Introduction
As one of the most prevalent malignant tumor types
worldwide, bladder cancer exhibited the fifth highest incidence
rate among new confirmed cases of cancer in men in the USA and the
estimated death toll reached 13,050, accounting for 4% of total
cancer-related deaths in 2020 (1). According to the European
Association of Urology guidelines, the therapeutic options for
bladder cancer include radical surgery, neo-adjuvant therapy and
traditional chemotherapy (2).
The high drug resistance to traditional chemotherapy is a challenge
for the treatment of bladder cancer. Although neo-chemotherapy
based on cisplatin was considered promising, 40% of patients
experienced therapeutic failure due to acquired chemoresistance
(3,4). Therefore, exploring the mechanism
underlying the development of cisplatin resistance in bladder
cancer cells is important for the optimization of therapeutic
strategies.
Sphingosine kinase 1 (SPHK1), which phosphorylates
sphingosine to generate sphingosine-1-phosphate (S1P) (5), has been demonstrated to exert
various functions at different stages of cancer progression
(6,7). SPHK1 exists in the cytoplasm and is
phosphorylated (8) to
participate in mediating the sensitivity of cancer cells to their
corresponding chemotherapy drugs (9). For instance, SPHK1 overexpression
contributes to drug resistant to oxaliplatin in the human colon
cancer RKO cell line, whereas SPHK1 downregulation increases drug
sensitivity to oxaliplatin in the human colon cancer HCT116 cell
line (10). A number of studies
have reported that SPHK1 could affect the chemoresistance of
colorectal cancer by regulating STAT3 expression, influencing
tumorigenesis, proliferation and progression (10,11). However, the effect of SPHK1 in
promoting cisplatin resistance in bladder cancer cells is not
completely understood.
The present study demonstrated that that SPHK1
inhibited apoptosis and promoted cisplatin resistance by forming a
complex with the RNA binding protein non-POU domain containing
octamer binding (NONO) in bladder cancer cells. Furthermore, NONO
modulated STAT3 activity and displayed a negative association with
cisplatin responsiveness. The identification of the
SPHK1/NONO/STAT3 axis may provide promising insight into the
biological characteristics of bladder cancer cells and aid with the
identification of alternative therapeutic strategies.
Materials and methods
Statistics acquisition
A total of 411 transcriptome profiling files from
430 samples were extracted from The Cancer Genome Atlas (TCGA;
https://portal.gdc.cancer.gov/) Genomic
Data Commons. Subsequently, two gene sets of chemotherapy-resistant
bladder cancer cells [GSE77883 (12) and GSE58624 (13)] and four gene sets of
cisplatin-resistant cancer types [GSE58624/bladder, GSE108214/lung
(14), GSE140996/ovarian
(15) and GSE77515/breast
(16)] were obtained from the
Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) of National Center
for Biotechnology Information.
The identification of differentially expressed genes
(DEGs) was performed by GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r). P<0.05 and
|logFC|>1.5 were set as the cut-off criteria. The Venn diagram
reflecting the differentially expressed genes was acquired using
bio-Venn software (https://www.biovenn.nl/). The Kaplan-Meier survival
curve analysis was performed using Gene Expression Profiling
Interactive Analysis (http://gepia.cancer-pku.cn/). The cut-off/threshold
value was Quartile: Cutoff-High (75%); Cutoff-Low (25%).
Patients and tissue samples
Bladder cancer and adjacent healthy tissue samples
were obtained from 10 patients admitted to the First Affiliated
Hospital of Chongqing Medical University (Chongqing, China;
Table I) between April 2020 and
January 2021. The fresh samples were washed with PBS and then
stored at −80°C prior to RNA extraction. Written informed consent
was obtained from patients. The present study was approved by
Ethics Committee of the First Affiliated Hospital of Chongqing
Medical University (ethical approval no. 2021086).
 | Table IPatient information. |
Table I
Patient information.
| ID | Age | Sex | Grade | T stage | N stage | M stage | Start | End |
|---|
| 1999981 | 78 | Male | Unknown | Unknown | Unknown | Unknown | 20.06.10 | 20.06.16 |
| 2023999 | 58 | Male | High | T1 | N1 | M0 | 20.04.05 | 20.04.16 |
| 1903064 | 63 | Female | High | T4 | N0 | M0 | 21.02.06 | 21.02.15 |
| 2030137 | 56 | Male | High | T2a | N0 | M0 | 20.09.16 | 20.09.29 |
| 2002228 | 74 | Male | Low | T2b | N0 | M0 | 20.10.11 | 20.10.20 |
| 2004550 | 73 | Male | High | T3 | N2 | M0 | 21.01.15 | 21.01.24 |
| 2006935 | 74 | Male | High | T4 | N2 | M0 | 21.01.15 | 21.01.26 |
| 2025171 | 65 | Male | High | T1 | N0 | M0 | 20.06.30 | 20.07.10 |
| 1984663 | 64 | Male | High | T1 | N0 | M0 | 20.08.31 | 20.09.08 |
| 1990615 | 51 | Male | High | T3b | N0 | M0 | 20.07.15 | 20.07.23 |
Cell lines and chemical reagents
Human bladder cancer cell lines (T24, UMUC-3 and
5637) and normal urothelial cells (SV-HUC-1) were purchased from
the American Type Culture Collection. Cells were cultured in
RPMI-1640 (Corning, Inc.; for T24 and 5637 cells), DMEM (Gibco;
Thermo Fisher Scientific, Inc.; for UMUC-3 cells) or F-12K (for
SV-HUC-1 cells) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified incubator containing 5%
CO2. Cisplatin was purchased from Beijing Solarbio
Science & Technology Co., Ltd. FTY-720 was purchased from
GLPBio. T24 cells (1.0 µg/ml) were exposed to increasing
concentrations of cisplatin (0.05, 0.1, 0.2, 0.4, 0.6, 0.8 or 1.0
µg/ml) for 6 months to generate cisplatin-resistant strains
(T24/DDP).
Small interfering RNA (siRNA) and
lentivirus transfection
All small interfering RNAs (siRNAs) targeting human
SPHK1 (siRNA1, siRNA2, siRNA3) and the scrambled negative control
(NC) were designed and synthetized by Shanghai GenePharma Co., Ltd.
For siRNA transfection, T24 and UMUC-3 cells were cultured in
6-well plates (5×103 cells/well). At 80% confluence, the
culture media was replaced with 1.5 ml basal medium containing 500
ml Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and 0.2 nmol siRNA. The sequences of the siRNAs
were as follows: si-SPHK1-1#, 5′-GCA GGC AUA UGG AGU AUG ATT UCA
UAC UCC AUA UGC CUG CTT -3′; si-SPHK1-2#, 5′-GCG UCAUGC AUC UGU UCU
ATT UAG AAC AGA UGC AUG ACG CTT -3′; si-SPHK1-3#, 5′-GAG GCU GAA
AUC UCC UUC ATT UGA AGG AGA UUU CAG CCU CTT -3′; si-NC, 5′-UUC UCC
GAA CGU GUC ACG UTT A CG UGA CAC GUU CGG AGA ATT -3.
To create cell lines overexpressing SPHK1 or a
control vector, a lentiviral system from Shanghai GenePharma Co.,
Ltd. was used. Empty vector was used as a negative control. For
lentivirus transduction, at 50% confluence, medium containing
lentivirus and polybrominated biphenyls was added to T24 cells at a
multiplicity of infection of 50, according to the manufacturer's
protocol. Polybrene was used to improve the infection efficiency.
The transfection temperature was 37°C for 24 h. At 24 h
post-transfection, the supernatant was replaced with RPMI-1640,
followed by culture for a further 24 h at 37°C. Puromycin (2 ng/ml)
was added to the medium for stable selection of transfected cell
lines.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from bladder cancer and
adjacent healthy tissue samples, as well as cell lines (at 70%
confluence) using TRIzol® reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. Total
RNA (1 µg) was reverse transcribed into cDNA using the
PrimeScript RT reagent kit (Takara Bio, Inc.) according to the
manufacturer's instructions. qPCR was performed using SYBR Green
assays (Takara Bio, Inc.) and an ABI 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.; 94°C 2 min,
one cycle; 94°C 40 sec, 50°C 40 sec, 72°C 1 min for 35 cycles and
72°C for 5 min and one cycle). Relative mRNA expression levels were
quantified using the 2−Δ∆Cq method (17) and normalized to the internal
reference gene GAPDH. The following primers were used for qPCR:
SPHK1 forward, 5′-GCG UCA UGC AUC UGU UCU ATT -3′ and reverse,
5′-UAG AAC AGA UGC AUG ACG CTT -3′; GAPDH forward, 5′-GCA GCG AGA
TCC CTC CAA AAT -3′ and reverse, 5′-GGC TGT TGT CAT ACT TCT CAT
GG-3′. The experiments were replicated three times.
Cell proliferation assay
Cell proliferation was analyzed by performing Cell
Counting Kit-8 (CCK-8) assays. At 48 h post-siRNA transfection,
UMUC-3 and T24 cells were seeded (5×103 cells/plate)
into 96-well plate. After incubation for 48 h, cells were incubated
with CCK-8 reagent for 1 h in the dark. The number of viable cells
was determined by measuring the absorbance at a wavelength of 450
nm by a microplate reader (Varioskan LUX; Thermo Fisher Scientific,
Inc.).
Flow cytometry
siRNA-transfected UMUC-3 and T24 cells were seeded
into 6-well plates and incubated with cisplatin (0, 5 or 10
µg/ml) for 48 h. After washing twice with PBS, cells
(~1×106/ml) were resuspended in 500 ml PBS. Cells were
transferred to 1.5 ml Eppendorf tubes and stained using the FITC
Annexin V Apoptosis Detection kit II (BD Biosciences, cat. no.
556570) and incubated in the dark for 10 min at room temperature
according to the manufacturer's protocol. Subsequently, 10
µl propidium iodide (PI, Beyotime Institute of
Biotechnology) was added to the stained cells and apoptotic cells
were distinguished by a fluorescence-activated cell sorting
analyzer (FACS; BD Biosciences). The results were analyzed by
Cytexpert V2.3 (Beckman Coulter, Inc.) software. The apoptotic rate
was calculated by the percentage of early + late apoptotic
cells.
Western blotting
Proteins were extracted from cell lines using RIPA
lysis buffer (Beyotime Institute of Biotechnology) containing 50
mmol/l Tris-HCl (pH 7.4), 150 mmol/l NaCl, 1% Triton X-100, 1
mmol/l EDTA (pH 8.0) and 1 mmol/l phenylmethylsulfonyl fluoride.
Total protein concentrations were measured using the BCA
quantitative kit (Beyotime Institute of Biotechnology). Proteins
(30 µg) were separated via 12% SDS-PAGE and transferred to
PVDF membranes. After blocking with 5% skimmed milk for 2 h at room
temperature, the membranes were incubated overnight at 4°C with the
following primary antibodies: SPHK1 (cat. no. 10670-1-AP;
ProteinTech Group, Inc.; 1:1,000), GAPDH (cat. no. AB0037; Shanghai
Abways Biotechnology Co., Ltd.; 1:5,000;), NONO (cat. no. 385171;
ZenBio, Inc.; 1:1,000), phosphorylated (p)-STAT3 (cat. no. 380906;
ZenBio, Inc.; 1:1,000), Bax (cat. no. 50599-2-Ig; Cell Signaling
Technology, Inc.; 1:1,000), Bcl2 (cat. no. 12789-1-AP; Cell
Signaling Technology, Inc.; 1:1,000) and cleaved caspase-3 (cat.
no. 9661; Cell Signaling Technology, Inc.; 1:1,000). After washing
three times with TBS-Tween (0.1%)-20 (TBST), the membranes were
incubated with HRP-conjugated secondary antibodies (Cell Signaling
Technology, Inc.) for 2 h at room temperature. After three washes
with TBST, the bands were visualized using an ECL system (Bio-Rad
Laboratories, Inc.). Protein expression was semi-quantified using
Quantity One software (v4.4.0.36; Bio-Rad Laboratories, Inc.).
Co-Immunoprecipitation (Co-IP)
After seeding and culture in a 10-cm culture dish
for 48 h, cells were washed with PBS and harvested with a specific
IP buffer at 4°C for 30 min. Cells were centrifuged at 12,000 × g
and 4°C for 15 min to collect the lysate. Subsequently, 2.5
µg SPHK1 and IgG primary antibodies were added to each
sample, followed by rotation at 4°C overnight with a rolling
incubator (cat. no. QB-128; Kylin-Bell Instruments Co., Ltd.).
Protein A/G beads were added to the cell lysate and then rotated
for 7 h at 4°C. The beads were washed three times with IP buffer on
a magnetic shelf and then boiled in 2X SDS loading buffer for 10
min. For mass spectrometry experiments, SPHK1-overexpression T24
cells were harvested and lysed at 4°C. Subsequently, the IP
experiment was performed as aforementioned. Tubes containing beads
were sent to Trump Biotechnology Co., Ltd. for mass spectrometry
and further analysis.
Statistical analysis
Data were expressed as the mean ± standard
deviation. GraphPad Prism 8 software (GraphPad Software, Inc.) and
SPSS 22.0 (IBM Corp.) were used for statistical analyses. For
statistical comparisons, a paired t-test was used when comparing
matched samples and an unpaired t-test for non-matched samples.
Dunnett test was the post hoc test used following two-way ANOVA for
multiple comparisons. For the Kaplan-Meier survival curve analysis,
log-rank was used to compare the curves. P<0.05 was considered
to indicate a statistically significant difference. Figures were
produced using Adobe Photoshop CC 2018 (Adobe Systems, Inc.).
Results
Identifying the target gene SHPK1
The gene sets related to chemoresistance in patients
with bladder cancer were demonstrated by the gene sets obtained
from the GEO database (Fig. 1A and
B). The Venn diagram revealed two cross-linked genes among the
885 upregulated genes in the GSE77883 gene set and 36 upregulated
genes in the GSE58624 gene set (Fig.
1C). There were six cross-link genes among the four gene sets
of selected cisplatin-resistant cancer types (Fig. 1D). Comprehensive analysis of the
results demonstrated that SPHK1 was the only gene contained in both
cohorts. Therefore, SPHK1 was selected as the target gene in the
present study.
SHPK1 is associated with the prognosis of
patients with bladder cancer
A higher expression level of SPHK1 was observed in
bladder cancer tissues compared with those in healthy adjacent
tissues both in the 19 paired samples (P=0.00; Fig. 2A) and TCGA database of 430
clinical samples (P=0.016; Fig.
2B). Kaplan-Meier curves revealed that patients in the high
expression SHPK1 group displayed a notably lower overall survival
rate compared with those in the low expression group in GEPIA
database (Fig. 2C).
Immunohistochemical staining images from the Human Protein Atlas
database (https://www.proteinatlas.org/) showed that SPHK1
intensity in bladder cancer tissues was stronger compared with that
in adjacent mucosa (Fig. 2D).
The RT-qPCR results demonstrated that the higher gene expression of
SPHK1 was not only observed in all three bladder cancer cell lines,
including T24, 5637 and UMUC-3 (Fig.
2E), but also in the tumor tissues of extracted patients
samples (Fig. 2F). Collectively,
the results suggested that SPHK1 was upregulated in bladder cancer
and was significantly correlated with the prognosis of
patients.
SPHK1 knockdown increases cisplatin
sensitivity and promoted the proliferation of bladder cancer cell
lines
The role of SPHK1 in the development of cisplatin
resistance in bladder cancer cells was investigated. In T24 and
UMUC-3 cell lines, SPHK1 knockdown (Fig. 3A and B) led to increased
sensitivity to cisplatin treatment compared with the negative
control (Fig. 3C and D). At a
concentration of 1 µg/ml, the two bladder cancer cells were
most sensitive to cisplatin treatment (P<0.001). In addition,
the flow cytometry results demonstrated that SPHK1 knockdown
displayed a higher rate of apoptosis compared with the negative
control (Fig. 3E and F). The
results demonstrated that SPHK1 knockdown promoted cell
proliferation, increased cisplatin sensitivity and induced
apoptosis in bladder cancer cells.
SPHK1 overexpression decreases apoptosis
in T24 bladder cancer cells
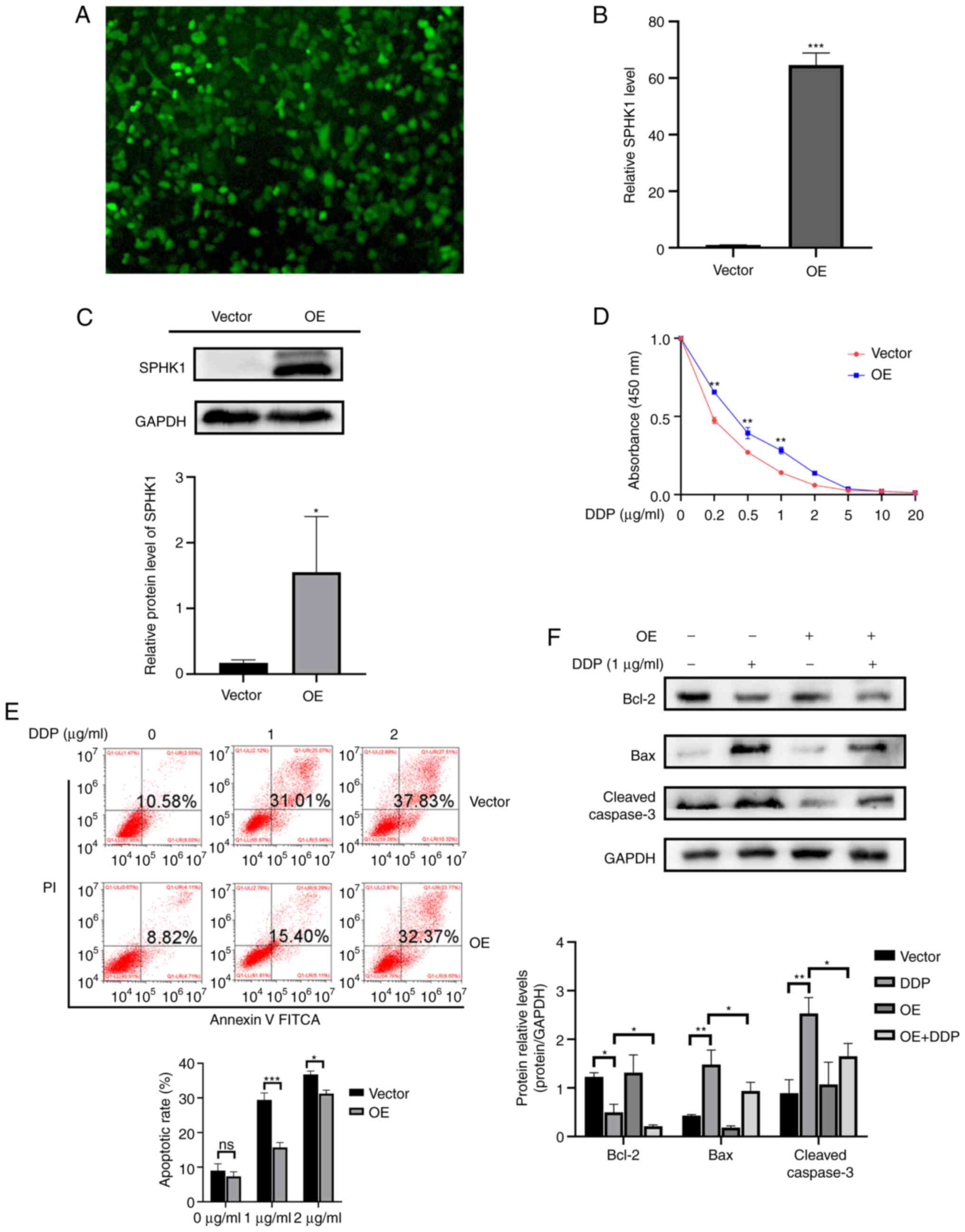
First, SPHK1-overexpression cell lines were
constructed by lentivirus transfection (Fig. 4A). The RT-qPCR and western
blotting results showed that the expression level of SPHK1 was
significantly increased after viral transduction (Fig. 4B and C). The CCK-8 assay results
displayed that SPHK1 overexpression significantly promoted cell
proliferation (Fig. 4D). Flow
cytometry results showed that SPHK1 overexpression also decreased
the apoptotic rate (Fig. 4E).
The expression of apoptosis-related proteins, including Bcl-2, Bax
and cleaved caspase-3, were detected using western blotting. In
SPHK1-overexpression cell lines, Bcl-2 was significantly
downregulated, whereas Bax and cleaved caspase-3 were upregulated.
Compared with normal T24 cells, SPHK1-overexpression T24 cell lines
exhibited higher expression levels of Bcl-2 and lower expression
levels of Bax and cleaved caspase-3 following treatment with
cisplatin (Fig. 4F). The
aforementioned results demonstrated that SPHK1 overexpression
contributed to cisplatin resistance and decreased apoptosis in
bladder cancer cells.
SPHK1 regulates STAT3 expression via
binding to NONO
Co-IP was performed to explore the mechanism
underlying SPHK1-mediated regulation of apoptosis in bladder cancer
cells (Fig. 5A). Based on the
mass spectrometry results, a novel protein, NONO, was identified
(Fig. 5B). The western blotting
results showed that SPHK1-overexpression cell lines displayed
higher expression levels of NONO and its downstream signaling
molecule p-STAT3 compared with those in normal T24 cells (Fig. 5C).
SPHK1 inhibition reverses antiapoptotic
effects in bladder cancer cells
Our previous results demonstrated that SPHK1 is
responsible for cisplatin resistance in bladder cancer cells.
Numerous studies have demonstrated that FTY-720 is an inhibitor of
SPHK1 (18,19) and may induce apoptosis in bladder
cancer cells (20). Hence,
whether inhibition of SPHK1 by FTY-720 could reverse antiapoptotic
effects and increase cisplatin sensitivity in bladder cancer cells
was investigated. The western blotting results demonstrated that
there was a significant decrease in SPHK1 expression after FTY-720
administration (Fig. 6A). In
addition, SPHK1 knockdown led to decreases in NONO and STAT3
expression (Fig. 6B), suggesting
that NONO may serve as a downstream regulator of SPHK1.
Furthermore, the expression levels of the apoptosis-related
proteins Bax and cleaved caspase-3 were increased, whereas Bcl-2
expression levels were decreased after SPHK1 inhibition (Fig. 6C), indicating that SPHK1-induced
antiapoptotic effects were dependent on NONO-regulated STAT3
activation.
SPHK1 and corresponding proteins in
establishing cisplatin-resistant bladder cancer cell lines
Cisplatin-resistant T24 bladder cancer cells
(T24/DDP) were established (Fig. 7A
and B). SPHK1, NONO and p-STAT3 were overexpressed in the
T24/DDP cell lines (Fig. 7C and
D). Following SPHK1 knockdown in T24/DDP cell line, the CCK-8
assay, flow cytometry and western blotting results displayed
similar results compared with previous studies (Fig. 7E-H). Therefore, the results
demonstrated that SPHK1 promoted cisplatin resistance in bladder
cancer cells via binding with NONO and regulating the activation of
STAT3.
 | Figure 7Validation of SPHK1 and its
corresponding proteins in the T24/DDP cell line. (A) DPP-resistant
T24 bladder cancer cells were established and drug resistance was
detected by performing the CCK-8 assay (1 µg/ml:
***P<0.001 T24/DDP vs. T24; 2 µg/ml:
***P<0.001 T24/DDP vs. T24). (B) Apoptosis was
analyzed in T24 vs. T24/DDP (1 µg/ml:
***P<0.001 T24/DDP vs. T24; 2 µg/ml:
***P<0.001 T24/DDP vs. T24). (C and D) SPHK1, NONO
and p-STAT3 were overexpressed in the T24/DDP cell line
(***P<0.001). Following SPHK1 knockdown in the
T24/DDP cell line, the results of (E and F) western blotting, (G)
flow cytometry and (H) CCK-8 assay were similar to those reported
in previous studies (*P<0.05, **P<0.01
and ***P<0.001). SPHK1, sphingosine kinase 1; DPP,
cisplatin; CCK-8, Cell Counting Kit-8; NONO, non-POU domain
containing octamer binding; p-, phosphorylated; OE,
overexpression. |
Discussion
Tumor progression is regulated by various mechanisms
(21-23). The present study demonstrated
that SPHK1 overexpression was significantly associated with the
development of chemoresistance in bladder cancer cells. Elevated
SPHK1 enhanced the chemoresistance to cisplatin and contributed to
poor survival rates in patients with bladder cancer. The
chemoresistance to cisplatin was reversed by SPHK1 knockdown via
siRNA transfection or FTY-720 treatment.
SPHK1 is reported to influence the biological
behaviors of cancer cells, including angiogenesis, cell
proliferation and motility, survival, autophagy and apoptosis
(9). Mediating apoptosis via
suppression of ceramide (24)
might be one of the most important oncological implications for
overcoming chemoresistance in various cancer cells, leading to an
improved prognosis of patients. SPHK1 can induce chemoresistance
and inhibit apoptosis in numerous types of cancer, such as acute
myeloid leukemia (25), chronic
myeloid leukemia (26) and
breast cancer (27) and is
regarded as a potential sensor to chemotherapy in prostate cancer
(28). However, the role of
SPHK1 in regulating cisplatin-induced apoptosis in bladder cancer
remains to be elucidated. To the best of the authors' knowledge,
the present study demonstrated for the first time that SPHK1
inhibition increased the expression of Bax and cleaved caspase-3,
but downregulated the expression of the antiapoptotic protein
Bcl-2. The CCK-8 assay results demonstrated that SPHK1 was
positively associated with bladder cancer cell proliferation.
SPHK1-overexpression T24 cells displayed a lower sensitivity to
cisplatin treatment. In addition, the present study indicated that
inhibition of SPHK1 by FTY-720 enhanced the apoptotic effect of
cisplatin treatment in bladder cancer cells.
RNA binding proteins (RBPs) have recently emerged as
critical factors that regulate multiple cellular activities,
including pre-mRNA splicing, translation and protein sub-cellular
localization (29,30). NONO is a novel RBP that is
located in the nucleus of numerous mammalian cells and distributed
in the sub-nuclear domain and participates in almost every step of
gene regulation (31). Studies
have found that NONO also serves an important role in
tumorigenesis, including proliferation regulation, DNA damage
repair, cell migration and apoptosis (32). For example, silencing NONO can
inhibit cell attachment to laminin, poly-l-lysine11 and the surface
of culture plates, thus reducing cell migration in melanoma
(32) and esophageal cancer
(33). Notably, NONO is found to
inhibit tumor metastasis in bladder cancer cells (34). Liang et al (35) identified that NONO directly
interacted with splicing factor proline/glutamine rich to regulate
the splicing of SET domain and mariner transposase fusion gene,
thus suppressing the invasion of bladder cancer cells. NONO is also
found to promote apoptosis by activating caspase-3 and Bax proteins
(36), which is consistent with
the results of the present study. In addition, the IP results
further indicated that NONO may serve as a pivotal regulator of
SPHK1-induced cisplatin resistance and SPHK1 could bind to NONO to
inhibit cisplatin-induced apoptosis.
Although the antiapoptotic effect of SPHK1 on
bladder cancer cells has been confirmed, the specific mechanism
remains unclear. It has been previously reported that SPHK1
phosphorylates sphingosine to generate S1P to activate STAT3
(37). The mechanisms underlying
STAT3-mediated induction of cisplatin resistance are complex,
including reducing uptake (38-40), cisplatin inactivation (41,42) and most importantly, increasing
DNA damage repair (43). It has
been reported that cisplatin can form inter- and intrastrand
crosslinked DNA adducts in cells and activate the proapoptotic
protein P73 (44) and the
accumulation of P37 could result in the release of cytochrome
c and ultimately lead to cell apoptosis (45,46). DNA mismatch repair (MMR) is a
process that corrects mismatched nucleotides by the recognition of
inter- and intrastrand DNA adducts and the consequent release of an
apoptotic signal. MMR is often impaired in cisplatin-resistant
cancer cells (47). MMR
deficiency usually results in downregulation of MutS homolog 3
(MSH3), which is associated with cisplatin sensitivity and patient
survival (47). Notably, STAT3
activation regulates MSH3 expression in colorectal cancer cells
(48). The present study
demonstrated that upregulation of p-STAT3 led to increased
apoptosis in bladder cancer cells and thus it was suggested that
one of the mechanisms underlying SPHK1-induced cisplatin resistance
might be regulation of STAT3 expression to activate the downstream
signature and inhibit the MMR pathway; however, further
investigations are required.
Furthermore, DNA damaged-induced cisplatin
resistance can also be reversed by homologous recombination (HR), a
non-STAT3-dependent pathway that repairs the double strand breaks
(49). Since the HR pathway has
been found to serve a role in platinum-based therapy resistance of
breast cancer, HR was inhibited by NONO in vivo (50) and the present study demonstrated
that SPHK1 could regulate the expression of NONO and it was
indicated that SPHK1 might induce cisplatin resistance through the
STAT3-dependent pathway, as well as the non-STAT3-dependent HR
pathway.
The present study had a number of limitations.
First, the lack of in vivo experiments may affect the
credibility of the present study. Second, although the present
study confirmed the existence of SPHK1/NONO/STAT3 axis, the
mechanisms underlying DNA double strands repair are still unclear
and require further research.
To the best of the authors' knowledge, the present
study demonstrated for the first time that SPHK1 may promote
cisplatin resistance and inhibit apoptosis in bladder cancer cells.
The antiapoptotic effect of SPHK1 was exerted via binding with the
DNA regulating protein NONO. In addition, the combination of SPHK1
and NONO may activate STAT3 to promote the survival of bladder
cancer cells.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available at TCGA dataset (https://portal.gdc.cancer.gov/) and GEO datasets
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58624;
https://www.ncbi.nlm.nih.gov/
geo/query/acc.cgi?acc= GSE108214; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140996;
and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77515.
Authors' contributions
HT and WH conceived the study. HT and ZQ collected
the data. TL, HC, JZ and SY performed the experiments. WH provided
project administration and resources. JZ provided software. ZQ
drafted the manuscript. HT reviewed and edited the manuscript. HT
and ZQ confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by Ethics Committee
of the First Affiliated Hospital of Chongqing Medical University
(Ethical approval number: 2021086-Ethics Committee of the First
Affiliated Hospital of Chongqing Medical University; 3 March
2021.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors thank the Laboratory Research Center,
The First Affiliated Hospital of Chongqing Medical University
(Chongqing, China) for technical support.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81874092).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar
|
|
2
|
Witjes JA, Bruins HM, Cathomas R, Compérat
EM, Cowan NC, Gakis G, Hernández V, Linares Espinós E, Lorch A,
Neuzillet Y, et al: European association of urology guidelines on
muscle-invasive and metastatic bladder cancer: Summary of the 2020
guidelines. Eur Urol. 79:82–104. 2021. View Article : Google Scholar
|
|
3
|
Choi W, Porten S, Kim S, Willis D, Plimack
ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL, et al:
Identification of distinct basal and luminal subtypes of
muscle-invasive bladder cancer with different sensitivities to
frontline chemotherapy. Cancer Cell. 25:152–165. 2014. View Article : Google Scholar
|
|
4
|
Shah JB, McConkey DJ and Dinney CP: New
strategies in muscle-invasive bladder cancer: On the road to
personalized medicine. Clin Cancer Res. 17:2608–2612. 2011.
View Article : Google Scholar
|
|
5
|
Nguyen AV, Wu YY and Lin EY: STAT3 and
sphingosine-1-phosphate in inflammation-associated colorectal
cancer. World J Gastroenterol. 20:10279–10287. 2014. View Article : Google Scholar
|
|
6
|
Maceyka M, Harikumar KB, Milstien S and
Spiegel S: Sphingosine-1-phosphate signaling and its role in
disease. Trends Cell Biol. 22:50–60. 2012. View Article : Google Scholar
|
|
7
|
Pyne S, Edwards J, Ohotski J and Pyne NJ:
Sphingosine 1-phosphate receptors and sphingosine kinase 1: Novel
biomarkers for clinical prognosis in breast, prostate, and
hematological cancers. Front Oncol. 2:1682012. View Article : Google Scholar
|
|
8
|
Pitson SM, Powell JA and Bonder CS:
Regulation of sphingosine kinase in hematological malignancies and
other cancers. Anticancer Agents Med Chem. 11:799–809. 2011.
View Article : Google Scholar
|
|
9
|
Vadas M, Xia P, McCaughan G and Gamble J:
The role of sphingosine kinase 1 in cancer: Oncogene or
non-oncogene addiction? Biochim Biophys Acta. 1781:442–447. 2008.
View Article : Google Scholar
|
|
10
|
Nemoto S, Nakamura M, Osawa Y, Kono S,
Itoh Y, Okano Y, Murate T, Hara A, Ueda H, Nozawa Y and Banno Y:
Sphingosine kinase isoforms regulate oxaliplatin sensitivity of
human colon cancer cells through ceramide accumulation and Akt
activation. J Biol Chem. 284:10422–10432. 2009. View Article : Google Scholar
|
|
11
|
Shen Z, Feng X, Fang Y, Li Y, Li Z, Zhan
Y, Lin M, Li G, Ding Y and Deng H: POTEE drives colorectal cancer
development via regulating SPHK1/p65 signaling. Cell Death Dis.
10:8632019. View Article : Google Scholar
|
|
12
|
Kameyama K, Horie K, Mizutani K, Kato T,
Fujita Y, Kawakami K, Kojima T, Miyazaki T, Deguchi T and Ito M:
Enzalutamide inhibits proliferation of gemcitabine-resistant
bladder cancer cells with increased androgen receptor expression.
Int J Oncol. 50:75–84. 2017. View Article : Google Scholar
|
|
13
|
Tanaka N, Kosaka T, Miyazaki Y, Mikami S,
Niwa N, Otsuka Y, Minamishima YA, Mizuno R, Kikuchi E, Miyajima A,
et al: Acquired platinum resistance involves epithelial to
mesenchymal transition through ubiquitin ligase FBXO32
dysregulation. JCI Insight. 1:e836542016. View Article : Google Scholar
|
|
14
|
Sarin N, Engel F, Rothweiler F, Cinatl J,
Michaelis M, Frötschl R, Fröhlich H and Kalayda GV: Key players of
cisplatin resistance: Towards a systems pharmacology approach. Int
J Mol Sci. 19:7672018. View Article : Google Scholar
|
|
15
|
Wantoch von Rekowski K, König P, Henze S,
Schlesinger M, Zawierucha P, Januchowski R and Bendas G: The impact
of integrin-mediated matrix adhesion on cisplatin resistance of W1
ovarian cancer cells. Biomolecules. 9:7882019. View Article : Google Scholar
|
|
16
|
Chisholm CL, Wang H, Wong AH,
Vazquez-Ortiz G, Chen W, Xu X and Deng CX: Ammonium
tetrathiomolybdate treatment targets the copper transporter ATP7A
and enhances sensitivity of breast cancer to cisplatin. Oncotarget.
7:84439–84452. 2016. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Huwiler A and Zangemeister-Wittke U: The
sphingosine 1-phosphate receptor modulator fingolimod as a
therapeutic agent: Recent findings and new perspectives. Pharmacol
Ther. 185:34–49. 2018. View Article : Google Scholar
|
|
19
|
Lim KG, Tonelli F, Li Z, Lu X, Bittman R,
Pyne S and Pyne NJ: FTY720 analogues as sphingosine kinase 1
inhibitors: Enzyme inhibition kinetics, allosterism, proteasomal
degradation, and actin rearrangement in MCF-7 breast cancer cells.
J Biol Chem. 286:18633–18640. 2011. View Article : Google Scholar
|
|
20
|
Azuma H, Takahara S, Horie S, Muto S,
Otsuki Y and Katsuoka Y: Induction of apoptosis in human bladder
cancer cells in vitro and in vivo caused by FTY720 treatment. J
Urol. 169:2372–2377. 2003. View Article : Google Scholar
|
|
21
|
Sun Z, Niu S, Xu F, Zhao W, Ma R and Chen
M: CircAMOTL1 promotes tumorigenesis through miR-526b/SIK2 axis in
cervical cancer. Front Cell Dev Biol. 8:5681902020. View Article : Google Scholar
|
|
22
|
Chen M, Zhuang C, Liu Y, Li J, Dai F, Xia
M, Zhan Y, Lin J, Chen Z, He A, et al: Tetracycline-inducible shRNA
targeting antisense long non-coding RNA HIF1A-AS2 represses the
malignant phenotypes of bladder cancer. Cancer Lett. 376:155–164.
2016. View Article : Google Scholar
|
|
23
|
Chen M, Wei X, Shi X, Lu L, Zhang G, Huang
Y and Hou J: LncRNA HIF1A-AS2 accelerates malignant phenotypes of
renal carcinoma by modulating miR-30a-5p/SOX4 axis as a ceRNA.
Cancer Biol Med. 18:587–603. 2021. View Article : Google Scholar
|
|
24
|
Cuvillier O, Pirianov G, Kleuser B, Vanek
PG, Coso OA, Gutkind S and Spiegel S: Suppression of
ceramide-mediated programmed cell death by sphingosine-1-phosphate.
Nature. 381:800–803. 1996. View
Article : Google Scholar
|
|
25
|
Powell JA, Lewis AC, Zhu W, Toubia J,
Pitman MR, Wallington-Beddoe CT, Moretti PA, Iarossi D, Samaraweera
SE, Cummings N, et al: Targeting sphingosine kinase 1 induces
MCL1-dependent cell death in acute myeloid leukemia. Blood.
129:771–782. 2017. View Article : Google Scholar
|
|
26
|
Bonhoure E, Lauret A, Barnes DJ, Martin C,
Malavaud B, Kohama T, Melo JV and Cuvillier O: Sphingosine kinase-1
is a downstream regulator of imatinib-induced apoptosis in chronic
myeloid leukemia cells. Leukemia. 22:971–979. 2008. View Article : Google Scholar
|
|
27
|
Ruckhäberle E, Rody A, Engels K, Gaetje R,
von Minckwitz G, Schiffmann S, Grösch S, Geisslinger G, Holtrich U,
Karn T and Kaufmann M: Microarray analysis of altered sphingolipid
metabolism reveals prognostic significance of sphingosine kinase 1
in breast cancer. Breast Cancer Res Treat. 112:41–52. 2008.
View Article : Google Scholar
|
|
28
|
Pchejetski D, Golzio M, Bonhoure E, Calvet
C, Doumerc N, Garcia V, Mazerolles C, Rischmann P, Teissié J,
Malavaud B and Cuvillier O: Sphingosine kinase-1 as a chemotherapy
sensor in prostate adenocarcinoma cell and mouse models. Cancer
Res. 65:11667–11675. 2005. View Article : Google Scholar
|
|
29
|
Gerstberger S, Hafner M and Tuschl T: A
census of human RNA-binding proteins. Nat Rev Genet. 15:829–845.
2014. View
Article : Google Scholar
|
|
30
|
Fox AH and Lamond AI: Paraspeckles. Cold
Spring Harb Perspect Biol. 2:a0006872010. View Article : Google Scholar
|
|
31
|
Feng P, Li L, Deng T, Liu Y, Ling N, Qiu
S, Zhang L, Peng B, Xiong W, Cao L, et al: NONO and tumorigenesis:
More than splicing. J Cell Mol Med. 24:4368–4376. 2020. View Article : Google Scholar
|
|
32
|
Schiffner S, Zimara N, Schmid R and
Bosserhoff AK: p54nrb is a new regulator of progression of
malignant melanoma. Carcinogenesis. 32:1176–1182. 2011. View Article : Google Scholar
|
|
33
|
Cheng R, Zhu S, Guo S, Min L, Xing J, Guo
Q, Li P and Zhang S: Downregulation of NONO induces apoptosis,
suppressing growth and invasion in esophageal squamous cell
carcinoma. Oncol Rep. 39:2575–2583. 2018.
|
|
34
|
Xie R, Chen X, Cheng L, Huang M, Zhou Q,
Zhang J, Chen Y, Peng S, Chen Z, Dong W, et al: NONO inhibits
lymphatic metastasis of bladder cancer via alternative splicing of
SETMAR. Mol Ther. 29:291–307. 2021. View Article : Google Scholar
|
|
35
|
Liang J, Nagahashi M, Kim EY, Harikumar
KB, Yamada A, Huang WC, Hait NC, Allegood JC, Price MM, Avni D, et
al: Sphingosine-1-phosphate links persistent STAT3 activation,
chronic intestinal inflammation, and development of
colitis-associated cancer. Cancer Cell. 23:107–120. 2013.
View Article : Google Scholar
|
|
36
|
Lee H, Deng J, Kujawski M, Yang C, Liu Y,
Herrmann A, Kortylewski M, Horne D, Somlo G, Forman S, et al:
STAT3-induced S1PR1 expression is crucial for persistent STAT3
activation in tumors. Nat Med. 16:1421–1428. 2010. View Article : Google Scholar
|
|
37
|
Bosch-Barrera J, Queralt B and Menendez
JA: Targeting STAT3 with silibinin to improve cancer therapeutics.
Cancer Treat Rev. 58:61–69. 2017. View Article : Google Scholar
|
|
38
|
Zhu H, Luo H, Zhang W, Shen Z, Hu X and
Zhu X: Molecular mechanisms of cisplatin resistance in cervical
cancer. Drug Des Devel Ther. 10:1885–1895. 2016. View Article : Google Scholar
|
|
39
|
Pabla N, Murphy RF, Liu K and Dong Z: The
copper transporter Ctr1 contributes to cisplatin uptake by renal
tubular cells during cisplatin nephrotoxicity. Am J Physiol Renal
Physiol. 296:F505–F511. 2009. View Article : Google Scholar
|
|
40
|
Galluzzi L, Vitale I, Michels J, Brenner
C, Szabadkai G, Harel-Bellan A, Castedo M and Kroemer G: Systems
biology of cisplatin resistance: Past, present and future. Cell
Death Dis. 5:e12572014. View Article : Google Scholar
|
|
41
|
Li S, Li C, Jin S, Liu J, Xue X, Eltahan
AS, Sun J, Tan J, Dong J and Liang XJ: Overcoming resistance to
cisplatin by inhibition of glutathione S-transferases (GSTs) with
ethacraplatin micelles in vitro and in vivo. Biomaterials.
144:119–129. 2017. View Article : Google Scholar
|
|
42
|
Henrique R, Nunes SP and Jerónimo C: MSH2
expression and resistance to cisplatin in muscle-invasive bladder
cancer: A mix of progress and challenges. Eur Urol. 75:251–252.
2019. View Article : Google Scholar
|
|
43
|
Tanida S, Mizoshita T, Ozeki K, Tsukamoto
H, Kamiya T, Kataoka H, Sakamuro D and Joh T: Mechanisms of
cisplatin-induced apoptosis and of cisplatin sensitivity: Potential
of BIN1 to Act as a potent predictor of cisplatin sensitivity in
gastric cancer treatment. Int J Surg Oncol. 2012:8628792012.
|
|
44
|
Viktorsson K, Ekedahl J, Lindebro MC,
Lewensohn R, Zhivotovsky B, Linder S and Shoshan MC: Defective
stress kinase and Bak activation in response to ionizing radiation
but not cisplatin in a non-small cell lung carcinoma cell line. Exp
Cell Res. 289:256–264. 2003. View Article : Google Scholar
|
|
45
|
Adams JM: Ways of dying: Multiple pathways
to apoptosis. Genes Dev. 17:2481–2495. 2003. View Article : Google Scholar
|
|
46
|
Kunkel TA and Erie DA: DNA mismatch
repair. Annu Rev Biochem. 74:681–710. 2005. View Article : Google Scholar
|
|
47
|
Vaisman A, Varchenko M, Umar A, Kunkel TA,
Risinger JI, Barrett JC, Hamilton TC and Chaney SG: The role of
hMLH1, hMSH3, and hMSH6 defects in cisplatin and oxaliplatin
resistance: Correlation with replicative bypass of platinum-DNA
adducts. Cancer Res. 58:3579–3585. 1998.
|
|
48
|
Tseng-Rogenski SS, Hamaya Y, Choi DY and
Carethers JM: Interleukin 6 alters localization of hMSH3, leading
to DNA mismatch repair defects in colorectal cancer cells.
Gastroenterology. 148:579–589. 2015. View Article : Google Scholar
|
|
49
|
Rocha C, Silva MM, Quinet A, Cabral-Neto
JB and Menck C: DNA repair pathways and cisplatin resistance: An
intimate relationship. Clinics (Sao Paulo). 73(Suppl 1): e478s2018.
View Article : Google Scholar
|
|
50
|
Johnson N, Johnson SF, Yao W, Li YC, Choi
YE, Bernhardy AJ, Wang Y, Capelletti M, Sarosiek KA, Moreau LA, et
al: Stabilization of mutant BRCA1 protein confers PARP inhibitor
and platinum resistance. Proc Natl Acad Sci USA. 110:17041–17046.
2013. View Article : Google Scholar
|