Introduction
Idiopathic pulmonary fibrosis (IPF) is a slow
progressive lung interstitial disease, with an unknown cause. The
risk factors for IPF include age, sex, exposure to metal and wood
chips, and a history of smoking (1). The histopathological manifestation
of IPF is a common type of interstitial pneumonia, characterized by
the remodeling of lung tissue, including the excessive
proliferation of fibroblasts and the deposition of large amounts of
collagen (2). IPF is also
considered to be an age-related disease. The age of the patients is
generally >60 years and the majority are male. In the absence of
lung transplantation, the median survival time of patients is 3-5
years from the date of diagnosis (3). At present, the specific
pathogenesis of IPF remains unclear; therefore, IPF is difficult to
diagnose and the identification of novel methods for the diagnosis
and treatment of this respiratory disease is a hot research
topic.
With the rapid development of whole-genome
next-generation sequencing, long non-coding RNAs (lncRNAs) have
been found to have a variety of biological functions, based on
their conserved secondary or tertiary structure (4). At present, lncRNAs are defined as a
large number of different types of transcriptional RNA molecules
that lack an open reading frame and are >200 nucleotides in
length (5). The expression level
of DLEU2 has been found to be increased in non-small cell lung
cancer (NSCLC) tissues and is associated with a poor survival and
tumor recurrence, while the downregulation of DLEU2 can partially
reverse tumorigenesis (6). The
DLEU2 expression level has been shown to be elevated in
osteosarcoma tissues and cells, and the downregulation of DLEU2
suppresses cell viability and migration (7). In cervical cancer tissues, DLEU2
expression has been found to be increased; the silencing of DLEU2
has been shown to block cell proliferation and cell cycle
progression, increased the rate of apoptosis in cervical cancer
cells in vitro, and inhibit tumor development in vivo
(8).
However, to the best of our knowledge, the role of
DLEU2 in IPF has not been reported to date. In the present study,
the Genome Expression Omnibus (GEO) database was used to download 3
IPF-related datasets (GSE10667, GSE24206 and GSE32537), and it was
observed that the DLEU2 expression level was increased in IPF lung
tissues.
The microRNAs (miRNAs/miRs) that were found to
interact with DLEU2 were predicted using the starBase data-base,
and the mRNAs that could bind to the aforementioned miRNAs were
identified using the TargetScan, miRTarBase and miRDB online tools.
The identified mRNAs and the upregulated mRNAs in the GSE10667,
GSE24206 and GSE32537 datasets were intersected to obtain the
target genes, tripartite motif containing 2 (TRIM2) and
prostaglandin F2 receptor inhibitor (PTGFRN). Following the
preliminary experiment, the effect of DLEU2 downregulation on the
TRIM2 mRNA expression level was more prominent compared with that
on PTGFRN mRNA expression. A previous study demonstrated that TRIM2
was predicted to regulate the ubiquitination of vimentin in lung
squamous cell carcinoma cell lines (9). Furthermore, TRIM2 has been shown to
promote the growth, invasion and metastasis of colorectal cancer
cells through epithelial-mesenchymal transition (EMT) (10). In addition, TRIM2 has been found
to promote the occurrence and metastasis of osteosarcoma through
the PI3K/KPB signaling pathway (11). Both DLEU2 and TRIM2 interact with
miR-369-3p. However, the mechanisms through which DLEU2 regulates
IPF through the miR-369-3p/DLEU2 pathway remain unknown and are
worthy of investigation.
Therefore, the aim of the present study was to
investigate the role of the DLEU2/miR-369-3p/TRIM2 axis in IPF.
Materials and methods
Data extraction
The microRNAs (miRNAs/miRs) that were found to
interact with DLEU2 were predicted using the starBase database
(http://starbase.sysu.edu.cn/index.php), and the mRNAs
that could bind to the aforementioned miRNAs were identified using
the TargetScan (http://www.targetscan.org/vert_72/), miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw/index.html) and
miRDB (http://www.mirdb.org/) online tools. The
raw microarray data were retrieved from the GEO database using the
accession numbers GSE10667, GSE24206 and GSE32537.
Xenograft model of pulmonary
fibrosis
A total of 40 male C57BL/6 mice (6-8 weeks old) were
provided from the Beijing Vital River Laboratory Animal Technology
Co., Ltd. Mice were housed in an environmentally controlled room
(22±2°C; 12 h light-12 h dark cycle; 50-60% humidity) and free
access to food and water. A mouse model of bleomycin (BLM)-induced
pulmonary fibrosis was established as described in a previous study
(12). Each group included 10
mice. Mice in the BLM group were stimulated by an intratracheal
instillation of BLM (1.5 U/kg body weight), while mice in the
control group were injected with an equal volume of physiological
saline. In the BLM + short hairpin (sh)RNA-negative control (NC)
and the BLM + shRNA-DLEU2 groups, the mice were injected
intratracheally with a lentivirus suspension of shRNA-NC or
shRNA-DLEU2 (3×107 transducing units/mouse),
respectively, 3 days following the administration of BLM.
Furthermore, mice in the BLM group were treated by injecting the
same volume of physiological saline into the trachea. During the
process of intratracheal instillation and intratracheal
transfection, mice were anesthetized by an intraperitoneal
injection of sodium pentobarbital (50 mg/kg). After 28 days, the
mice were euthanized by an intraperitoneal injection of sodium
pentobarbital (200 mg/kg), and the lung tissues were collected for
subsequent analysis. All the experiments were approved and
supervised by the Animal Care and Use Committee and the Animal
Ethics Committee at University of South China.
Cells, cell culture and induction
The human A549 alveolar epithelial cell line
(ATCC® CCL-185™) was purchased from the American Type
Culture Collection (ATTC) and routinely cultured in high-glucose
DMEM, containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillium and 100 g/ml streptomycin in a cell incubator
(37°C and 5% CO2). When the cell confluency reached 80%,
the A549 cells were stimulated with transforming growth factor
(TGF)-β1 (10 ng/ml) for 48 h.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from the lung tissues and
the A549 cells using TRIzol® reagent (Thermo Fisher
Scientific, Inc.). The lung tissue was ground into a suspension
using a homogenizer. A total of 50 mg/ml TRIzol® was
added to the suspension and centrifuged at 3,000 × g for 15 min at
4°C to obtain the supernatant. The A549 cells were digested with 1
ml TRIzol® at room temperature for 5 min, then
centrifuged at 3,000 × g for 15 min at 4°C to obtain the
supernatant. The supernatant was then added to isopropyl alcohol
with sufficient mixing, then centrifuged at 3,000 × g for 10 min at
4°C to discard the supernatant. The precipitate was washed with
anhydrous ethyl alcohol, then added to diethyl pyrocarbonate water
to measure the RNA concentration (ng/µl). Reverse
transcription was performed using the cDNA RT kit (Roche Molecular
Diagnostics) and qPCR was performed using the SYBR-Green Fast qPCR
mix (Beyotime Institute of Biotechnology). The thermocycling
condition were as follows: Initial denaturation at 95°C for 25 sec;
followed by 40 cycles of 90°C for 30 sec, 55°C for 35 sec and 72°C
for 35 sec. The following primer pairs were used for the qPCR:
DLEU2 forward, 5′-TCC GAG AGT ATA GCG CCA CT-3′ and reverse, 5′-ACT
GCC CTT TGC TCC AAG TA-3′; TRIM2 forward, 5′-TGG AGA AGG AAA TGG
GCA TG-3′ and reverse, 5′-CTG CAA CCA CAA CAT GCA CCA-3′;PTGFRN
forward, 5′-CTT CAG CAG GAT GCC TGA CA-3′ and reverse, 5′-CAC CAG
GGA ATC ACG GTC AA-3′; GAPDH forward, 5′-AAG GTG AAG GTC GGA GTC
AAC-3′ and reverse, 5′-GGG GTC ATT GAT GGC AAC AAT A-3′; miR-369-3p
5′-GGG ACC CAG TTC AAG TAA TTC AGG-3′ and reverse, 5′-TTT GGC ACT
AGC ACA TT-3′; U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′ and
reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3′. RNA levels were
quantified using the 2−ΔΔCq method (13).
Cell transfection
A549 cells in the logarithmic growth phase were
cultured overnight in 6-well plates. Subsequently, 5 nM shRNA-NC, 5
nM shRNA-DLEU2-1/2, 5 nM mimic-NC, 5 nM miR-369-3p mimic, 5 nM
inhibitor-NC and 5 nM miR-369-3p inhibitor were transfected into
the A549 cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientifc, Inc.), and incubated for 24 h at 37°C. The
culture medium was changed every 6 h. The supplier of the shRNAs,
controls, mimics and inhibitors were RiboBio (https://www.ribobio.com/).
Cell Counting jkt (CCK)-8 assay
The experiment was performed using a 96-well cell
culture plate (3,000 cells/well), and the A549 cells in each group
were measured at 24, 48 and 72 h. Following stimulation with TGF-β1
induction and/or cell transfection for 48 h, 10 µl CCK-8
solution were added to each well followed by incubation at 37°C for
2 h. The absorbance of each group was determined using a Multiskan
FC photometer (Thermo Fisher Scientific, Inc.) at 450 nm.
Wound healing assay
After TGF-β1 induction and/or cell transfection, the
A549 cells were cultured in 6-well plates overnight. The cell
monolayers were scratched vertically with a 200-µl
micropipette tip. Serum-free medium was then added to the cells,
which were incubated in an incubator at 37°C with 5% CO2
for 24 h. Images were then captured under a light microscope
(Olympus Corporation) at a magnification of ×100 and analyzed using
ImageJ software (v1.8.0; National Institutes of Health).
Immunofluorescence assay
A549 cells in the logarithmic growth phase were
collected and ~1×105 cells/per well were placed in
immunofluorescence dishes until they had adhered to the plate. The
old culture medium was discarded. Following TGF-β1 stimulation
and/or cell transfection for 48 h, the A549 cells were washed with
pre-cooled PBS and fixed with 4% paraformaldehyde for 10 min at
room temperature. After washing with PBS, the A549 cells were
permeated for 10 min with 0.5% Triton X-100 at room temperature.
Normal goat serum was added into the dish and incubated at room
temperature for 30 min. After the blocking reagent was discarded,
the appropriate primary antibodies collagen I (ab260043; 1:250;
Abcam), α-SMA (ab124964; 1:250; Abcam) and E-cadherin (ab40772;
1:500; Abcam) were added to each dish and placed in a wet box for
incubation at 4°C overnight. The cells were then incubated at room
temperature for 1 h in a wet box with a fluorescent secondary
antibody (goat anti-rabbit IgG H&L; Alexa Fluor®
488; dilution, 1:500; ab150077; Abcam). DAPI was added to the A549
cells and incubated for 5 min in the dark to stain the cell nuclei.
Anti-fluorescence quenching agent was added to the dishes, and
images were then captured using an inverted fluorescence microscope
(Olympus Corporation) at a magnification of ×200.
Western blot analysis
Total protein was extracted from the lung tissue and
the A549 cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology) with protease inhibitor, then centrifuged at 3,000 ×
g at 4°C for 10 min to obtain the supernatant, which was
subsequently stored at −20°C until further use. BCA assay (Abcam)
was used for protein determination. Protein lysates (30 µg)
of each group were separated using 10% SDS-PAGE, transferred to a
PVDF membrane and blocked with TBS-Tween-20 (0.1%), containing 5%
bovine serum albumin (Gibco; Thermo Fisher Scientifc, Inc.) for 1 h
at room temperature. TBST was used to dilute the primary antibodies
containing collagen I (ab34710; Abcam), α-SMA (#19245; Cell
Signaling Technology, Inc.), E-cadherin (ab76055; Abcam), TRIM2
(ab3942; Abcam), PTGFRN (ab97567; Abcam) and GAPDH (ab8245; Abcam)
at a 1:1,000 dilution, which were then added to the membrane for
overnight incubation at 4°C. The PVDF membrane was washed with TBST
3 times, for 10 min each time. Subsequently, anti-rabbit IgG-HRP
secondary antibody (#7074; dilution,1:1,000; Cell Signaling
Technology, Inc.), diluted with TBST at 1:1,000, was added to the
membrane and incubated at room temperature for 1 h. The PVDF
membrane was then washed with TBST and the protein bands were
visualized using an enhanced chemiluminescence reagent. The protein
bands were imaged using an iBright CL1500 Imaging System
(Invitrogen; Thermo Fisher Scientific, Inc.). Image-Pro Plus
software (version 6.0; Media Cybernetics, Inc.) was used for
densitometry.
Hematoxylin and eosin (H&E) and
Masson's trichrome staining
Following euthanasia, the left lung was dissected,
washed with iso-osmotic saline and pressed between 2 filter papers.
The dried lungs were preserved in a 10% formalin, dehydrated with a
gradient concentration of ethanol and embedded in paraffin. The
paraffin block was fixed on the slicer and cut into
5-µm-thick sections, affixed to the slides, and placed in a
45°C thermostat to dry. Pathological staining of the lung tissue
was performed according to the instructions of the H&E
(Beyotime Institue of Biotechnology) and Masson kits (Beyotime
Institue of Biotechnology) at a magnification of ×400 (14).
Immunohistochemistry
Lung tissue samples embedded in paraffin were
rehydrated and incubated with the anti-collagen I primary antibody
(dilution, 1:100; ab270993; Abcam) at 4°C overnight. After washing,
the slides were incubated with the anti-mouse IgG-HRP secondary
antibody (dilution, 1:100; ab6728; Abcam) at 37°C for 20 min. Lung
tissue specimens were counterstained with hematoxylin at room
temperature for 1 min and mounted with neutral balsam. The staining
extent and intensity were observed under an optical microscope
(Olympus Corporation) at a magnification of ×400 and evaluated as
previously described (15).
Dual-luciferase reporter assay
To verify whether DLEU2 can bind to miR-369-3p, the
A549 cells were co-transfected with the DLEU2 3′-untranslated
region (UTR) pmirGLO plasmid [containing wild-type (WT) DLEU2
3′-UTR or mutant (MUT) DLEU2 3′-UTR] and miR-369-3p mimic or
mimic-NC using Lipofectamine® 2000. In addition, to
verify whether miR-369-3p can bind to TRIM2, the A549 cells were
co-transfected with TRIM2 3′-UTR pmirGLO plasmid (containing WT
TRIM2 3′UTR or MUT TRIM2 3′-UTR) and miR-369-3p mimic or mimic-NC
using Lipofectamine® 2000. The relative luciferase
(R-Luc) activity was detected using a dual-luciferase assay system
(Promega Corporation), which was normalized to Renilla
luciferase activity following transfection for 24 h. The pmirGLO
plasmid, mimics and inhibitors were obtained from Guangzhou RiboBio
Co., Ltd.
Statistical analysis
SPSS v19.0 software (SPSS, Inc.) was used for
statistical analysis, and the data are presented as the means ±
standard deviation. The differences between multiple groups were
analyzed using one-way ANOVA followed by Tukey's post hoc test. The
differences between 2 groups were analyzed using a Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
DLEU2 expression is increased in
pulmonary fibrosis
The IPF datasets (GSE10667, GSE24206 and GSE32537)
were downloaded from the GEO database and used to identify the
differentially expressed lncRNAs (log fold change, 0.5) (Fig. 1A). One of the differentially
expressed lncRNAs identified from the 3 datasets was DLEU2
(Fig. 1B). The DLEU2 expression
level was increased in the IPF group, which was found in the
GSE24206 and GSE32537 datasets (Fig.
1C). The differentially expressed mRNAs were also identified
from the 3 datasets (Fig. 1D).
The miRNAs that were found to interact with DLEU2 were predicted
using the star-Base database, and the mRNAs that could bind to the
identified miRNAs were predicted using the TargetScan, miRTarBase
and miRDB databases. The identified mRNAs and upregulated mRNAs in
the 3 datasets were intersected, and the target genes TRIM2 and
PTGFRN were identified (Fig.
1E). The expression level of TRIM2 and PTGFRN was increased in
the IPF group, which was found in all 3 datasets (Fig. 1F).
Knockdown of DLEU2 inhibits the
TGF-β1-induced proliferation, migration and EMT of A549 cells
DLEU2 expression was increased in the
TGF-β1-stimulated A549 cells (Fig.
2A). Following transfection, DLEU2 expression was decreased in
the A549 cells transfected with shRNA-DLEU2-1/2 and DLEU2
expression was lower in the shRNA-DLEU2-1-transfected A549 cells.
Therefore, shRNA-DLEU2-1 was selected for use in further
experiments (Fig. 2B). TGF-β1
promoted the proliferation (Fig.
2C) and migration (Fig. 2D and
E) of the A549 cells, which was suppressed by the knockdown of
DLEU2. TGF-β1 increased the expression levels of collagen I and
α-SMA, and decreased the expression levels of E-cadherin; these
effects were reversed by the knockdown of DLEU2 (Fig. 2G). The results of
immunofluorescence assay (Fig.
2F) were consistent with those of the western blot
analysis.
Knockdown of DLEU2 attenuates BLM-induced
pulmonary fibrosis in mice
BLM contributed to inflammation and pulmonary
fibrosis, and increased the expression level of collagen I in the
lung tissues, which was alleviated by the knockdown of DLEU2
(Fig. 3A). The protein
expression of collagen I and α-SMA was also increased, while the
expression of E-cadherin was decreased in the lung tissues from
mice with BLM-induced fibrosis. The knockdown of DLEU2 was found to
reverse these effects (Fig. 3B).
BLM also increased the expression levels of DLEU2, TRIM2 and
PTGFRN, which were decreased following the knockdown of DLEU2 in
lung tissues (Fig. 3C and D).
The change in TRIM2 mRNA expression was more significant compared
with that in PTGFRN expression following the knockdown of DLEU2
(Fig. 3D). The protein
expression levels of TRIM2 and PTGFRN were increased in the lung
tissues from mice BLM-induced fibrosis, which were decreased
following the knockdown of DLEU2 (Fig. 3E). TRIM2 was selected for
analysis in the following experiments. BLM was also found to be
associated with a decrease in the mRNA expression level of
miR-369-3p in the lung tissues of mice with BLM-induced fibrosis
and the knockdown of DLEU2 increased the miR-369-3p expression
level (Fig. 3F).
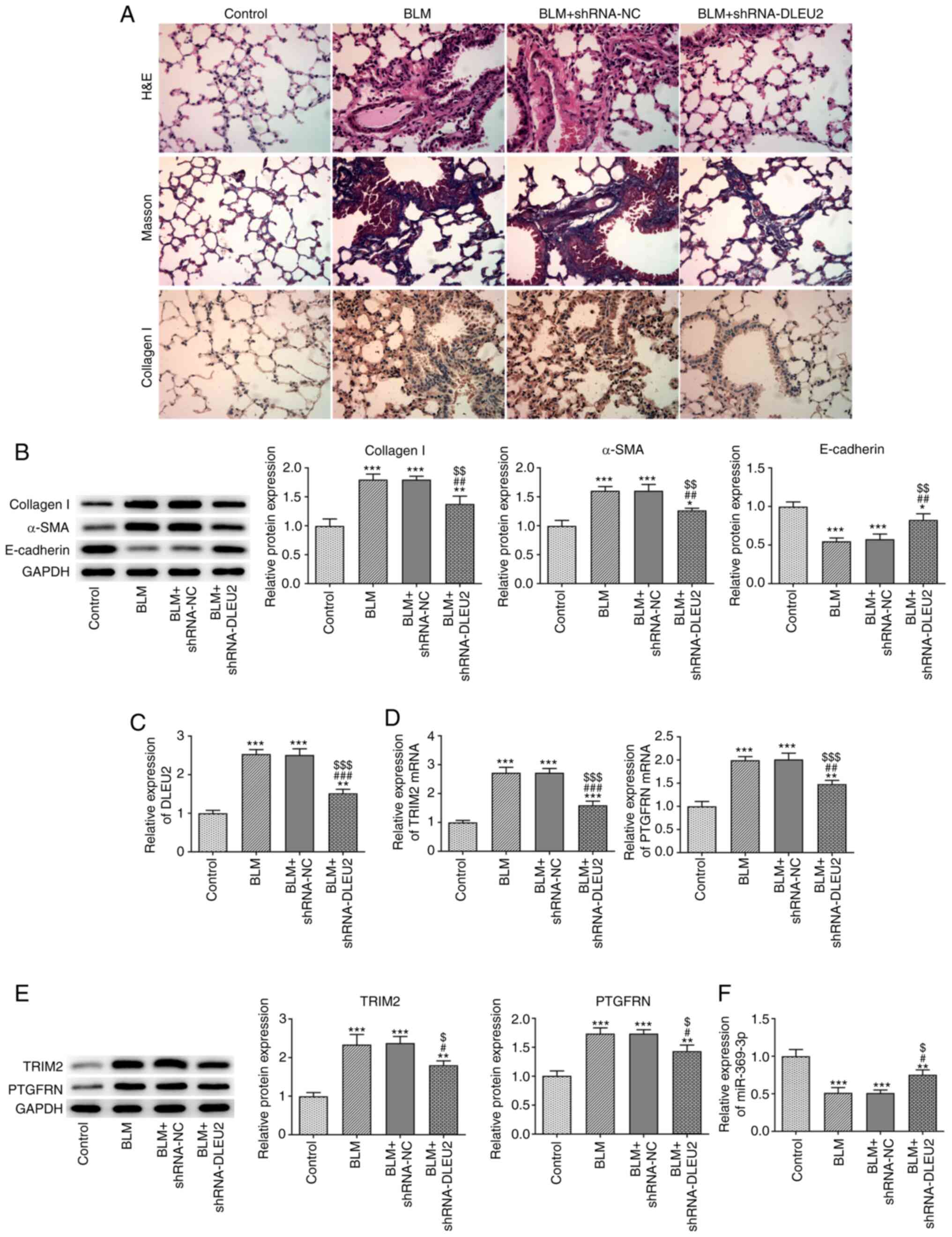 | Figure 3Knockdown of DLEU2 attenuates
BLM-induced pulmonary fibrosis in mice. (A) The pathological
changes, pulmonary fibrosis and collagen I expression in lung
tissues were in turn detected by H&E staining, Masson's
staining and immunohistochemistry. (B) The expression of collagen
I, α-SMA and E-cadherin in lung tissues of mice with BLM-induced
fibrosis following transfection was detected by western blot
analysis. (C) DLEU2 expression in lung tissues of mice with
BLM-induced fibrosis following transfection was detected by
RT-qPCR. (D) The mRNA expression of TRIM2 and PTGFRN in lung
tissues of mice with BLM-induced fibrosis following transfection
was detected by RT-qPCR. (E) The protein expression of TRIM2 and
PTGFRN in lung tissues of mice with BLM-induced fibrosis following
transfection was detected by western blot analysis. (F) miR-369-3p
expression in lung tissues of mice with BLM-induced fibrosis
following transfection was detected by RT-qPCR.
*P<0.05, **P<0.01 and
***P<0.001 vs. control group; #P<0.05,
##P<0.01 and ###P<0.001 vs. BLM group;
$P<0.05, $$P<0.01 and
$$$P<0.001 vs. BLM + shRNA-NC group. BLM, bleomycin;
TGF-β1, transforming growth factor β1; α-SMA, α-smooth muscle
actin; TRIM2, tripartite motif containing 2; PTGFRN, prostaglandin
F2 receptor inhibitor. |
DLEU2 directly targets miR-369-3p
The binding sites between DLEU2 and miR-369-3p are
presented in Fig. 4A. TGF-β1
decreased the miR-369-3p expression level in A549 cells (Fig. 4B). The miR-369-3p expression
level was increased in the A549 cells transfected with miR-369-3p
mimic (Fig. 4C) and decreased in
the A549 cells transfected with miR-369-3p inhibitor (Fig. 4D). The Luc/R-Luc value was
decreased in the A549 cells co-transfected with miR-369-3p mimics
and DLEU2 WT vector (Fig. 4E).
In addition, the miR-369-3p expression level was increased in the
A549 cells transfected with shRNA-DLEU2-1 (Fig. 4F).
Knockdown of DLEU2 attenuates the growth
and invasion of TGF-β1-stimulated A549 cells by regulating
miR-369-3p
The knockdown of DLEU2 decreased the proliferation
(Fig. 5A) and migration
(Fig. 5B and C) of the
TGF-β1-stimulated A549 cells, which was reversed following
transfection with the miR-369-3p inhibitor. The knockdown of DLEU2
suppressed the expression levels of collagen I and α-SMA, and
increased the expression level of E-cadherin in the
TGF-β1-stimulated A549 cells; these effects were reversed following
transfection with the miR-369-3p inhibitor (Fig. 5D). In addition, the knockdown of
DLEU2 decreased the mRNA (Fig.
5E) and protein (Fig. 5F)
expression level of TRIM2 in the TGF-β1-stimulted A549 cells; these
effects were reversed following transfection with the miR-369-3p
inhibitor.
TRIM2 is the target gene of
miR-369-3p
The binding sites between TRIM2 and miR-369-3p are
shown in Fig. 6A. When the A549
cells were co-transfected with miR-369-3p mimics and TRIM2 WT
vector, the Luc/R-Luc value was decreased (Fig. 6B). In addition, miR-369-3p
overexpression decreased the mRNA (Fig. 6C) and protein (Fig. 6D) expression levels of TRIM2.
Discussion
IPF is a chronic and progressive pulmonary disease,
characterized by the gradual increase of dyspnea, leading to
respiratory failure, and the mortality rate is increasing (16,17). A previous retrospective study
analyzed a database of 135 patients with interstitial pneumonia
confirmed by surgical lung biopsy, and reported that age was an
important independent predictor (18). Previous studies have demonstrated
that the regulation of gene expression plays an important role in
the aging process (19,20). lncRNAs have been found to be
associated with both IPF and senescence (21,22).
In the present study, DLEU2 expression was found to
be increased in IPF tissues by analyzing 3 datasets from the GEO
database. DLEU2 has been previously investigated in cancer. DLEU2
expression has been found to be increased in glioma tissues and
cell lines, and the downregulation of DLEU2 has been shown to
suppress the colony formation, migration and invasion of glioma
cells (23). DLEU2 has also been
found to be highly expressed in hepatocellular carcinoma (HCC)
tissues, and to be associated with tumor size, vascular invasion
and worse tumor stage, whereas the knockdown of DLEU2 reduces the
proliferative, migratory and invasive abilities of HCC cells
(24). In addition, DLEU2
expression has been shown to be increased in esophageal cancer
tissues and DLEU2 knockdown suppresses the proliferation, migration
and invasion of esophageal cancer cells, and promotes apoptosis by
regulating the Bcl-2/Bax axis (25). In the present study,
TGF-β1-stimulated A549 cells were established as an in vitro
model of IPF. Similar with the change and role of DLEU2 in a number
of types of cancer, DLEU2 expression was also found to be increased
in TGF-β1-stimulated A549 cells and lung tissues from mice with
BLM-induced fibrosis. The knockdown of DLEU2 inhibited the
TGF-β1-induced proliferation, migration and EMT of A549 cells, and
attenuated BLM-induced pulmonary fibrosis in mice.
lncRNA is a competitive endogenous RNA with a miRNA
recognition element, which can compete with mRNAs for the same
miRNA response element, thus inhibiting the binding of miRNAs to
mRNAs and affecting the regulatory effect of miRNAs on target genes
(26). miR-369-3p expression has
been found to be decreased in endometrioid adenocarcinoma tissues
and miR-369-3p overexpression suppresses cell proliferation and
migration in endometrioid adenocarcinoma (27). miR-369-3p expression has been
shown to be notably increased in Hirschsprung's disease and the
dysregulation of miR-369-3p has been shown to decrease the
proliferation and migration of the SH-SY5Y and 293T cells (28). In addition, in a previous study,
the expression of miR-369-3p was to be decreased and to be
associated with a poor prognosis in patients with HCC.
Functionally, miR-369-3p overexpression inhibited the viability and
motility of HCC cells (29).
miR-369-3p expression was decreased in the inflamed intestinal
regions obtained from patients with inflammatory bowel disease and
the overexpression of miR-369-3p alleviated the inflammatory
response in response to lipopolysaccharide (30). Consistent with the findings of
previous studies, the expression levels of miR-369-3p were also
decreased in TGF-β1-stimulated A549 cells and lung tissues from
mice with BLM-induced fibrosis in the present study. The
downregulation of miR-369-3p weakened the effect of DLEU2 knockdown
on TGF-β1-stimulated A549 cells and further promoted the
TGF-β1-induced proliferation, migration and EMT of A549 cells. In
addition, TRIM2 was the target gene of miR-369-3p and TRIM2
expression was increased in TGF-β1-stimulated A549 cells and lung
tissues from mice with BLM-induced fibrosis.
A previous study indicated that TRIM2 promoted the
growth, invasion and metastasis of colorectal cancer cells through
EMT (10). In the present study,
it was demonstrated that DLEU2 regulated TRIM2 by miR-369-3p, and
it was thus hypothesized that TRIM2 may affect tumor metastasis
through EMT. Herein, it was found that the downregulation of DLEU2
improved E-cadherin expression in TGF-β1-stimulated A549 cells and
lung tissues from mice with BLM-induced fibrosis.
In conclusion, DLEU2 expression was found to be
increased in IPF tissues from the analysis of expression data
downloaded from the GEO database. The expression levels of DLEU2
and TRIM2 were increased, while the expression levels of miR-369-3p
were decreased in TGF-β1-stimulated A549 cells and lung tissues
from mice with BLM-induced fibrosis. The knockdown of DLEU2
suppressed IPD by increasing and decreasing the miR-369-3p and
TRIM2 expression levels, respectively. These findings indicated
that the negative modulation of DLEU2 may be a novel treatment
strategy for IPF. In the future, the authors aim to confirm DLEU2
expression in lung tissues of patients with IPF and to explore the
role of TRIM2 in IPF. In addition, the regulatory role of TRIM2 in
IPF warrants further investigation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DJ consulted the literature, and conceived and
designed the study. HY conducted all the experiments and DL
assisted HY in collecting the raw data. YX assisted HY in
performing the statistical analysis. HY drafted the manuscript,
which was reviewed and corrected by LJ. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All the animal experiments were approved and
supervised by the Animal Care and Use Committee and the Animal
Ethics Committee at University of South China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This study was funded by the Natural Science Foundation of Hunan
Province (grant no. 2019JJ80044).
References
|
1
|
Caminati A, Madotto F, Cesana G, Conti S
and Harari S: Epidemiological studies in idiopathic pulmonary
fibrosis: Pitfalls in methodologies and data interpretation. Eur
Respir Rev. 24:436–444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chanda D, Otoupalova E, Smith SR,
Volckaert T, De Langhe SP and Thannickal VJ: Developmental pathways
in the pathogenesis of lung fibrosis. Mol Aspects Med. 65:56–69.
2019. View Article : Google Scholar :
|
|
3
|
Meyer KC: Pulmonary fibrosis, part I:
Epidemiology, pathogenesis, and diagnosis. Expert Rev Respir Med.
11:343–359. 2017.PubMed/NCBI
|
|
4
|
Johnsson P, Lipovich L, Grandér D and
Morris KV: Evolutionary conservation of long non-coding RNAs;
Sequence, structure, function. Biochim Biophys Acta.
1840:1063–1071. 2014. View Article : Google Scholar
|
|
5
|
Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang
X and Cui Y: Long non-coding RNA MIAT in development and disease: A
new player in an old game. J Biomed Sci. 25:232018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu W, Zhao Y, Gao E, Li Y, Guo X, Zhao T,
He W and Zhang H: LncRNA DLEU2 accelerates the tumorigenesis and
invasion of non-small cell lung cancer by sponging miR-30a-5p. J
Cell Mol Med. 24:441–450. 2020. View Article : Google Scholar
|
|
7
|
Liu W, Liu PC, Ma K, Wang YY, Chi QB and
Yan M: LncRNA DLEU2 promotes tumour growth by sponging miR-337-3p
in human osteosarcoma. Cell Biochem Funct. 38:886–894. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang B, Hang J, Li W and Yuan W: Knockdown
of LncRNA DLEU2 inhibits cervical cancer progression via targeting
miR-128-3p. Onco Targets Ther. 13:10173–10184. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu M, Chen W, Zhuang W and Zhan X:
Label-free quantitative identification of abnormally ubiquitinated
proteins as useful biomarkers for human lung squamous cell
carcinomas. EPMA J. 11:73–94. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cao H, Fang Y, Liang Q, Wang J, Luo B,
Zeng G, Zhang T, Jing X and Wang X: TRIM2 is a novel promoter of
human colorectal cancer. Scand J Gastroenterol. 54:210–218. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qin Y, Ye J, Zhao F, Hu S and Wang S:
TRIM2 regulates the development and metastasis of tumorous cells of
osteosarcoma. Int J Oncol. 53:1643–1656. 2018.PubMed/NCBI
|
|
12
|
Dong R, Liu M, Huang XX, Liu Z, Jiang DY,
Xiao HJ, Geng J, Ren YH and Dai HP: Water-Soluble C60
protects against bleomycin-induced pulmonary fibrosis in mice. Int
J Nanomedicine. 15:2269–2276. 2020. View Article : Google Scholar :
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
14
|
Tang F, Li R, Xue J, Lan J, Xu H, Liu Y,
Zhou L and Lu Y: Azithromycin attenuates acute radiation-induced
lung injury in mice. Oncol Lett. 14:5211–5220. 2017.PubMed/NCBI
|
|
15
|
Abdelfattah MS, Elmallah MIY, Ebrahim HY,
Almeer RS, Eltanany RMA and Abdel Moneim AE: Prodigiosins from a
marine sponge-associated actinomycete attenuate HCl/ethanol-induced
gastric lesion via antioxidant and anti-inflammatory mechanisms.
PLoS One. 14:e02167372019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cookson WO and Moffatt MF: Bedside to gene
and back in idiopathic pulmonary fibrosis. N Engl J Med.
368:2228–2230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fernandez IE and Eickelberg O: New
cellular and molecular mechanisms of lung injury and fibrosis in
idiopathic pulmonary fibrosis. Lancet. 380:680–688. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fell CD, Martinez FJ, Liu LX, Murray S,
Han MK, Kazerooni EA, Gross BH, Myers J, Travis WD, Colby TV, et
al: Clinical predictors of a diagnosis of idiopathic pulmonary
fibrosis. Am J Respir Crit Care Med. 181:832–837. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baumgart M, Groth M, Priebe S, Savino A,
Testa G, Dix A, Ripa R, Spallotta F, Gaetano C, Ori M, et al:
RNA-seq of the aging brain in the short-lived fish N.
furzeri-conserved pathways and novel genes associated with
neurogenesis. Aging Cell. 13:965–974. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benayoun BA, Pollina EA and Brunet A:
Epigenetic regulation of ageing: Linking environmental inputs to
genomic stability. Nat Rev Mol Cell Biol. 16:593–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grammatikakis I, Panda AC, Abdelmohsen K
and Gorospe M: Long noncoding RNAs(lncRNAs) and the molecular
hallmarks of aging. Aging (Albany NY). 6:992–1009. 2014. View Article : Google Scholar
|
|
22
|
Huang C, Yang Y and Liu L: Interaction of
long noncoding RNAs and microRNAs in the pathogenesis of idiopathic
pulmonary fibrosis. Physiol Genomics. 47:463–469. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie Z, Li X, Chen H, Zeng A, Shi Y and
Tang Y: The lncRNA-DLEU2/miR-186-5p/PDK3 axis promotes the progress
of glioma cells. Am J Transl Res. 11:4922–4934. 2019.PubMed/NCBI
|
|
24
|
Guo Y, Bai M, Lin L, Huang J, An Y, Liang
L, Liu Y and Huang W: LncRNA DLEU2 aggravates the progression of
hepatocellular carcinoma through binding. Biomed Pharmacother.
118:1092722019. View Article : Google Scholar
|
|
25
|
Lu T, Wang R, Cai H and Cui Y: Long
non-coding RNA DLEU2 promotes the progression of esophageal cancer
through miR-30e-5p/E2F7 axis. Biomed Pharmacother. 123:1096502020.
View Article : Google Scholar
|
|
26
|
Li C, Wang Z, Zhang J, Zhao X, Xu P, Liu
X, Li M, Lv C and Song X: Crosstalk of mRNA, miRNA, lncRNA, and
circRNA and their regulatory pattern in pulmonary fibrosis. Mol
Ther Nucleic Acids. 18:204–218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu P, Ma C, Wu Q, Zhang W, Wang C, Yuan L
and Xi X: MiR-369-3p participates in endometrioid adenocarcinoma
via the regulation of autophagy. Cancer Cell Int. 19:1782019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan W, Yu H, Zheng B, Gao Y, Li P, Huang
Q, Xie C and Ge X: Upregulation of MiR-369-3p suppresses cell
migration and proliferation by targeting SOX4 in Hirschsprung's
disease. J Pediatr Surg. 52:1363–1370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu Q and Liu K: MiR-369-3p inhibits
tumorigenesis of hepatocellular carcinoma by binding to PAX6. J
Biol Regul Homeost Agents. 34:917–926. 2020.PubMed/NCBI
|
|
30
|
Scalavino V, Liso M, Cavalcanti E, Gigante
I, Lippolis A, Mastronardi M, Chieppa M and Serino G: miR-369-3p
modulates inducible nitric oxide synthase and is involved in
regulation of chronic inflammatory response. Sci Rep. 10:159422020.
View Article : Google Scholar : PubMed/NCBI
|




















