be aid
in the investigation of potential treatment strategies for the
patients with OS.
Supplementary Data
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
AH, SJ and WH were involved in the study design,
data curation, formal analysis and methodology. AH, YW, SM, ZW, KL
and QZ were involved in data curation and investigation. AH, JZ, ZL
and LC were responsible for supervising and tracking the progress
of the experiments, and assisting in the smooth progress of the
research. JZ, ZL and LC were also involved in the search for
references required for the study. All authors agree that this is
the final submission.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Eighth Affiliated Hospital, Sun Yat-Sen
University. Written informed consent was obtained from all patients
for the use of human clinical tissues in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors acknowledge the support provided by the
members of the laboratory of the Eighth Affiliated Hospital, Sun
Yat-Sen University.
References
|
1
|
Smolle MA and Pichler M: The role of long
non-coding RNAs in osteosarcoma. Noncoding RNA. 4:72018.
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meyers PA, Schwartz CL, Krailo M,
Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W,
Gebhardt M, Goorin AM, et al: Osteosarcoma: A randomized,
prospective trial of the addition of ifosfamide and/or muramyl
tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J
Clin Oncol. 23:2004–2011. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bishop MW, Janeway KA and Gorlick R:
Future directions in the treatment of osteosarcoma. Curr Opin
Pediatr. 28:26–33. 2016. View Article : Google Scholar :
|
|
5
|
Biazzo A and De Paolis M:
Multidisciplinary approach to osteosarcoma. Acta Orthop Belg.
82:690–698. 2016.
|
|
6
|
Reed DR, Hayashi M, Wagner L, Binitie O,
Steppan DA, Brohl AS, Shinohara ET, Bridge JA, Loeb DM, Borinstein
SC and Isakoff MS: Treatment pathway of bone sarcoma in children,
adolescents, and young adults. Cancer. 123:2206–2218. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dean DC, Shen S, Hornicek FJ and Duan Z:
From genomics to metabolomics: Emerging metastatic biomarkers in
osteosarcoma. Cancer Metastasis Rev. 37:719–731. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo X, Wei Y, Wang Z, Liu W, Yang Y, Yu X
and He J: LncRNA LINC00163 upregulation suppresses lung cancer
development though transcriptionally increasing TCF21 expression.
Am J Cancer Res. 8:2494–2506. 2018.
|
|
11
|
Jing H, Xia H, Qian M and Lv X: Long
noncoding RNA CRNDE promotes non-small cell lung cancer progression
via sponging microRNA-338-3p. Biomed Pharmacother. 110:825–833.
2019. View Article : Google Scholar
|
|
12
|
Chen Z, Hu X, Wu Y, Cong L, He X, Lu J,
Feng J and Liu D: Long non-coding RNA XIST promotes the development
of esophageal cancer by sponging miR-494 to regulate CDK6
expression. Biomed Pharmacother. 109:2228–2236. 2019. View Article : Google Scholar
|
|
13
|
Bian Y, Gao G, Zhang Q, Qian H, Yu L, Yao
N, Qian J, Liu B and Qian X: KCNQ1OT1/miR-217/ZEB1 feedback loop
facili-tates cell migration and epithelial-mesenchymal transition
in colorectal cancer. Cancer Biol Ther. 20:886–896. 2019.
View Article : Google Scholar :
|
|
14
|
Dong Z, Yang P, Qiu X, Liang S, Guan B,
Yang H, Li F, Sun L, Liu H, Zou G and Zhao K: KCNQ1OT1 facilitates
progression of non-small-cell lung carcinoma via modulating
miRNA-27b-3p/HSP90AA1 axis. J Cell Physiol. 234:11304–11314. 2019.
View Article : Google Scholar
|
|
15
|
Li Y, Li C, Li D, Yang L, Jin J and Zhang
B: lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in
colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets
Ther. 12:2649–2660. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei R, Ding C, Rodriguez RA and Del Mar
Requena Mullor M: The SOX2OT/miR-194-5p axis regulates cell
proliferation and mobility of gastric cancer through suppressing
epithelial-mesenchymal transition. Oncol Lett. 16:6361–6368.
2018.PubMed/NCBI
|
|
17
|
Luo Y, Tan W, Jia W, Liu Z, Ye P, Fu Z, Lu
F, Xiang W, Tang L, Yao L, et al: The long non-coding RNA LINC01606
contributes to the metastasis and invasion of human gastric cancer
and is associated with Wnt/β-catenin signaling. Int J Biochem Cell
Biol. 103:125–134. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Gao X, Ge J, Li W, Zhou W and Xu L: LncRNA
KCNQ1OT1 promotes osteogenic differentiation to relieve osteolysis
via Wnt/β-catenin activation. Cell Biosci. 8:192018. View Article : Google Scholar
|
|
20
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Z, Wang Y, Hu R, Xu R and Xu W: LncRNA
B4GALT1-AS1 recruits HuR to promote osteosarcoma cells stemness and
migration via enhancing YAP transcriptional activity. Cell Prolif.
51:e125042018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu R, Feng F, Yu X, Liu Z and Lao L:
LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and
predicts poor survival and recurrence in human osteosarcoma. Cell
Prolif. 51:e125152018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Srivastava SK, Bhardwaj A, Leavesley SJ,
Grizzle WE, Singh S and Singh AP: MicroRNAs as potential clinical
biomarkers: Emerging approaches for their detection. Biotech
Histochem. 88:373–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J,
Tang Z, Liao QX, Zhang H, Zeng LS and Cui SZ: LINC01133 as ceRNA
inhibits gastric cancer progression by sponging miR-106a-3p to
regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer.
17:1262018. View Article : Google Scholar
|
|
28
|
Zhan Y, Chen Z, Li Y, He A, He S, Gong Y,
Li X and Zhou L: Long non-coding RNA DANCR promotes malignant
phenotypes of bladder cancer cells by modulating the miR-149/MSI2
axis as a ceRNA. J Exp Clin Cancer Res. 37:2732018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu B, Pan S, Xiao Y, Liu Q, Xu J and Jia
L: LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer
progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway.
J Exp Clin Cancer Res. 37:3162018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fei D, Zhang X, Liu J, Tan L, Xing J, Zhao
D and Zhang Y: Long noncoding RNA FER1L4 suppresses tumorigenesis
by regulating the expression of PTEN targeting miR-18a-5p in
osteosarcoma. Cell Physiol Biochem. 51:1364–1375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pan L, Tang Z, Pan L and Tang R:
MicroRNA-3666 inhibits lung cancer cell proliferation, migration,
and invasiveness by targeting BPTF. Biochem Cell Biol. 97:415–422.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi H, Ji Y, Zhang D, Liu Y and Fang P:
MicroRNA-3666-induced suppression of SIRT7 inhibits the growth of
non-small cell lung cancer cells. Oncol Rep. 36:3051–3057. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao L, Zhang Y, Liu J, Yin W, Jin D, Wang
D and Zhang W: MiR-185 inhibits cell proliferation and invasion of
non-small cell lung cancer by targeting KLF7. Oncol Res.
27:1015–1023. 2019. View Article : Google Scholar
|
|
34
|
An YX, Shang YJ, Xu ZW, Zhang QC, Wang Z,
Xuan WX and Zhang XJ: STAT3-induced long noncoding RNA LINC00668
promotes migration and invasion of non-small cell lung cancer via
the miR-193a/KLF7 axis. Biomed Pharmacother. 116:1090232019.
View Article : Google Scholar : PubMed/NCBI
|