Introduction
Hepatic carcinoma is the sixth most common cancer
(4.7% of all registered cancer cases) and the fourth leading cause
of cancer-related mortality (8.2% of the all cancer-related deaths)
worldwide according to findings from 2018 (1). Due to the absence of typical
symptoms in the early stage, most patients are diagnosed at an
advanced stage, thus having a poor prognosis (2). Uncured hepatic carcinoma in
terminal stages is often combined with severe ascites (3). Understanding the molecular
mechanisms of hepatic carcinoma is vital for identifying new
therapeutic targets.
MicroRNAs (miRNAs or miRs) are short-chain (19-25
nucleotides in length) non-coding RNA that have an important role
in a variety of human diseases, including cardiovascular disease,
immune response and cancer (4,5).
miR-29b is a key tumor regulator (6) that can inhibit tumor cell
viability, prompt cell apoptosis, suppress tumor invasion and
migration and delay tumor progression (7,8).
miR-29b frequently acts as a tumor suppressor and is suppressed in
multiple tumors such as non-small cell lung cancer, hepatocellular
carcinoma and colorectal cancer (9-11). Our previous studies revealed an
increased level of miR-29b in hepatoma 22 (H22) cells in ascites
tumor-bearing mice treated with supercritical-carbon dioxide fluid
or scutellarin administration. The treatment further decreased
ascites and improved survival (12,13).
Numerous malignant tumor mediators have been
associated with miR-29b. The protein p53 is a key cell division
checkpoint-monitoring DNA mutant (14). The function of p53 in cancer is
usually impaired or mutated, which enables abnormal cell
proliferation (15). Our
previous studies reported that p53 may induce apoptosis in H22
cells (13,16). Additionally, both miR-29b and p53
had a consistent trend in H22 ascites, in accordance with cell
apoptotic degree (12). Numerous
studies have indicated a regulatory circuitry between miR-29b and
p53 (10,17). miR-29b can reduce the levels of
the SET domain bifurcated histone lysine methyltransferase 1
(SETDB1) and ultimately upregulate p53. Similarly, p53 can also
increase the expression of miR-29b, thus forming an antitumor
circle (10). However, to date,
to the best of our knowledge, no studies have reported the
regulation between p53 and miR-29b of malignant ascites.
Transforming growth factor-β1 (TGF-β1) is a
pleiotropic cytokine that regulates cell growth, differentiation,
migration and apoptosis (18).
It is another malignant tumor mediator associated with miR-29b
(19). TGF-β1 signaling is a
double-edged sword in the regulation of cancer (20). In the initial stages of cancer,
TGF-β1 often has an antitumor effect, inhibiting cell
proliferation, cell cycle arrest in G1 and inducing apoptosis
(21). However, in advanced
tumors, deletion or mutation of TGF-β1 or Smads has been revealed
(22,23). When mutated, TGF-β1 loses its
antitumor function and enhances tumor cell epithelial-mesenchymal
transition (EMT), leading to invasion and metastasis (24,25). Interestingly, TGF-β1 can improve
p53 expression in vascular senescence and apoptosis of hepatic
stellate cells (26,27). Moreover, a previous study has
demonstrated a crosstalk between TGF-β1 and p53 (28).
The aim of this study was to investigate the effect
of miR-29b on proliferation and apoptosis of hepatocellular
carcinoma ascites H22 cells and its association with the TGF-β1
signaling pathway and p53-mediated apoptotic pathway. The crosstalk
between TGF-β1 and p53 in hepatocellular carcinoma ascites H22
cells was also explored. The findings of the present study provided
further understanding of molecular mechanisms of hepatic
carcinoma.
Materials and methods
Materials
miR-29b-3p (hereinafter referred to as miR-29b)
mimic [sense strand (5′ to 3′): UAG CAC CAU UUG AAA UCA GUG UU and
antisense strand (3′ to 5′): AUC GUG GUA AAC UUU AGU CAC AA;
product no. miR10000127-1-5], mimic negative control (product no.
miR1N0000002-1-5), miR-29b inhibitor (5′ to 3′: AUC GUG GUA AAC UUU
AGU CAC AA; product no. miR20000127-1-5), inhibitor negative
control (product no. miR2N0000002-1-5), TGF-β1 small interfering
RNA (siRNA) (siTGF-β1, 5′ to 3′: CCA GAA ATA TAG CAA CAA T), and
control siRNA (product no. siP0000003-1-5) were purchased from
Guangzhou RiboBio Co., Ltd. Recombinant human TGF-β1 protein was
obtained from PeproTech, Inc. Recombinant human TGF-β1 was
dissolved in citric acid provided by Multi Sciences (Lianke)
Biotech, Co., Ltd. TGF-β1 negative control (TGF-β1 NC) was 1 mM
citric acid which was the solvent of the recombinant human TGF-β1.
TGF-β1 has cross-reactivity towards mouse according to the
manufacturer's instructions. Lipofectamine™ RNAiMAX Transfection
Reagent was provided by Thermo Fisher Scientific, Inc. MTS
CellTiter 96 AQueous one solution cell proliferation assay solution
was acquired from Promega Corporation. Annexin V-FITC/PI double
staining apoptosis detection kit was supplied by BestBio
Biotechnology Co., Ltd. Dulbecco's modified Eagle's medium (DMEM),
Roswell Park Memorial Institute-1640 (RPMI-1640) medium, Opti-MEM™
I reduced serum medium (Opti-MEM), fetal bovine serum (FBS),
penicillin/streptomycin solution and TRIzol reagent were obtained
from Thermo Fisher Scientific, Inc. Reverse transcription primers
were designed by Sangon Biotech Co., Ltd. Specific antibodies for
TGF-β1 (cat. no. AF1027), p53 (cat. no. AF0865), phosphorylated
(p)-Smad3 (cat. no. AF3362), Smad7 (cat. no. AF5147), B-cell
lymphoma-2 (Bcl-2) (cat. no. AF6139), Bcl-2-Associated X protein
(Bax) (cat. no. AF0120), and β-actin (cat. no. AF7018) were
obtained from Affinity Biosciences Pty Ltd. RIPA lysis buffer,
phenylmethylsulfonyl fluoride (PMSF), cocktail protease inhibitor,
and phosphatase inhibitors A and B were purchased from Servicebio
Technology Co., Ltd. Horseradish peroxidase (HRP) and goat
anti-rabbit immunoglobulin G (H+L) (cat. no. E030120-01) were
obtained from EarthOx Life Sciences. Fluorescein isothiocyanate
(FITC; product no. GB22301), Cyanine3 (CY3; product no. GB21303)
and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from
Servicebio Technology Co., Ltd. Chloroform, isopropanol and
anhydrous ethanol were at least of the analytic grade.
Cell culture
NCTC1469, Hepa1-6 and H22 cells were all provided by
iCell Bioscience, Inc. NCTC1469 and Hepa1-6 were cultured in DMEM
medium, while H22 was cultured in RPMI-1640 medium containing 10%
(v/v) FBS and 1% (v/v) penicillin/streptomycin. The cells were
cultured in a humidified atmosphere containing 5% CO2 at
37°C.
RNA and protein extraction
NCTC1469, Hepa1-6, and H22 cells were seeded in
6-well plates at a concentration of 2x105 cells/ml at 2
ml per well and cultured for 48 h. TRIzol reagent and chloroform
(15,000 x g, at 4°C for 15 min), isopropanol (12,000 x g, at 4°C
for 5 min) and anhydrous ethanol (7,500 x g, at 4°C for 5 min) were
utilized to extract and purify total RNA. Cell disruption reagent
containing RIPA, PMSF, cocktail and phosphatase inhibitors A and B
at ratios of 100:1:2:1:1 was used to extract the total protein
(14,000 x g, at 4°C for 10 min).
Transfection and interference in H22
cells
H22 cells were diluted with RPMI-1640 and seeded at
a concentration of 2x105 cells/ml in 96-well or 6-well
plates at a volume of 100 µl or 1 ml per well, respectively.
Cells were first starved for 12 h. Lipofectamine™ RNAiMAX
Transfection Reagent was used for transfection according to the
manufacturer's instructions; specific steps were as follows:
Opti-MEM containing transfection reagent (at a ratio of 50:3) or
gene sequence complex (at a ratio of 50:0.5) (10 nM) was prepared.
Transfection concentrations of miR-29b mimic, mimic negative
control, miR-29b inhibitor, inhibitor negative control, siTGF-β1
and control siRNA were 50, 50, 100, 100, 80 and 80 nM,
respectively. Opti-MEM-transfection reagent complex was mixed with
Opti-MEM-gene sequence complex and incubated for 5 min at room
temperature. The complex was then added to H22 cells (96- or 6-well
plate) at 10 or 100 µl/well for 48 h. After 6 h of
transfection at 37°C, 10 ng/ml recombinant human TGF-β1 was added
to the wells, and cells were cultured for 48 h in the TGF-β1
interference groups. Cells were harvested for further
experimentation.
RNA and protein extraction from H22 cell
interference with miR-29b or TGF-β1
H22 cells were transfected with or without miR-29b
mimic, mimic negative control, miR-29b inhibitor, or inhibitor
negative control. H22 cells were also transfected with miR-29b
mimic or mimic negative control, as well as siTGF-β1 or control
siRNA. Total RNA and protein were extracted as aforementioned.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA extracted from NCTC1469, Hepa1-6 and H22
cells or from H22 cells with transfection or interference was
reversely transcribed to cDNA using the miRNA 1st Strand cDNA
Synthesis Kit (by stem-loop) (product serial no. MR101-01) or
HiScript II Q RT SuperMix (product no. R223-01) (both from Vazyme
Biotech Co., Ltd.) according to the manufacturer's instructions.
The cDNA was amplified and quantified using the miRNA Universal
SYBR qPCR Master Mix (product no. MQ101-01) or SYBR quantitative
polymerase chain reaction (qPCR) Master Mix kit (product no.
Q711-02) (Vazyme Biotech Co., Ltd.) according to the manufacturer's
instructions. The thermocycling conditions were as follows: cDNA
was subjected to a temperature of 95°C for 30 sec (or 5 min for
miRNA); 40 cycles at 95°C for 10 sec and 60°C for 30 sec; and then
95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec. Primer
sequences are presented in Table
I. U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
employed as an internal reference. The gene expression was
quantified using the following formulas:
 | Table IPrimer sequences used for reverse
transcription-quantitative qPCR. |
Table I
Primer sequences used for reverse
transcription-quantitative qPCR.
| Gene name | | Primer (5′-3′) |
|---|
| U6 | RT-primer |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAAT |
| Forward |
AGAGAAGATTAGCATGGCCCCTG |
| Reverse |
AGTGCAGGGTCCGAGGTATT |
| miR-29b | RT-primer |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACACT |
| Forward C |
GCGTAGCACCATTTGAAATC |
| Reverse |
AGTGCAGGGTCCGAGGTATT |
| GAPDH | Forward |
AATGGTGAAGGTCGGTGTGAACG |
| Reverse |
TCGCTCCTGGAAGATGGTGATGG |
| TGF-β1 | Forward |
GCAGTGGCTGAACCAAGGAGAC |
| Reverse |
GCCGTGAGCTGTGCAGGTG |
| Smad3 | Forward C |
ACAGCATGGACGCAGGTTCTC |
| Reverse C |
ACTGAGGCACTCCGCAAAGAC |
| Smad7 | Forward |
GCAAGATCGGCTGTGGCATCC |
| Reverse |
GCTGATGAACTGGCGGGTGTAG |
| p53 | Forward CC |
TGGGAGAGACCGCCGTAC |
| Reverse C |
TAGGCTGGAGGCTGGAGTGAG |
| Bax | Forward |
GCGAGTGTCTCCGGCGAATTG |
| Reverse |
TGGTGAGCGAGGCGGTGAG |
| Bcl-2 | Forward CC |
GTCGTGACTTCGCAGAGATG |
| Reverse |
GGTGTGCAGATGCCGGTTCAG |
Cellular viability assay
H22 cells were seeded in 96-well plates and
transfected as aforementioned. Then, 20 µl MTS reagent was
added to each well for 4 h. The optical density (OD) value at 490
nm was determined using a microplate reader (Thermo Fisher
Scientific, Inc.). The cell viability was calculated as follows:
[(OD control-OD blank)-(OD transfection-OD blank)]/(OD control-OD
blank) x100%. The experiment was conducted in triplicate.
Cell apoptosis
H22 cells were seeded at a concentration of
2x105 cells/ml in 6-well plates and transfected as
aforementioned. Annexin V-FITC/PI double staining apoptosis
detection kit (cat. no. BB-4101; BestBio Biotechnology Co., Ltd.)
was then applied according to the manufacturer's instructions.
Specific steps were as follows: 400 µl 1X Annexin V was used
to bind the suspension cells. Then, 5 µl Annexin V-FITC was
added to stain the cells with incubation for 15 min at 2-8°C in a
dark place. Finally, 10 µl PI was added to stain the cells
with incubation for 15 min at 2-8°C in a dark place. BD LSRFortessa
(BD Biosciences) was used to detect the apoptotic rate. FlowJo
(version 10.0; BD Biosciences) was used for analysis.
Western blot analysis
The protein content of the samples was determined by
bicinchoninic acid (BCA) method. Proteins were loaded at 50
µg per lane. Proteins collected were separated by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto polyvinylidene fluoride (PVDF) membranes. The
membranes were blocked for 1 h at room temperature in 5% skimmed
milk diluted in Tris-buffered solution-Tween-20 (0.1%, TBST).
Subsequently, samples were incubated with specific primary
antibodies (1:2,000 for TGF-β1 and p53; 1:1,000 for p-Smad3, Smad7,
Bcl-2, Bax and β-actin) overnight at 4°C and then with the
appropriate secondary antibody (1:3,000 in TBST) for 1 h at room
temperature. The protein bands were observed using an enhanced
chemiluminescence advanced kit according to the instructions of the
manufacturer (product no. RPN2232; GE Healthcare; Cytiva). Quantity
one analysis software (version 4.6.2; Bio-Rad Laboratories, Inc.)
was used to analyze band intensity.
Immunofluorescence
H22 cells were seeded in 6-well plates and
transfected as aforementioned. After being washed 3 times with
phosphate-buffered saline (PBS), cells were collected, and 4%
paraformaldehyde was added to fix the cells at room temperature for
24 h. H22 cell pellets were obtained using centrifugation at 800 x
g for 5 min at room temperature. Samples were then embedded in
paraffin and cut into 4-µm sections. The sections were
incubated in 5% bovine serum albumin solution (BSA) at room
temperature for 30 min, then incubated with specific primary
antibodies (1:1,000 for TGF-β1) overnight at 4°C, and finally
incubated with secondary antibody (1:500) conjugated with
horseradish peroxidase (HRP) at room temperature for 1 h. Next,
sections were stained with FITC-conjugated antibody (1:300) at room
temperature for 10 min. Sections were incubated with another
primary (1:2,500 for p53) overnight at 4°C and secondary antibody
(HRP-conjugated goat anti-rabbit IgG (H+L); cat. no. GB23303;
Servicebio Technology Co., Ltd.; 1:500) for 1 h at room temperature
and stained with CY3 antibody (1:300) at room temperature for 10
min. Finally, DAPI (2 µg/ml) was used to stain the cell
nucleus at room temperature for 10 min. A fluorescence microscope
(Nikon Corporation) was used to observe the intensity of
fluorescence at a magnification of x200.
Statistical analysis
Data analysis was performed by SPSS software
(version 23.0; IBM Corp.). The significance of different groups was
assessed by one-way analysis of variance (ANOVA) and Fisher's least
significant difference (LSD) post hoc test following ANOVA.
P<0.05 was considered to indicate a statistically significant
difference. Graphs were drawn by GraphPad Prism software version 7
(GraphPad Software, Inc.).
Results
Expression of miR-29b, TGF-β1 and p53 in
NCTC1469, Hepa1-6 and H22 cells
RT-qPCR and western blot analysis were used to
evaluate the different expression levels of miR-29b, TGF-β1 and p53
in NCTC1469, Hepa1-6 and H22 cells. As demonstrated in Fig. 1, the expression of miR-29b in
Hepa1-6 and H22 was lower compared with NCTC1469 cells (P<0.01).
Moreover, lower protein expression levels of TGF-β1 and p53 were
revealed in Hepa1-6 and H22 compared with NCTC1469 cells
(P<0.05).
miR29b expression with transfection of
miR29b mimic and inhibitor in H22 cells
RT-qPCR was applied to detect the miR-29b expression
in H22 cells transfected with miR-29b mimic or inhibitor. The
results revealed that with the transfection of miR-29b mimic, the
expression of miR-29b significantly increased (P<0.01) (Fig. 2A). Concurrently, the expression
of miR-29b was significantly downregulated with miR29b inhibitor
transfection (P<0.05) (Fig.
2A).
Effect of miR-29b on cell proliferation
in H22 cells
To investigate the effect of miR-29b on
hepatocellular carcinoma ascites cell proliferation, H22 cells were
transfected with miR-29b mimic or miR-29b inhibitor. Transfection
with miR-29b mimic in H22 cells inhibited cell proliferation,
compared with the control group (P<0.01) (Fig. 2B). Conversely, miR-29b inhibitor
increased cell proliferation compared with the control group
(P<0.05) (Fig. 2B).
Effect of miR-29b on cell apoptosis in
H22 cells
Flow cytometry was used to verify whether miR-29b
had an effect on hepatocellular carcinoma ascites cell apoptosis.
As revealed in Fig. 3, miR-29b
mimic could significantly increase cell apoptosis of H22 cells
compared with the control mimic group (P<0.01), while the
opposite effect was observed when using miR-29b inhibitor
(P<0.05).
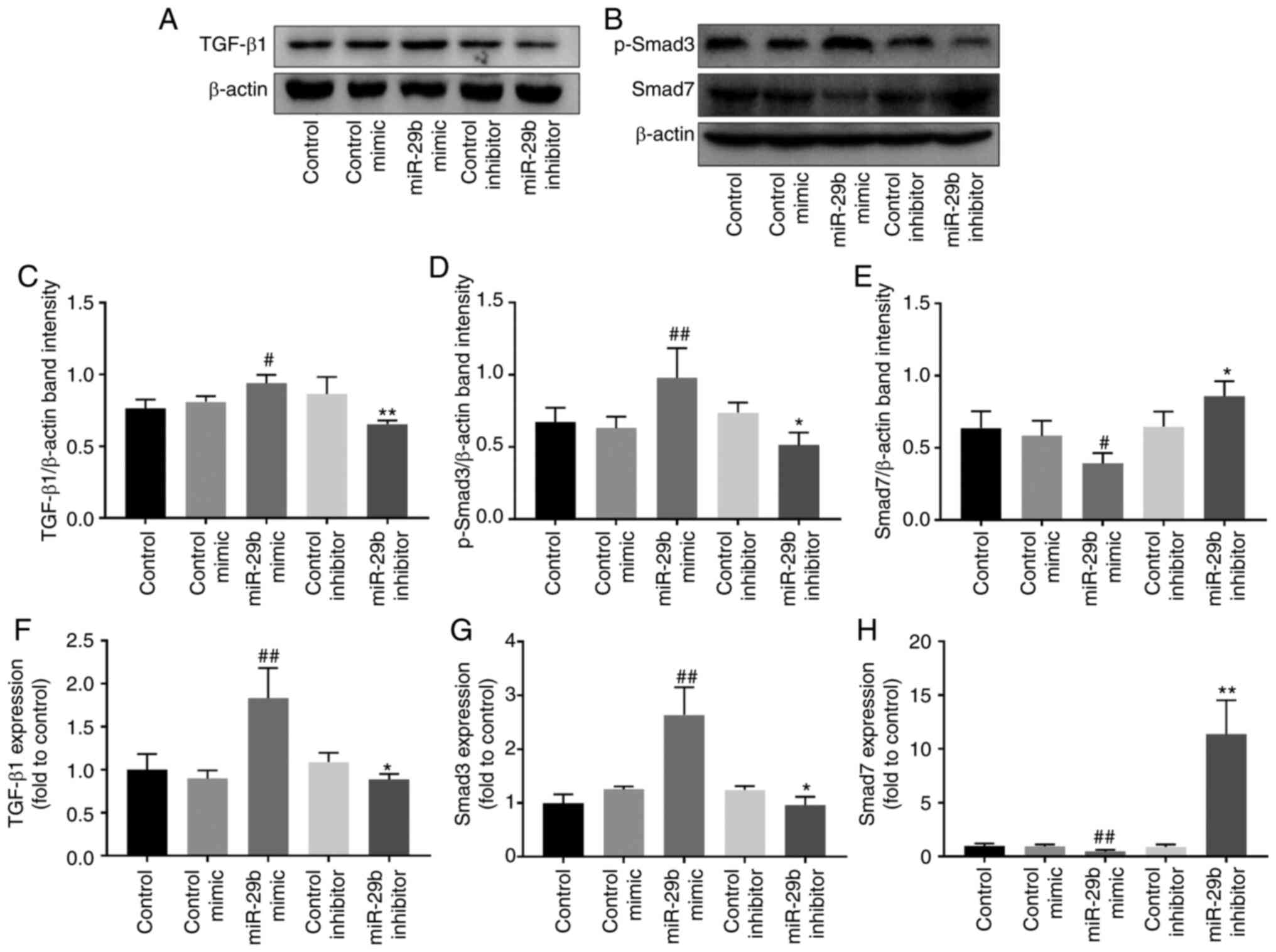
Effect of miR-29b on the TGF-β1 signaling
pathway in H22 cells
Western blotting and RT-qPCR were used to further
explore the regulation of miR-29b on the TGF-β1 signaling pathway.
As revealed in Fig. 4, when
compared with the control mimic group, miR-29b overexpression
increased the protein and mRNA levels of TGF-β1 (P<0.05 and
P<0.01, respectively). Concurrently, p-Smad3 protein and Smad3
mRNA levels were increased (P<0.01), while Smad7 protein and
mRNA levels were decreased (P<0.05 and P<0.01, respectively).
The opposite effects were observed when using miR-29b inhibitor
(P<0.05 and P<0.01).
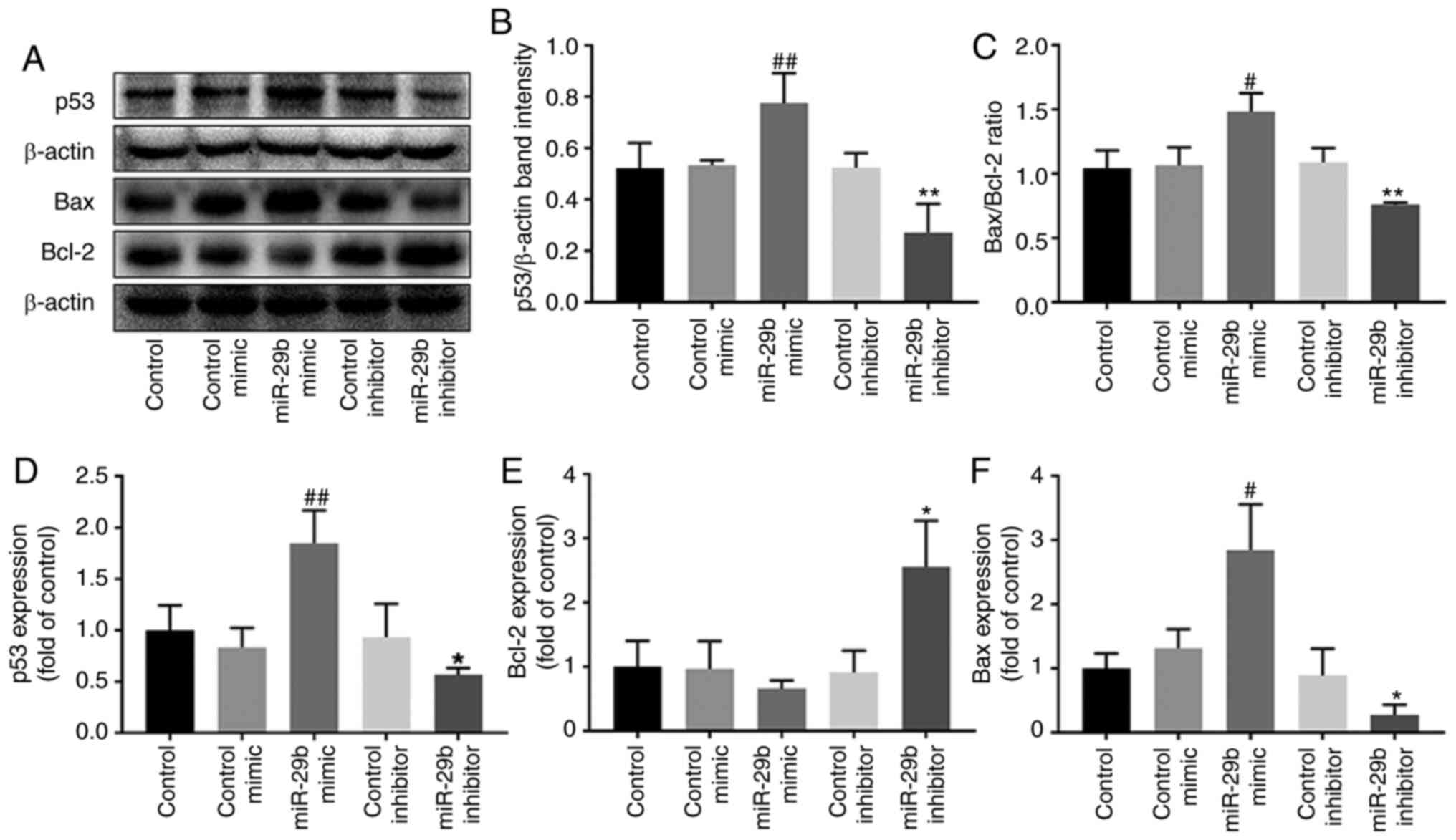
Effect of miR-29b on p53-mediated
apoptotic pathway in H22 cells
Western blotting and RT-qPCR were used to verify the
effect of miR-29b on the p53-mediated apoptotic pathway. miR-29b
mimic increased the expression p53 protein and mRNA levels
(P<0.01), the protein ratio of Bax/Bcl-2 and mRNA level of Bax,
compared with the control group (P<0.05). Furthermore, it
decreased the mRNA level of Bcl-2. Conversely, miR-29b inhibitor
induced the opposite effects (P<0.05 and P<0.01) (Fig. 5).
Effect of TGF-β1 on miR-29b-regulated
cell proliferation in H22 cells
Exogenous TGF-β1 and siTGF-β1 were used to
investigate whether the effect of miR-29b on H22 cell proliferation
inhibition and apoptotic promotion was associated with the TGF-β1
signaling pathway. The results revealed that under miR-29b mimic
and exogenous TGF-β1 stimulation, the proliferation of H22 cells
was significantly inhibited compared with miR-29b mimic and TGF-β1
NC stimulation group (P<0.01). However, in cells transfected
with miR-29b mimic and siTGF-β1, the proliferation of H22 cells was
significantly increased compared with cells transfected with
miR-29b mimic and control siRNA (P<0.01) (Fig. 6).
Effects of miR-29b overexpression and
TGF-β1 suppression on the TGF-β1 signaling pathway
Transfection of siTGF-β1 in H22 cells reduced the
mRNA level of TGF-β1 by 43.75% compared with the group without any
transfection (P<0.01). Furthermore, siTGF-β1 transfection could
reverse TGF-β1 mRNA increase induced by miR-29b overexpression
(P<0.01). Smad3 also increased with the miR-29b overexpression
(P<0.01); however, siTGF-β1 significantly reduced this effect
(P<0.05). Smad7 mRNA was inhibited by miR-29b overexpression
(P<0.01); whereas siTGF-β1 reversed this inhibition (P<0.05)
(Fig. 7).
Effects of miR-29b overexpression and
TGF-β1 suppression on TGF-β1 and p53 expression
Immunofluorescence was utilized to analyze the
interaction between TGF-β1 and p53. The results revealed that
inhibition of TGF-β1 could reduce the fluorescence intensity of
TGF-β1 and p53. Concurrently, overexpression of miR-29b along with
inhibition of TGF-β1 (siTGF-β1) significantly reduced the
fluorescence intensity of TGF-β1 and p53 compared with cells
overexpressing miR-29b and treated with control siRNA (Fig. 8).
Effects of miR-29b overexpression and
TGF-β1 suppression on the p53-mediated apoptotic pathway
RT-qPCR was used to investigate whether TGF-β1 could
impact the expression of p53 and thus jointly regulate the cell
growth process downstream of miR-29b in H22 cells. The mRNA levels
of p53 and Bax demonstrated a significant downward trend (P<0.01
and P<0.05, respectively), while the levels of Bcl-2 mRNA
increased in cells overexpressing miR-29b and treated with TGF-β1
siRNA compared with cells overexpressing miR-29b and treated with
control siRNA (P<0.05) (Fig.
9).
Discussion
The protein p53 is considered one of the most
popular tumor suppressor factors (14). As a cell cycle checkpoint, p53
can recognize cells with aberrant mutation, arresting the cell
cycle, inhibiting cell growth and inducing cell apoptosis (30). Several p53 mutations or
deficiencies have been revealed in malignant cells, enabling cancer
cells to escape and proliferate (15). TGF-β1 is a pleiotropic cytokine
that has been regarded as a double-edged sword in the progression
of cancer (20). In the early
stage of tumor development, TGF-β1 inhibits aberrant cell
proliferation, arrests the cell cycle in G1, and induces apoptosis
(21). However, in an
advanced-stage tumor, TGF-β1 prompts cancer evolution by enhancing
EMT, invasion, and metastasis (24,25).
The effect of TGF-β1 on H22 cells remains to be
elucidated. In the present study, the protein expression levels of
TGF-β1 and p53 were detected in normal hepatocytes and hepatoma
cells. Compared with mouse normal hepatocytes NCTC1469 cells,
hepatoma cells Hepa1-6 and H22 cells demonstrated low expression
levels of TGF-β1 and p53 proteins. These results indicated that low
levels of TGF-β1 and p53 may prompt hepatoma development.
Meanwhile, the RT-qPCR results revealed low expression of miR-29b
in Hepa1-6 and H22 cells compared with NCTC1469 cells, indicating
that miR-29b may act as a hepatoma suppressor.
miRNAs have been considered as diagnostic and
prognostic targets for cancer (31). A tumor-suppressing effect of
miR-29b has been reported in non-small cell lung cancer,
hepatocellular carcinoma and colorectal cancer (9-11). The present study demonstrated
that overexpression of miR-29b could decrease cell viability and
increase cell apoptosis in H22 cells. Conversely, inhibition of
miR-29b caused the opposite effects, indicating that miR-29b is a
suppressor in H22 cells. Furthermore, these results were consistent
with our previous studies that found an increased level of miR-29b
in H22 cells in ascites tumor-bearing mice treated with
supercritical-carbon dioxide fluid or scutellarin administration
(12,13).
Proliferation and apoptotic disorder have been
widely recognized as important mechanisms in the course of a
malignant tumor (32). Bcl-2 is
the most common anti-apoptotic protein, which can form a
heterodimer with the pro-apoptotic protein Bax, block the
transmission of apoptosis signals, and promote the survival and
growth of the cells. Bax can form dimers on its own and transmit
apoptosis signals to the caspase family, eventually inducing cell
apoptosis (33). Both Bcl-2 and
Bax are regulators of the mitochondrial pathway (33). It is noteworthy that p53 can
mediate Bcl-2 and Bax gene transcription to interact with the
mitochondrial pathway and regulate apoptosis together (34,35). Moreover, it has been reported
that miR-29b can induce apoptosis by directly targeting the
prototypical anti-apoptotic molecules Bcl-2 in hepatocellular
carcinoma cells or improving the level of Bax (11,36). Our data indicated that miR-29b
overexpression could facilitate transcription and translation of
p53, transcription of Bax and improve protein ratio of Bax/Bcl-2,
while inhibiting the transcription of Bcl-2. An opposite effect was
observed when using miR-29b inhibitor. The results indicated that
miR-29b has an important role in the activation of the p53-mediated
apoptotic pathway. Additionally, prompting heterodimer formation
between Bax and Bcl-2 may contribute to the activation of miR-29b
to the p53-mediated apoptotic pathway.
TGF-β1 is a pleiotropic cytokine regulating cell
growth, differentiation, migration and apoptosis (18). The classic TGF-β1 signaling
pathway is transduced by the Smad family (37). Smad3 transmits signals by binding
to the TGF-β1 receptor from the cytoplasm to the nucleus; Smad7
negatively regulates the TGF-β1 signaling pathway and is considered
as a candidate oncogene (38).
In the present study, miR-29b overexpression activated the TGF-β1
signal, facilitating the transcription of Smad3 and phosphorylation
of Smad3, and inhibiting transcription and translation of Smad7.
Thus, it was hypothesized that the effect of miR-29b on cell
proliferation and apoptosis in H22 cells may be regulated by the
activation of the TGF-β1 signaling pathway and the p53-mediated
apoptotic pathway. Furthermore, it was revealed that the inhibition
of miR-29b expression could inactivate p53 and TGF-β1 pathways to a
certain extent. However, this inactivation effect was modest
compared with miR-29b overexpression. Thus, miR-29b overexpression
was only focused on in further analysis.
Crosstalk between p53 and the TGF-β1 signaling
pathway has been observed in multiple diseases, including cancer
(26). TGF-β1 can activate p53,
induce the combination between p53 and Smad and further regulate
cell progression with TGF-β1 (39). Moreover, TGF-β1 can improve p53
expression in vascular senescence and apoptosis of hepatic stellate
cells (24,25). To further determine whether
miR-29b regulates H22 cells via the TGF-β1 signaling pathway and
the crosstalk between p53 and TGF-β1 in H22 cells, miR-29b was
overexpressed and cells were treated with exogenous TGF-β1. The
results demonstrated that the cell viability was significantly
reduced compared with cells transfected with miR-29b mimic but not
treated with exogenous TGF-β1, whereas the proliferation inhibited
by miR-29b overexpression was reversed with TGF-β1 inhibition and
the expression of a key element of the TGF-β1 signaling pathway.
These data indicated that TGF-β1 possessed an antitumor effect.
Additionally, the inhibitory effect of miR-29b on H22 cell
proliferation may be accomplished by TGF-β1 signaling pathway
activation.
Next, the expression of a key element of the p53
regulated apoptotic pathway was further detected. Immunofluorescent
results revealed that the p53 protein expression was in agreement
with TGF-β1, i.e., both were reduced by siTGF-β1 transfection.
Moreover, the improvement of p53 expression by miR-29b
overexpression was reversed once the TGF-β1 was inhibited.
Furthermore, the mRNA levels of p53 and Bax were decreased, and
Bcl-2 was increased by TGF-β1 inhibition even when miR-29b was
overexpressed. These data indicated that siTGF-β1 could reverse the
activation of TGF-β1 signal pathway which was induced by miR-29b
overexpression, as well as the p53-dependent apoptotic pathway.
These data also indicated that a crosstalk existed between TGF-β1
and p53 under the regulation of miR-29b. Moreover, it was possible
that miR-29b may inhibit H22 cell proliferation by regulating the
TGF-β1 signaling pathway, the p53-dependent apoptotic pathway, and
the crosstalk between TGF-β1 and p53.
However, there are some limitations in the present
study that need to be further assessed. Firstly, all the data in
the present study were obtained from H22 cells, miR-29b regulation
of TGF-β1 and p53 signaling pathways should be investigated with
additional cell lines in a future study. Secondly, as important
endogenous regulatory molecules, miRNAs inhibit the translation of
genes or directly accelerate mRNA degradation at the
post-transcriptional level (40). Nevertheless, in the present
study, when miR-29b was overexpressed, the level of TGF-β1 was
significantly increased. There is no direct report on how miR29b
upregulates TGF-β1 in cancer. Hence, the mechanism of miR29b
upregulation of the expression level of TGF-β1 requires thorough
investigation in a future study. Thirdly, the invasion assays were
carried out to determine the effect of miR29b on H22 cell invasion.
However, there was no significant difference between the control
group and the miR-29b intervention group (data not shown). The
aforementioned finding may be related to the cancer suppressive
effect of TGF-β1 in the present study. As a result, further
functional experiments also need to be carried out.
In conclusion, as a short-chain non-coding RNA,
miR-29b has an important role in H22 cells. It can reduce
proliferation and induce apoptosis of H22 cells by regulating the
TGF-β1 signaling pathway, the p53-dependent apoptotic pathway and
the crosstalk between TGF-β1 and p53. The present study enhanced
the functional knowledge of miR-29b and the understanding of
hepatic carcinoma molecular mechanisms. It may provide novel
insight into possible targets for hepatic carcinoma therapy.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, DJ and JNC made substantial contributions to the
conception and design of the study. YLL, BYC and JN acquired and
interpreted the data of the study. WHY, JNZ, STG analyzed the data
of the study. YL, DJ, ZRS drafted the study and revised it
critically for important intellectual content. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
You LN, Tai QW, Xu L, Hao Y, Guo WJ, Zhang
Q, Tong Q, Zhang H and Huang WK: Exosomal LINC00161 promotes
angiogenesis and metastasis via regulating miR-590-3p/ROCK axis in
hepatocellular carcinoma. Cancer Gene Ther. 28:719–736. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith EM and Jayson GC: The current and
future management of malignant ascites. Clin Oncol (R Coll Radiol).
15:59–72. 2003. View Article : Google Scholar
|
|
4
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Croce CM: 37 causes and consequences of
microRNA dysregulation in cancer. Eur J Cancer. 48(Suppl 5): S8–S9.
2012. View Article : Google Scholar
|
|
6
|
Yan B, Guo Q, Fu FJ, Wang Z, Yin Z, Wei YB
and Yang JR: The role of miR-29b in cancer: Regulation, function,
and signaling. Onco Targets Ther. 8:539–548. 2015.PubMed/NCBI
|
|
7
|
Wang LH, Huang J, Wu CR, Huang LY, Cui J,
Xing ZZ and Zhao CY: Downregulation of miR-29b targets DNMT3b to
suppress cellular apoptosis and enhance proliferation in pancreatic
cancer. Mol Med Rep. 17:2113–2120. 2018.
|
|
8
|
Cui H, Wang L, Gong P, Zhao C, Zhang S,
Zhang K, Zhou R, Zhao Z and Fan H: Deregulation between miR-29b/c
and DNMT3A is associated with epigenetic silencing of the CDH1
gene, affecting cell migration and invasion in gastric cancer. PLoS
One. 10:e01239262015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang B, Li W, Liu H, Yang L, Liao Q, Cui
S, Wang H and Zhao L: miR-29b suppresses tumor growth and
metastasis in colorectal cancer via downregulating Tiam1 expression
and inhibiting epithelial-mesenchymal transition. Cell Death Dis.
5:e13352014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen B, Wang J, Wang J, Wang H, Gu X, Tang
L and Feng X: A regulatory circuitry comprising TP53, miR-29
family, and SETDB1 in non-small cell lung cancer. Biosci Rep.
38:BSR201806782018. View Article : Google Scholar :
|
|
11
|
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y,
Jia WH and Zhuang SM: Effects of microRNA-29 on apoptosis,
tumorigenicity, and prognosis of hepatocellular carcinoma.
Hepatology. 51:836–845. 2010.
|
|
12
|
Yang HM, Sun CY, Liang JL, Xu LQ, Zhang
ZB, Luo DD, Chen HB, Huang YZ, Wang Q, Lee DY, et al:
Supercritical-carbon dioxide fluid extract from chrysanthemum
indicum enhances anti-tumor effect and reduces toxicity of
bleomycin in tumor-bearing mice. Int J Mol Sci. 18:4652017.
View Article : Google Scholar :
|
|
13
|
Nie J, Yang HM, Sun CY, Liu YL, Zhuo JY,
Zhang ZB, Lai XP, Su ZR and Li YC: Scutellarin enhances antitumor
effects and attenuates the toxicity of bleomycin in H22 ascites
tumor-bearing mice. Front Pharmacol. 9:6152018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He XX, Zhang YN, Yan JW, Yan JJ, Wu Q and
Song YH: CP-31398 inhibits the growth of p53-mutated liver cancer
cells in vitro and in vivo. Tumour Biol. 37:807–815. 2016.
View Article : Google Scholar
|
|
15
|
Liu L, Yu ZY, Yu TT, Cui SH, Yang L, Chang
H, Qu YH, Lv XF, Zhang XA and Ren CC: A Slug-dependent mechanism is
responsible for tumor suppression of p53-stabilizing compound
CP-31398 in p53-mutated endometrial carcinoma. J Cell Physiol.
235:8768–8778. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo H, Zhang Z, Su Z, Sun C, Zhang X, Zhao
X, Lai X, Su Z, Li Y and Zhan JY: Enhanced anti-tumor activity and
reduced toxicity by combination andrographolide and bleomycin in
ascitic tumor-bearing mice. Eur J Pharmacol. 776:52–63. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Avasarala S, Van Scoyk M, Wang J, Sechler
M, Vandervest K, Brzezinski C, Weekes C, Edwards MG, Arcaroli J,
Davis RE, et al: Hsa-miR29b, a critical downstream target of
non-canonical Wnt signaling, plays an anti-proliferative role in
non-small cell lung cancer cells via targeting MDM2 expression.
Biol Open. 2:675–685. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bierie B and Moses HL: Tumour
microenvironment: TGFbeta: The molecular Jekyll and Hyde of cancer.
Nat Rev Cancer. 6:506–520. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou H, Wang K, Hu Z and Wen J: TGF-β1
alters microRNA profile in human gastric cancer cells. Chin J
Cancer Res. 25:102–111. 2013.PubMed/NCBI
|
|
20
|
Zhenye L, Chuzhong L, Youtu W, Xiaolei L,
Lei C, Lichuan H, Hongyun W, Yonggang W, Fei W and Yazhuo Z: The
expression of TGF-β1, Smad3, phospho-Smad3 and Smad7 is correlated
with the development and invasion of nonfunctioning pituitary
adenomas. J Transl Med. 12:712014. View Article : Google Scholar
|
|
21
|
Buenemann CL, Willy C, Buchmann A,
Schmiechen A and Schwarz M: Transforming growth
factor-beta1-induced Smad signaling, cell-cycle arrest and
apoptosis in hepatoma cells. Carcinogenesis. 22:447–452. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Johansson J, Tabor V, Wikell A, Jalkanen S
and Fuxe J: TGF-β1-induced epithelial-mesenchymal transition
promotes monocyte/macrophage properties in breast cancer cells.
Front Oncol. 5:32015. View Article : Google Scholar
|
|
23
|
Han G and Wang XJ: Roles of TGFβ signaling
Smads in squamous cell carcinoma. Cell Biosci. 1:412011. View Article : Google Scholar
|
|
24
|
Zhang G, Feng W and Wu J: Down-regulation
of SEPT9 inhibits glioma progression through suppressing
TGF-β-induced epithelial-mesenchymal transition (EMT). Biomed
Pharmacother. 125:1097682020. View Article : Google Scholar
|
|
25
|
Takahashi K, Menju T, Nishikawa S, Miyata
R, Tanaka S, Yutaka Y, Yamada Y, Nakajima D, Hamaji M, Ohsumi A, et
al: Tranilast inhibits TGF-β1-induced epithelial-mesenchymal
transition and invasion/metastasis via the suppression of Smad4 in
human lung cancer cell lines. Anticancer Res. 40:3287–3296. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Samarakoon R, Higgins SP, Higgins CE and
Higgins PJ: The TGF-β1/p53/PAI-1 signaling axis in vascular
senescence: Role of caveolin-1. Biomolecules. 9:3412019. View Article : Google Scholar
|
|
27
|
Saile B, Matthes N, El Armouche H,
Neubauer K and Ramadori G: The bcl, NFkappaB and p53/p21WAF1
systems are involved in spontaneous apoptosis and in the
anti-apoptotic effect of TGF-beta or TNF-alpha on activated hepatic
stellate cells. Eur J Cell Biol. 80:554–561. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun Y, Xia P, Zhang H, Liu B and Shi Y:
P53 is required for Doxorubicin-induced apoptosis via the TGF-beta
signaling pathway in osteosarcoma-derived cells. Am J Cancer Res.
6:114–125. 2016.PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Kim S, Lee JH, Kang I, Hyun S, Yu J and
Shin C: An amphiphilic peptide induces apoptosis through the
miR29b-p53 pathway in cancer cells. Mol Ther Nucleic Acids.
5:e3302016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tu HC, Jacobs SC, Borkowski A and
Kyprianou N: Incidence of apoptosis and cell proliferation in
prostate cancer: Relationship with TGF-beta1 and bcl-2 expression.
Int J Cancer. 69:357–363. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oltvai ZN, Milliman CL and Korsmeyer SJ:
Bcl-2 heterodimerizes in vivo with a conserved homolog, bax, that
accelerates programmed cell death. Cell. 74:609–619. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fries KL, Miller WE and Raab-Traub N:
Epstein-Barr virus latent membrane protein 1 blocks p53-mediated
apoptosis through the induction of the A20 gene. J Virol.
70:8653–8659. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miyashita T and Reed JC: Tumor suppressor
p53 is a direct transcriptional activator of the human bax gene.
Cell. 80:293–299. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu S, Wang T, Luo F, Li H, Jia Q, He T,
Wu H and Zou T: Astaxanthin inhibits proliferation and induces
apoptosis of LX-2 cells by regulating the miR-29b/Bcl-2 pathway.
Mol Med Rep. 19:3537–3547. 2019.PubMed/NCBI
|
|
37
|
Liang H, Wang Q, Wang D, Zheng H,
Kalvakolanu DV, Lu H, Wen N, Chen X, Xu L, Ren J, et al: RGFP966, a
histone deacetylase 3 inhibitor, promotes glioma stem cell
differentiation by blocking TGF-β signaling via SMAD7. Biochem
Pharmacol. 180:1141182020. View Article : Google Scholar
|
|
38
|
Li B, Yin GF, Wang YL, Tan YM, Huang CL
and Fan XM: Impact of fecal microbiota transplantation on
TGF-β1/Smads/ERK signaling pathway of endotoxic acute lung injury
in rats. 3 Biotech. 10:522020. View Article : Google Scholar
|
|
39
|
Sivadas VP and Kannan S: The microRNA
networks of TGFβ signaling in cancer. Tumour Biol. 35:2857–2869.
2014. View Article : Google Scholar
|
|
40
|
Aure MR, Jernstrom S, Krohn M; Due Oslo
Breast Cancer Consortium E; Mills GB, Borresen-Dale AL, Sahlberg
KK, Lingjaerde OC and Kristensen VN: 331: Integrative analysis
reveals extensive association between microRNA expression and
mRNA-protein translation. Eur J Cancer. 50(Suppl 5): S792014.
View Article : Google Scholar
|