Introduction
Breast cancer (BC) is the most common type of cancer
among women worldwide, and the leading cause of cancer-associated
mortality. According to the global cancer burden data released by
the International Agency for Cancer Research of the World Health
Organization, the number of new BC cases worldwide reached 2.26
million in 2020; this was the first time that BC cases surpassed
the number of lung cancer cases (2.21 million) to become the most
common type of cancer in the world (1). Therefore, it is necessary to
determine the underlying molecular mechanisms of the occurrence and
development of BC to develop effective methods for early diagnosis
and treatment.
Dickkopf (DKK) protein family members, which consist
of DKK1-4, as well as DKK3-related proteins, are secretory proteins
(2). The DKK proteins are
glycoproteins composed of 255–355 amino acids, which contain a
signal sequence and two conserved cysteine-rich domains (3). DKK3 has been discovered to act as a
tumor suppressor gene (4), and
physiologically functions as an antagonist of the Wnt receptor
complex in the Wnt signaling pathway; therefore, it can inhibit the
Wnt signaling pathway (5). When
DKK3 is inactivated, the Wnt protein binds to its receptor and
excessive activation of this signaling pathway can lead to
tumorigenesis (5,6). The expression levels of DKK3 have
been found to be downregulated in numerous types of cancer cells,
including melanoma, gallbladder cancer and BC (7,8), and
its overexpression was discovered to inhibit the proliferation of
cancer cells. However, the upstream regulatory mechanism of DKK3 in
BC remains to be clarified, to the best of our knowledge.
MicroRNAs (miRNAs/miRs) are a type of non-protein
coding RNA, 19-25 nucleotides in length, which play a variety of
regulatory roles in the processes of cell proliferation and
development (9). miRNAs bind with
the 3′-untranslated regions of their target mRNA in a complete or
incomplete complementary manner, resulting in a reduction in
translation or cleavage of the target mRNA (10). Multiple studies have shown that the
expression levels of miR-25 were significantly upregulated in a
variety of tumor tissues and cells, which could promote cell
invasion and proliferation by targeting DKK3 (11,12).
However, whether miR-25 can target DKK3 in BC has not been
reported, to the best of our knowledge.
Long non-coding RNAs (lncRNAs) are another type of
non-coding RNA that are >200 nucleotides in length and lack
protein-coding ability. Previous studies have shown that lncRNAs
are associated with the occurrence and development of numerous
types of human disease, including cancer and Alzheimer's disease
(13,14). In addition, studies have
demonstrated that lncRNA acts as a competitive endogenous RNA
(ceRNA) (15–17). For example, hepatocellular
carcinoma upregulated lncRNA HULC was found to act as an endogenous
sponge of miR-372, downregulating its expression, thereby reducing
the inhibitory effect on the translation of its target genes
(18).
The present study aimed to investigate the role of
the lnc-MICAL2-1/miR-25/DKK3 signaling axis in BC. First, the
expression levels of lnc-MICAL2-1 and DKK3 in BC tissues were
analyzed using data obtained from The Cancer Genome Atlas (TCGA).
Cell proliferation, invasion and migration were determined
following the knockdown or overexpression of lnc-MICAL2-1. RNA
pull-down and dual-luciferase reporter assays verified the
targeting relationship between lnc-MICAL2-1 and miR-25, and between
miR-25 and DKK3. In addition, the effects of the overexpression or
knockdown of lnc-MICAL2-1 on DKK3 expression and the Wnt signaling
pathway were further evaluated in a nude mouse xenograft model.
Taken together, the results of the present study indicated that the
lnc-MICAL2-1/miR-25/DKK3 signaling axis may play an important role
in BC, suggesting that the activation of lnc-MICAL2-1/DKK3 or
inhibition of miR-25 may be beneficial to the treatment of BC.
Materials and methods
Bioinformatics analysis
GeneCards (genecards.org/) database was used to
explore the upstream regulatory mechanism of DKK3 in tumor cells.
Prediction of lncRNA and miRNA, and miRNA and target gene binding
sites was performed using TargetScan (targetscan.org/vert_72/) and
miRanda (microrna.org/microrna/home.do) databases. The Atlas of
non-coding RNAs in Cancer (TANRIC) database
(ibl.mdanderson.org/tanric/design/basic/download.html), which is an
interactive data analysis and visualization platform (19), was used to analyze the expression
of lnc-MICAL2-1 in BC tissues and calculate the correlation
coefficient between lnc-MICAL2-1 and DKK3 expression, which was
subsequently displayed as a scatter diagram. In addition, TANRIC
was used to study the relationship between lnc-MICAL2-1 expression,
clinical characteristics and overall survival in BC. The
corresponding clinical data of the patients in TANRIC were obtained
from The Cancer Genome Atlas (TCGA) data portal
(portal.gdc.cancer.gov/; normal, n=105; tumor, n=837).
Cell lines and culture
MDA-MB-231, MCF-7 and 293T cells cell lines were
purchased from the American Type Culture Collection and cultured as
described previously (20).
MDA-MB-231 and MCF-7 cells were cultured in DMEM (Invitrogen;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.). 293T cells were cultured in DMEM
containing 10% FBS, 2 mM L-glutamine and 50 µg/ml gentamicin. All
cells were cultured at 37°C in a humidified incubator with 5%
CO2.
Cell transfection
miR-25 mimics (5′-AGGCGGAGACUUGGGCAAUUG-3′), miR-25
inhibitor (5′-CAAUUGCCCAAGUCUCCGCCU-3′), miR-25 mimics control
(5′-UUGUACUACACAAAAGUACUG-3′) and miR-25 inhibitor control
(5′-CAGUACU-UUUGUGUAGUACAA-3′) were purchased from Sangon Biotech
Co., Ltd. Cells were seeded (4×105 cells/well) into
6-well cell culture plates containing 2 ml complete medium. Ay
50–80% confluence, MCF-7 cells were transfected with miR-25 mimics
(50 nM), miR-25 mimics control (50 nM), miR-25 inhibitor (100 nM)
or miR-25 inhibitor control (100 nM) using riboFECT CP Transfection
kit (Guangzhou RiboBio Co., Ltd.) at 37°C for 48 h.
pcDNA3.1/V5-His-TOPO vector (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to construct the lnc-MICAL2-1 overexpression and
knockdown vectors. pcDNA3.1/V5-His-TOPO empty vector was used as
the overexpression negative control. The sequences of the shRNAs
were as follows: sh-lnc-MICAL2-1 (2 µg;
5′-AATGGCAAACACTGAGCTGCT-3′), sh-NC (2 µg;
5′-TTCTCCGAACGTGTCACGT-3′). Vectors (2 µg) were transfected into
cells using FuGENE® HD transfection reagent (Roche
Diagnostics GmbH) at 37°C for 48 h, according to the manufacturer's
protocol. After 48 h of transfection, transfection efficiencies
were determined using reverse transcription-quantitative PCR
(RT-qPCR).
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay (Dojindo Molecular Technologies, Inc.)
was used to study cell viability. A total of 2×103
cells/well were plated in 96-well plates and cultured for 24, 48,
72 and 96 h at 37°C. Subsequently, 10 µl CCK-8 solution was added
to each well and incubated with the cells at 37°C with 5%
CO2 for 4 h. The absorbance was measured at a wavelength
of 450 nm.
Wound healing assay
Cells were cultured for 24 h until the cell
confluence reached ~90%. A scratch was then made in the cell
monolayer using a pipette tip to create an artificial wound. The
cells were washed twice with PBS and incubated with serum-free DMEM
at 37°C. The cells were visualized in the same position after 0, 24
and 48 h to determine the migratory distance. Images were obtained
under a light microscope. Wound healing was calculated according to
the following formula: Relative wound healing (%)=(0 h scratch
distance-24/48 h scratch distance)/0 h scratch distance ×100.
Transwell assays
The Transwell assays were performed as described by
Li et al (21). Briefly,
the cells were cultured in serum-free DMEM, then trypsinized. The
cell density was adjusted to 1×106 cells/ml, and cells
were seeded into the upper chamber of a 24-well Transwell culture
plate. DMEM supplemented with 10% FBS was added to the lower
chamber of the Transwell plate. The cells were then incubated for
24 h for evaluation of migration. After 24 h, the cells on the
surface of the lower chamber membrane were fixed with 5%
glutaraldehyde for 15 min at room temperature and then stained with
0.1% crystal violet for 20 min at room temperature. Stained cells
were visualized using a light microscope in three randomly selected
fields of view. To evaluate the invasive ability of cells, 200 µl
cells (1×106 cells/ml) were seeded into the upper
chambers of Transwell plates with 50 mg/l Matrigel (8 µm; cat. no.
CLS3374; Corning, Inc.) and serum-free medium after Matrigel
precoating at 37°C for 1 h. 500 µl DMEM supplemented with 10% FBS
was added to the lower chamber. Following incubation at 37°C for 24
h, the cells remaining in the upper chamber were removed, whereas
cells in the lower chamber were fixed with 5% glutaraldehyde for 15
min at 4°C, washed with PBS and stained with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. Stained
cells were visualized using a light microscope in three randomly
selected fields of view.
Colony formation assay
After 24 h of transfection, cells were digested with
0.25% trypsin to prepare a single cell suspension. The cells were
then seeded into a 6-well culture plate at a density of 500
cells/well (three repeats/experimental group) and cultured at 37°C
in a 5% CO2 incubator. The culture medium was changed
every 3 days during the culture period. After 15 days, the culture
was terminated upon the appearance of visible colonies in the
culture dish. The medium was subsequently discarded, the cells were
washed with pre-cooled (4°C) PBS 2–3 times, and then fixed with 5
ml pure methanol for 15 min at room temperature. The cells were
washed another three times with PBS and subsequently stained with
Giemsa staining solution for 15 min at room temperature. Stained
cells were visualized using a light microscope (magnification, ×5)
to count the number of colonies with >10 cells.
RNA pull-down assay
Biotinylated miR-25-5p probe
(5′-AGGCGGAGACUUGGGCAAUUG-Biotin-3′) and control probe
(5′-GTTAACGGGTTCAGAGGCGGA-Biotin-3′) were synthesized by Sangon
Biotech Co., Ltd. First, cells (1×107) were lysed with
500 µl RIP lysis buffer centrifuged at 4°C and 12,000 × g for 15
min, and the cell lysate in the supernatant was collected. The
Pierce Magnetic RNA-Protein Pull-Down kit (Thermo Fisher
Scientific, Inc.) was used for RNA pull-down experiments.
Biotinylated RNA probes (miR-25-5p probe and control probe) and 400
µl streptavidin magnetic beads (Thermo Fisher Scientific, Inc.)
were incubated with cell lysates overnight at 4°C. After proteinase
K digestion at 55°C for 20 min, the RNA attached to the magnetic
beads was separated twice with washing buffer, and then centrifuged
at 5,000 × g for 30 sec at 4°C to collect the Bio-miR/mRNA mixture,
after which RT-qPCR was performed.
Dual-luciferase reporter assay
TargetScan (targetscan.org/vert_72/) and miRanda
(microrna.org/microrna/home.do) databases were used to predict the
target genes of miR-25. The reporter vector pmirGLO-DKK3-wild-type
(wt), pmirGLO-DKK3-mutant (mut), pmirGLO-MICAL2-1-wt and
pmirGLO-MICAL2-1-mut plasmids, or synthetic miR-NC mimics (forward,
5′-UUGUACUACACAAAAGUACUG-3′ and reverse,
5′-GUACUUUUGUGUAGUACAAUU-3′) or miR-25 mimics (forward,
5′-AGGCGGAGACUUGGGCAAUUG-3′ and reverse,
5′-AUUGCCCAAGUCUCCGCCUUU-3′) were obtained from Sangon Biotech Co.,
Ltd. Upon cell confluence reaching 70–80%, each reporter plasmid
was co-transfected into 293T cells alongside miR-NC mimics or
miR-25 mimics using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). A total of 24 h after
transfection, the relative firefly luciferase activity was detected
using a Dual-Glo Luciferase Reporter assay system, according to the
manufacturer's protocol (Promega Corporation). The relative
luciferase activity was normalized to Renilla luciferase
activity.
Fluorescence colocalization
analysis
Cells transfected with the lnc-MICAL2-1
overexpression or control NC vector for 48 h, or transfected with
lnc-MICAL2-1 knockdown or control NC vector were washed with PBS
once and fixed with methanol for 15 min at room temperature.
Subsequently, cells were incubated with an anti-β-catenin primary
antibody (1:500; cat. no. ab32572; Abcam) overnight at 4°C. After
primary antibody incubation, the cells were incubated with a
TRITC-conjugated goat anti-rabbit IgG antibody (cat. no. TA130015
1:200; OriGene Technologies, Inc.) for 1 h at room temperature. The
nuclei were counterstained with DAPI for 5 min at room temperature.
Stained cells were visualized using a fluorescence microscope.
Cell cycle analysis
After 72 h of transfection, cells were trypsinized
and 2×105 cells/ml were plated in a 6-well culture
plate. The cells were fixed with 75% ice-cold ethanol at 4°C for at
least 30 min, then washed with PBS to remove the fixative solution.
Following the addition of 100 µl RNase, the cells were incubated in
a water bath at 37°C for 30 min. Cells were subsequently incubated
with 400 µl propidium iodide (PI; Sigma-Aldrich; Merck KGaA) at 4°C
in the dark for 30 min. A ACEA NovoCyte flow cytometer (ACEA
Bioscience, Inc.) with NovoExpress Software (version 1.0.2; ACEA
Biosciences, Inc.) was used for cell cycle distribution
analysis.
Flow cytometry analysis of
apoptosis
Cells in the logarithmic growth phase were collected
and seeded into a 6-well plate at a density of 1×105
cells/well, then incubated with 5% CO2 at 37°C for 18 h.
Cells were then transfected with lnc-MICAL2-1 overexpression,
knockdown or NC vectors. After 48 h, 1–5×105 cells were
harvested and resuspended in 1X PBS. A total of 5 µl Annexin V-FITC
and 5 µl PI was added and incubated at room temperature for 5 min
in the dark. Apoptotic cells were analyzed using a flow cytometer
(FACSCalibur; BD Biosciences) with BD Cell Quest Pro Software
(version 3.3; Becton, Dickinson and Company).
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The purity of total RNA was detected and quantified using an
UV spectrophotometer (260 nm to calculate RNA concentration and
260/280 nm to evaluate RNA purity). Then, total RNA (1 µg) was
reverse transcribed into cDNA using a reverse transcription system
(cat. no. A3500; Promega Corporation), according to the
manufacturer's protocol. Primer3web software (version 4.1.0;
http://primer3.ut.ee/) was used to design the
following primers for qPCR: GAPDH (NM_001256799.3) forward,
5′-GAAAGCCTGCCGGTGACTAA-3′ and reverse, 5′-TTCCCGTTCTCAGCCTTGAC-3′;
DKK3 (NM_001018057.2) forward, 5′-CCTGGCAAACTTACCTCCCA-3′ and
reverse, 5′-CATTTTGGTGCAGTGACCCC-3′; lnc-MICAL2-1 forward,
5′-TCAGCCTGCCTGCCAATATG-3′ and reverse, 5′-AAGCAGTGCTGCAAGGAAGA-3′;
miR-25-5p forward, 5′-AGGCGGAGACTTGGGCAATTG-3′ and reverse,
5′-GTGCGTGTCGTGGAGTCG-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACACAATTG-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. qPCR was performed on an ABI 7500
Real-Time PCR Detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The thermocycling conditions were as follows:
Initial denaturation at 95°C for 10 min; 40 cycles of denaturation
at 95°C for 10 sec, annealing and extension at 60°C for 1 min. mRNA
and miRNA expression levels were determined using the
2−ΔΔCq method (22).
GAPDH was used as the mRNA housekeeping gene and U6 was used as the
miRNA housekeeping gene.
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (Beyotime Institute of Biotechnology). Total protein
was quantified using a BCA protein assay kit (Bio-Rad Laboratories,
Inc.), loaded on a 12% SDS gel and resolved using 10% SDS-PAGE
(NuPAGE; Invitrogen; Thermo Fisher Scientific, Inc.). The separated
proteins were subsequently transferred to PVDF membranes (Whatman
plc; Cytiva) and blocked with 5% non-fat dry milk powder (Merck
KGaA) at room temperature for 2 h. The PVDF membrane was then
incubated with DKK3 (1:1,000; cat. no. ab187532; Abcam) primary
antibodies at 4°C overnight. After the primary antibody incubation,
the membranes were washed with TBS-Tween 20 (0.05%) three times and
incubated with a goat anti-rabbit secondary antibody (cat. no.
bs-0295G-HRP; 1:3,000; BIOSS) for 2 h at room temperature. Protein
bands were visualized using chemiluminescence reagent on a gel
imaging system (Thermo Fisher Scientific, Inc.). The band densities
were analyzed using ImageJ software (version 4.6; National
Institutes of Health). The gray value of the target protein was
normalized to the gray value of the internal reference protein,
GAPDH, and the data are presented as the relative content of the
target protein in a sample.
Establishment of a xenograft
model
A total of 63 male BALB/c-nude mice (5-week-old;
weight 18–20 g; Chengdu Dashuo Laboratory Animal Co., Ltd.) were
maintained in aseptic conditions at 25°C and 40–60% humidity, with
12-h light/dark cycles, and free access to food and water. Mice
were subcutaneously injected with 1×107 MCF-7 cells
stably transfected with lnc-MICAL2-1 overexpression, knockdown or
NC. After inoculation, the tumor volume was measured once a week.
After 6 weeks, the mice (n=6 per group) were euthanized via
cervical dislocation, the tumor tissue was harvested, and the
weight and volume were measured. The volume of the tumor was
calculated using the following equation: Volume=length ×
width2 ×0.5, where the length represented the longest
diameter and the width represented the shortest diameter of the
tumor. The maximum tumor diameter observed in the present study was
1.84 cm and the maximum tumor volume was 1,935.3 mm3. To
evaluate the survival of mice, 15 mice from each group were
observed until the 10th week. If ulceration, infection or necrosis
occurred, or mice were on the verge of death, the experiment was
terminated and the mice were euthanized. The survival rate of
tumor-bearing mice after inoculation with tumor cells was analyzed
using the Kaplan-Meier method. The log-rank (Mantel-Cox) test was
used to determine statistical differences in the survival curves.
All animal procedures and experimental methods were approved by the
Committee on the Ethics of Animal Experiments of Southern Medical
University (approval no. SYXK 2020–0012). This study was performed
in strict accordance with the recommendations in the Guide for the
Care and Use of Laboratory Animals of the National Institutes of
Health (23), and in accordance
with the Animal Research: Reporting In Vivo Experiments
guidelines (24).
Hematoxylin and eosin (H&E)
staining
Tumor tissues were fixed with 4% neutral
formaldehyde for 24 h at room temperature, routinely embedded in
paraffin and cut into 4-µm slices. After conventional dewaxing, the
sections were stained with hematoxylin at room temperature for 5
min and eosin for 3 min at room temperature. Then, graded alcohol
dehydration, xylene clearing and neutral gum sealing were
performed. The morphological changes of tumor cells were observed
under a light microscope.
Immunohistochemistry
Tumor tissues were embedded in paraffin, sectioned
into 5-µm slices, heated for 2 h at 60°C, Subsequently, the tissues
were deparaffinized with xylene I and II (each for 10 min), and
dehydrated in a gradient alcohol series. Heat-mediated antigen
retrieval was performed with citrate buffer (pH 6), then the
sections were immersed in 3% hydrogen peroxide for 10 min to block
endogenous peroxidase, followed by blocking with 5% bovine serum
albumin (Wuhan Servicebio Technology Co., Ltd.) for 30 min at room
temperature. The sections were subsequently incubated with 50 µl
anti-DKK3 rabbit monoclonal antibody (1:100; cat. no. ab187532;
Abcam), MMP-25 (1:100; cat. no. ab56309; Abcam) overnight at 4°C.
Following the primary antibody incubation, the sections were washed
with PBS for 2 min and incubated with 50 µl biotinylated goat
anti-rabbit IgG (1:100; cat. no. bs-0295G-HRP; BIOSS) at 37°C for
30 min. Sections were then washed with PBS three times (15 min
each), and then incubated with high sensitivity streptavidin-HRP
conjugate (Biosynthesis Biotechnology Co., Ltd.) for 30 min at
37°C. Sections were stained with DAB (Biosynthesis Biotechnology
Co., Ltd.) for 8 min at room temperature and hematoxylin for 5 min
at room temperature, then dehydrated in gradient ethanol for 1 min,
made transparent with xylene and sealed with neutral gum. The
immunostaining images were captured using a light microscope
(magnification, ×400). The integrated optical density in the
selected area was calculated in five non-overlapping fields of view
of each section as the statistical area. Image-Pro Plus 6.0
software (Media Cybernetics, Inc.) was used for the analysis.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5.0 software (GraphPad Software, Inc.). Statistical
differences between two groups were compared using the
non-parametric Mann Whitney-U test, whereas a Kruskal-Wallis test
and Dunn's post hoc test was used for the comparisons between more
than two groups. The differential expression of lncRNAs was
analyzed using an unpaired Student's t-test between tumor and
adjacent normal tissues. Spearman's rank correlation analysis was
used to determine the correlation between lncRNA and mRNA
expression. The nonparametric Kaplan-Meier method was used for
survival analysis. The log-rank (Mantel-Cox) test was used to
compare the difference between survival curves. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression levels of lnc-MICAL2-1 in
BC, and association with DKK3 expression and clinicopathological
parameters
To determine the underlying mechanism of
downregulated DKK3 expression in BC, the GeneCards bioinformatics
tool was used. The results revealed that lnc-MICAL2-1 may regulate
DKK3 expression via modulation of miR-25. The expression levels of
lnc-MICAL2-1 in the tumor tissues of patients with breast invasive
carcinoma (BRCA) obtained from TCGA database were analyzed. The
results demonstrated that the expression levels of lnc-MICAL2-1
were downregulated in human BRCA tissues compared with those in
normal tissues The results also found that the expression levels of
lnc-MICAL2-1 were significantly downregulated in tumor tissues
(n=837) compared with in adjacent normal tissues (n=105) (Fig. 1A). Breast cancer is divided
according to the expression matrix by molecular profiling (GEP).
For example, PAM50-based GEP can divide breast cancer into
different subtypes, including normal-like, basal, luminal A,
luminal B and Her2. Analysis of lnc-MICAL2-1 expression in the
PAM50 subtype of BRCA samples identified significant differences
between the basal and luminal A subtypes (Fig. 1B). Correlation analysis in basal,
luminal B and human epidermal growth factor receptor 2 (Her2)
subtypes of BC identified a positive correlation between
lnc-MICAL2-1 and DKK3 mRNA expression in the basal (correlation,
0.3569581; P<0.05; n=139), luminal B (correlation, 0.4869612;
P<0.05; n=191) and Her2 (correlation, 0.4401873; P<0.05;
n=67) subtypes (Fig. 1C-E).
According to the median expression of lnc-MICAL2-1 in all patients,
the patients were divided into high expression and low expression
groups. Survival analysis showed that high expression levels of
lnc-MICAL2-1 decreased the overall survival rate of BC and was thus
indicative of a poorer prognosis (Fig.
1F). The association between the expression of lnc-MICAL2-1 and
the estrogen receptor, progesterone receptor and therapy of
patients with BC was statistically significant (P<0.05), but
there was no association with staging and HER2 expression
(P>0.05) (Fig. 1G-K).
 | Figure 1.lnc-MICAL2-1 expression is
downregulated in BC and is associated with DKK3 expression. (A)
TANRIC database analysis of the expression of lnc-MICAL2-1 in BC
tissues: Normal tissues, n=105; tumor tissues, n=837. P<0.05.
(B) Expression of lnc-MICAL2-1 in the PAM50 subtype of BC. TANRIC
database analysis of the association between the expression of
lnc-MICAL2-1 and DKK3 mRNA in (C) basal, (D) LumB and (E) Her2
subtypes of breast invasive carcinoma. P<0.05. (F) Relationship
between the expression levels of lnc-MICAL2-1 and the overall
survival rate of patients with BC. TANRIC database analysis of the
association between the expression of lnc-MICAL2-1 and (G) staging,
(H) ER, (I) PR, (J) HER2 and (K) therapy. *P<0.05,
***P<0.001. Num, Number; Lum A, luminal A; Lum B, luminal B;
DKK3, Dickkopf 3; ER, estrogen receptor; PR, progesterone receptor;
HER2, human epidermal growth factor receptor 2; lnc-MICAL2-1, long
non-coding RNA MICAL2-1; BC/BRCA, breast cancer; TANRIC, The Atlas
of ncRNA in Cancer. |
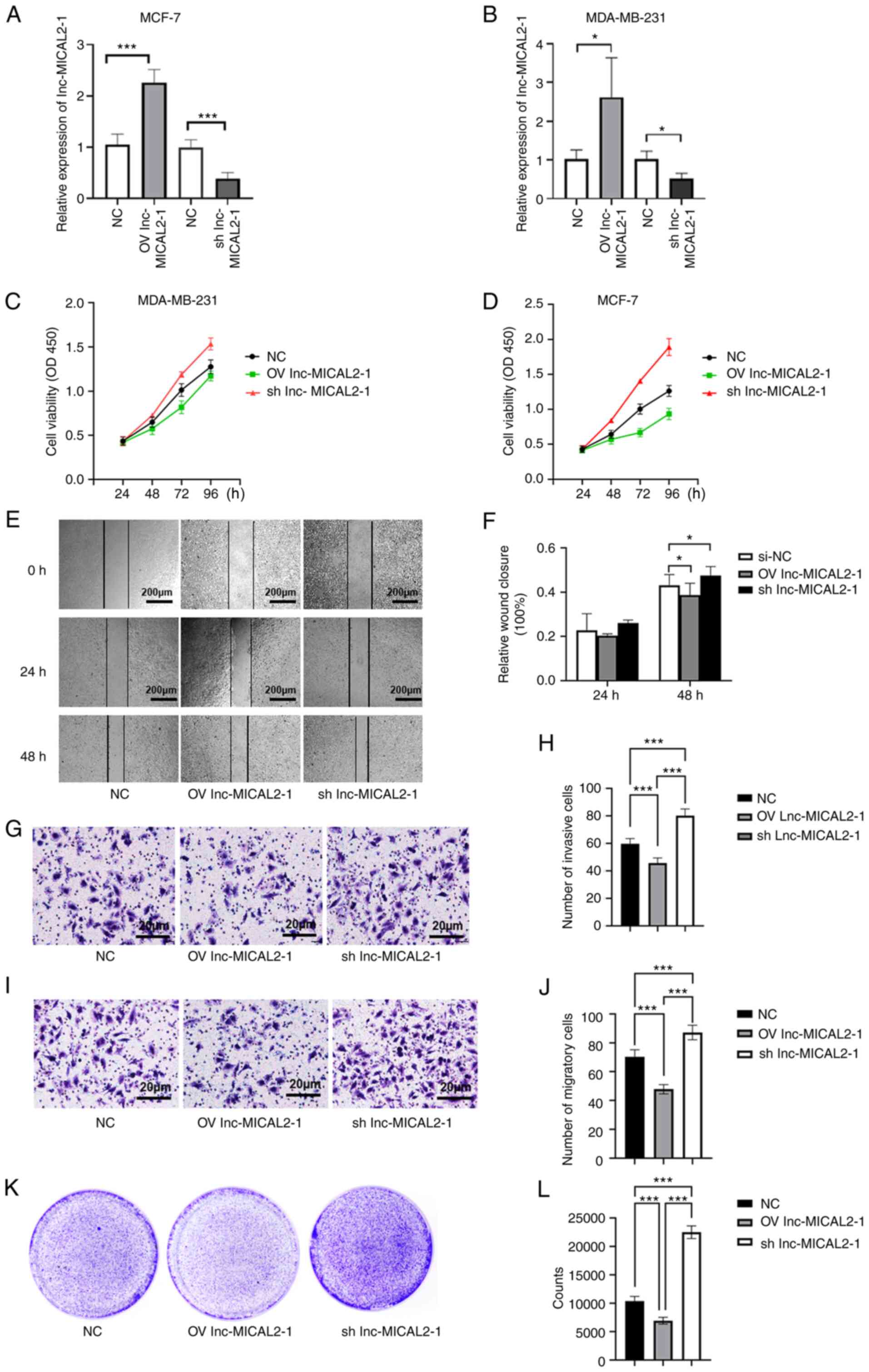
Effects of the knockdown and
overexpression of lnc-MICAL2-1 on BC cell proliferation, invasion
and migration
To further understand the effects of lnc-MICAL2-1 on
BC cells, lnc-MICAL2-1 overexpression and knockdown vectors were
constructed and transfected into the BC cell lines, MCF-7 and
MBA-MD-231 (Fig. 2A and B). The
results revealed that the optical density value was decreased
following the overexpression of lnc-MICAL2-1 in BC cells,
suggesting that cell proliferation was inhibited. Conversely, the
proliferative ability of cells with lnc-MICAL2-1 knockdown was
significantly increased (Fig. 2C and
D). In addition, a wound healing assay was performed using
transfected MCF-7 cells to determine the effects on cell migration
(Fig. 2E). The results
demonstrated that cell migration was inhibited following the
overexpression of lnc-MICAL2-1, whereas the knockdown of
lnc-MICAL2-1 promoted cell migration (Fig. 2F). The results of the Transwell
assays indicated that the overexpression of lnc-MICAL2-1 inhibited
the invasive and migratory ability of MCF-7 cells (Fig. 2G-J). The data from the colony
formation assay experiments further validated that the
overexpression of lnc-MICAL2-1 inhibited cell proliferation
(Fig. 2K and L).
lnc-MICAL2-1 upregulates DKK3
expression via sponging miR-25
A positive correlation was identified between
lnc-MICAL2-1 and DKK3 mRNA expression in BC tissues (correlation,
0.632; n=47; P<0.05; Fig. 3A).
Subsequently, lnc-MICAL2-1 was overexpressed or knocked down in
MCF-7 and MDA-MB-231 cells. The results revealed that in MCF-7
cells overexpressing lnc-MICAL2-1, the mRNA and protein expression
levels of DKK3 were significantly upregulated. Conversely,
following the knockdown of lnc-MICAL2-1 expression in MCF-7 cells,
the mRNA and protein expression levels of DKK3 were significantly
downregulated (Fig. 3B-D).
Therefore, it was hypothesized that lnc-MICAL2-1 may exert a
positive regulatory effect on DKK3 expression. Bioinformatics
analysis identified 13 complementary base pairings between miR-25
and lnc-MICAL2-1 (Fig. 3E).
Similarly, complementary base pairings were identified between
miR-25 and DKK3 (Fig. 3F). A
previous study reported that miR-25 targeted DKK3 and regulated the
Wnt/β-catenin signaling pathway (12). Therefore, dual-luciferase reporter
and RNA pull-down assays were performed to validate the binding.
The results of dual-luciferase reporter showed that compared with
miRNA NC, the luciferase activity in wt-lnc-MICAL2-1 cells
transfected with miRNA-25 mimic was significantly decreased
(P<0.01), whereas the luciferase activity in mut-lnc-MICAL2-1
cells was not affected (P>0.05) (Fig. 3G). The results revealed that
lnc-MICAL2-1 was able to sponge miR-25. This relationship was
further determined using the RNA pull-down assay. The results
indicated that lnc-MICAL2-1 could be pulled down by miRNA-25
(Fig. 3H). The results of the
dual-luciferase reporter assay showed that compared with the miRNA
NC group, the luciferase activity in wt-DKK3 cells transfected with
miRNA-25 mimic was significantly decreased (P<0.01), but the
luciferase activity in mut-DKK3 cells was not affected (P>0.05)
(Fig. 3I). This relationship was
further determined using the RNA pull-down assay, whereby DKK3 was
pulled down by miRNA-25 (Fig. 3J).
These findings suggested that lnc-MICAL2-1 may have reduced the
degradation of DKK3 mRNA by sponging miR-25, thereby exerting an
indirect positive regulatory effect on DKK3.
 | Figure 3.lnc-MICAL2-1 sponges miR-25 as a
competing endogenous RNA, indirectly enhancing DKK3 gene
transcription. (A) Analysis of data obtained from The Atlas of
non-coding RNA in Cancer, assessing the relationship between
lnc-MICAL2-1 and DKK3 expression in breast cancer. (B) Reverse
transcription-quantitative PCR was used to determine the effect of
lnc-MICAL2-1 OV or knockdown on DKK3 mRNA expression. (C and D)
Western blotting was used to assess the effect of lnc-MICAL2-1 OV
or knockdown on DKK3 expression levels. The predicted binding sites
between (E) miR-25 and lnc-MICAL2-1, and (F) miR-25 and DKK3. (G)
The target relationship between lnc-MICAL2-1 and miR-25 was
verified using a dual-luciferase reporter assay. (H) RNA pull-down
assay detected the molecular interaction between lnc-MICAL2-1 and
miR-25. (I) The target relationship between DKK3 and miR-25 was
verified by dual-luciferase reporter assay. (J) An RNA pull-down
assay detected the molecular interaction between DKK3 and miR-25.
Data are presented as the mean ± standard deviation. *P<0.05,
**P<0.01, ***P<0.001. DKK3, Dickkopf 3; lnc-MICAL2-1, long
non-coding RNA MICAL2-1; OV, overexpression; miR, microRNA; WT,
wild-type; MUT, mutant; sh, short hairpin. |
lnc-MICAL2-1 upregulates DKK3
expression and downregulates the Wnt signaling pathway
To explore the regulatory effect of lnc-MICAL2-1 on
the DKK3-mediated Wnt signaling pathway, a lnc-MICAL2-1
overexpression plasmid and miR-25 mimic were co-transfected into
MCF-7 cells, and shRNA lnc-MICAL2-1 and miR-25 inhibitor were
co-transfected into MCF-7 cells. Transfection efficiency of miR-25
mimic and inhibitor were determined by RT-qPCR in MCF-7 cells, and
the results showed that the transfection was successful (Fig. 4A). Furthermore, the results
revealed that the mRNA and protein expression levels of DKK3 in
cells co-transfected with the lnc-MICAL2-1 overexpression vector
and miR-25 mimic were downregulated compared with in cells only
transfected with the lnc-MICAL2-1 overexpression vector (Fig. 4B-D). Conversely, the mRNA and
protein expression levels of DKK3 in cells co-transfected with
shRNA lnc-MICAL2-1 and miR-25 inhibitor were upregulated compared
with in cells transfected with shRNA lnc-MICAL2-1 only (Fig. 4E-G). In addition, the
overexpression of lnc-MICAL2-1 decreased β-catenin nuclear
localization, whereas shRNA lnc-MICAL2-1 increased the
translocation of β-catenin into the nucleus (Fig. 4H). Next, the effect of lnc-MICAL2-1
on the β-catenin signaling pathway-mediated apoptosis of MCF-7
cells was explored. Activation of the Wnt/β-catenin pathway affects
cell apoptosis and cell cycle progression (25). The data revealed that in the
lnc-MICAL2-1 knockdown cells, the number of cells in the S phase of
the cell cycle was significantly increased compared with that in
the control cells (Fig. 4I and J),
whereas the levels of apoptosis were significantly increased
following the overexpression of lnc-MICAL2-1 in MCF-7 cells
compared with in the control cells (Fig. 4K and L).
 | Figure 4.lnc-MICAL2-1 enhances DKK3 gene
transcription and downregulates the Wnt signaling pathway. (A)
Transfection efficiency of miR-25 mimic and inhibitor determined by
reverse transcription-quantitative PCR in MCF-7 cells.
Overexpression of lnc-MICAL2-1 competitively binds and absorbs
miR-25, increasing DKK3 (B) mRNA and (C and D) protein expression
levels. Knockdown of lnc-MICAL2-1 results in increased miR-25
levels, thus reducing DKK3 (E) mRNA and (F and G) protein
expression. (H) Immunofluorescence staining of β-catenin and DAPI
staining of the nuclei. (I and J) Cell cycle distribution analysis
was performed to determine the effect of overexpression or
knockdown of lnc-MICAL2-1 on cell cycle progression. (K and L) Flow
cytometric analysis of the effect of OV or knockdown of
lnc-MICAL2-1 on cell apoptosis. Data are presented as the mean ±
standard deviation. *P<0.05, **P<0.01, ***P<0.001. DKK3,
Dickkopf 3; OV, overexpression; miR, microRNA; sh, short hairpin;
NC, negative control; lnc-MICAL2-1, long non-coding RNA
MICAL2-1. |
lnc-MICAL2-1 inhibits tumor growth in
vivo via regulation of the Wnt signaling pathway
To further explore the inhibitory function of
lnc-MICAL2-1 on BC cells, a BC xenograft model was established. The
overexpression of lnc-MICAL2-1 significantly reduced tumor weight
(Fig. 5A) and volume (Fig. 5B) compared with in the control and
lnc-MICAL2-1 knockdown groups. The survival curve also revealed
that the overexpression of lnc-MICAL2-1 significantly prolonged the
survival time of mice compared with in the control group (Fig. 5C).
H&E staining showed that NC group tumors grew
rapidly with a different morphology. Inflammatory infiltration was
observed around the edges of the tumor tissues, as well as a small
number of necrotizing cells. The tumors in the lnc-MICAL2-1 group
displayed a reduced number of tumor cells, increased cell
degeneration and necrosis, and an enhanced tissue response
(Fig. 5D). The results of
immunohistochemistry showed that the knockdown of lnc-MICAL2-1
downregulated the expression levels of DKK3, whereas the expression
levels of β-catenin and MMP-25 were upregulated (Fig. 5D and E). These findings suggested
that lnc-MICAL2-1 expression may be positively associated with DKK3
expression, whereas DKK3 expression may be negatively associated
with the expression of β-catenin and MMP-25 (Fig. 5D and E).
Discussion
DKK3 is a tumor suppressor gene and its expression
has been reported to be downregulated in numerous types of cancer,
including prostate (26), gastric
(27), pancreatic (28), lung and renal cell (29) cancer. A previous study reported
that DKK3 could inhibit the Wnt signaling pathway by blocking
planar cell polarity (30).
However, the upstream regulatory mechanism of DKK3 in BC requires
further investigation.
lncRNAs are known to act as ceRNAs (endogenous
sponges) to downregulate miRNA expression and thereby reduce the
translational inhibition of miRNA target genes (18). The abnormal expression of lncRNAs
has been discovered to play an important role in the progression of
cancer, including BC (21). It has
been shown that lncRNA MEG3 can regulate E-cadherin expression by
adsorbing miR-421 in BC cells (31). lncRNA HOTAIR has also been reported
to regulate HMGA2 through sponging miR-20a-5p to affect BC cell
proliferation, migration, invasion and apoptosis (32). However, the upstream lncRNA
involved in the regulation of DKK3 was unknown, to the best of our
knowledge.
The present study first predicted the upstream
regulatory miRNA and lncRNA of DKK3 through bioinformatics
analysis. The results demonstrated that lnc-MICAL2-1 upregulated
the expression levels of DKK3 in invasive BRCA, suggesting a
positive association between lnc-MICAL2-1 and DKK3. The expression
levels of lnc-MICAL2-1 had a positive relationship with the
clinical characteristics and overall survival rate of patients with
BC. In order to confirm the effect of lnc-MICAL2-1 on BC cells,
lnc-MICAL2-1 knockdown and overexpression vectors were transfected
into BC cells, and cellular behavior experiments were performed.
The results showed that lnc-MICAL2-1 inhibited BC cell
proliferation, migration and invasion. Previous studies have shown
that the expression levels of miR-25 were significantly upregulated
in human tumor tissues and cells, such as melanoma, glioma and
non-small cell lung cancer, which promoted cell invasion and
proliferation by targeting DKK3 (11,12).
However, the targeting relationship between lnc-MICAL2-1 and miR-25
is unclear. Using the GeneCards database, it was further predicted
that lnc-MICAL2-1 and miR-25, as well as miR-25 and DKK3 possess
complementary binding sites with each other, respectively. To
further understand the relationship between lnc-MICAL2-1, miR-25
and DKK3, bioinformatics analysis was used to determine the binding
site between lnc-MICAL2-1 and miR-25, then dual-luciferase reporter
and RNA pull-down assays were used to validate the competitive
binding relationship between lnc-MICAL2-1 and miR-25. The results
of the dual luciferase reporter assay and RNA pull-down assays
further confirmed that DKK3 was a target gene of miR-25.
DKK3 is an antagonist of the Wnt signaling pathway.
The inactivation of DKK3 has been shown to be related to the poor
prognosis of various solid tumors and hematological malignancies
(12). The Wnt signaling pathway
is a conserved and complex signaling pathway, which regulates stem
cell self-renewal, cell proliferation, differentiation and
apoptosis, and participates in embryonic development, tissue
homeostasis and carcinogenesis (33,34).
DKK3 was discovered to bind to the Wnt-signaling member LDL
receptor-related 5/6 or kringle containing transmembrane protein
1/2 to negatively regulate the Wnt signaling pathway (35). The activation of the Wnt signaling
pathway causes a reduction in β-catenin degradation and the
accumulation of β-catenin in the cytoplasm. β-catenin can enter the
nucleus to form complexes, affecting the proliferation, cell cycle
and apoptosis of tumor cells, amongst other behaviors (36). To further understand the
involvement of lnc-MICAL2-1 in the downstream signaling cascade,
the localization of β-catenin in BC cells was analyzed.
Overexpression of lnc-MICAL2-1 decreased β-catenin nuclear
localization, whereas shRNA lnc-MICAL2-1 increased the nuclear
translocation of β-catenin, suggesting that lnc-MICAL2-1
downregulated the Wnt signaling pathway. The results of the present
study also discovered that the knockdown of lnc-MICAL2-1 increased
the number of cells in the S phase of the cell cycle and
To further explore the inhibitory effect of
lnc-MICAL2-1 on BC cells, a BC xenograft model was established. The
results revealed that the knockdown of lnc-MICAL2-1 downregulated
the expression levels of DKK3, whereas the expression levels of
β-catenin were upregulated.
In conclusion, the present study discovered that
lnc-MICAL2-1 may be a good prognostic marker, since it may act as a
tumor suppressor in BC. lnc-MICAL2-1, as a ceRNA, was demonstrated
to sponge miR-25 and further stabilize DKK3, thereby reducing
Wnt/β-catenin signaling pathway activation. These findings
uncovered novel potential targets for BC, which provides a new
research direction for molecular targeted therapy and further
investigations into the underlying pathogenesis of BC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Hainan Province (grant no. 818QN3143).
Availability of data and materials
All data generated or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JY, GL, ML and SY conceived and designed the study.
HS performed the experiments. JY, GL, ML and CY performed the data
analysis. JY, GL and ML wrote the manuscript. JY and SY confirm the
authenticity of all the raw data. All authors edited the
manuscript. All authors have read and approved the manuscript.
Ethics approval and consent to
participate
The animal experiments performed in the present
study were approved by the Committee on the Ethics of Animal
Experiments of Southern Medical University (approval no. SYXK
2020-0012). The authors declare that this study was performed in
strict accordance with the recommendations in the Guide for the
Care and Use of Laboratory Animals of the National Institutes of
Health, and in accordance with the Animal Research: Reporting In
Vivo Experiments guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization (WHO), . Global
tuberculosis report 2020. WHO; Geneva: pp. 1–232. 2020
|
|
2
|
Niehrs C: Function and biological roles of
the Dickkopf family of Wnt modulators. Oncogene. 25:7469–7481.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bee C, Abdiche YN, Stone DM, Collier S,
Lindquist KC, Pinkerton AC, Pons J and Rajpal A: Exploring the
dynamic range of the kinetic exclusion assay in characterizing
antigen-antibody interactions. PLoS One. 7:e362612012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim MS, Lee HN, Kim HJ and Myung SC:
Single nucleotide polymorphisms in DKK3 gene are associated with
prostate cancer risk and progression. Int Braz J Urol. 41:869–897.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shao YC, Wei Y, Liu JF and Xu XY: The role
of Dickkopf family in cancers: From bench to bedside. Am J Cancer
Res. 7:1754–1768. 2017.PubMed/NCBI
|
|
6
|
Xu J, Sadahira T, Kinoshita R, Li SA,
Huang P, Wada K, Araki M, Ochiai K, Noguchi H, Sakaguchi M, et al:
Exogenous DKK-3/REIC inhibits Wnt/β-catenin signaling and cell
proliferation in human kidney cancer KPK1. Oncol Lett.
14:5638–5642. 2017.PubMed/NCBI
|
|
7
|
Yang Y, Xu W, Zheng Z and Cao Z: LINC00459
sponging miR-218 to elevate DKK3 inhibits proliferation and
invasion in melanoma. Sci Rep. 9:191392019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khan Z, Arafah M, Shaik JP, Mahale A and
Alanazi MS: High-frequency deregulated expression of Wnt signaling
pathway members in breast carcinomas. Onco Targets Ther.
11:323–335. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lei SL, Zhao H, Yao HL, Chen Y, Lei ZD,
Liu KJ and Yang Q: Regulatory roles of microRNA-708 and microRNA-31
in proliferation, apoptosis and invasion of colorectal cancer
cells. Oncol Lett. 8:1768–1774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peng G, Yang C, Liu Y and Shen C:
miR-25-3p promotes glioma cell proliferation and migration by
targeting FBXW7 and DKK3. Exp Ther Med. 18:769–778. 2019.PubMed/NCBI
|
|
12
|
Huo J, Zhang Y, Li R, Wang Y, Wu J and
Zhang D: Upregulated MicroRNA-25 mediates the migration of melanoma
cells by targeting DKK3 through the WNT/β-catenin pathway. Int J
Mol Sci. 17:11242016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A,
Egranov SD, Zhang Y, Xia W, Gong J, et al: Oncogenic lncRNA
downregulates cancer cell antigen presentation and intrinsic tumor
suppression. Nat Immunol. 20:835–851. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Fu Q, Li S, Liang N, Li F, Li C,
Sui C, Dionigi G and Sun H: LncRNA FOXD2-AS1 functions as a
competing endogenous RNA to regulate TERT expression by sponging
miR-7-5p in thyroid cancer. Front Endocrinol (Lausanne).
10:2072019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang XF, Ye Y and Zhao SJ: LncRNA Gas5
acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p
in papillary thyroid carcinoma. Oncotarget. 9:3519–3530. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu K, Liu C and Zhang Z: lncRNA GAS5 acts
as a ceRNA for miR-21 in suppressing PDGF-bb-induced proliferation
and migration in vascular smooth muscle cells. J Cell Biochem.
120:15233–15240. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Han L, Roebuck P, Diao L, Liu L,
Yuan Y, Weinstein JN and Liang H: TANRIC: An interactive open
platform to explore the function of lncRNAs in cancer. Cancer Res.
75:3728–3737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nokin MJ, Durieux F, Peixoto P, Chiavarina
B, Peulen O, Blomme A, Turtoi A, Costanza B, Smargiasso N, Baiwir
D, et al: Methylglyoxal, a glycolysis side-product, induces Hsp90
glycation and YAP-mediated tumor growth and metastasis. Elife.
5:e193752016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Li H and Wei X: Long noncoding RNA
LINC00261 suppresses prostate cancer tumorigenesis through
upregulation of GATA6-mediated DKK3. Cancer Cell Int. 20:4742020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Institute of Laboratory Animal Resources
(U.S.), Committee on Care and Use of Laboratory Animals, National
Institutes of Health (U.S.), Division of Research Resources, .
Guide for the Care and Use of Laboratory Animals. U.S Dept of
Health and Human Services, Public Health Service, National
Institutes of Health; Bethesda, MD: 1985
|
|
24
|
Kilkenny C, Browne W, Cuthill IC, Emerson
M and Altman DG; National Centre for the Replacement, Refinement
and Reduction of Amimals in Research, . Animal research: Reporting
in vivo experiments-the ARRIVE guidelines. J Cereb Blood Flow
Metab. 31:991–993. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lodygin D, Epanchintsev A, Menssen A,
Diebold J and Hermeking H: Functional epigenomics identifies genes
frequently silenced in prostate cancer. Cancer Res. 65:4218–4227.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sato H, Suzuki H, Toyota M, Nojima M,
Maruyama R, Sasaki S, Takagi H, Sogabe Y, Sasaki Y, Idogawa M, et
al: Frequent epigenetic inactivation of DICKKOPF family genes in
human gastrointestinal tumors. Carcinogenesis. 28:2459–2466. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsieh SY, Hsieh PS, Chiu CT and Chen WY:
Dickkopf-3/REIC functions as a suppressor gene of tumor growth.
Oncogene. 23:9183–9189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kurose K, Sakaguchi M, Nasu Y, Ebara S,
Kaku H, Kariyama R, Arao Y, Miyazaki M, Tsushima T, Namba M, et al:
Decreased expression of REIC/Dkk-3 in human renal clear cell
carcinoma. J Urol. 171:1314–1318. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mao B, Wu W, Li Y, Hoppe D, Stannek P,
Glinka A and Niehrs C: LDL-receptor-related protein 6 is a receptor
for Dickkopf proteins. Nature. 411:321–325. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang W, Shi S, Jiang J, Li X, Lu H and
Ren F: LncRNA MEG3 inhibits cell epithelial-mesenchymal transition
by sponging miR-421 targeting E-cadherin in breast cancer. Biomed
Pharmacother. 91:312–319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao W, Geng D, Li S, Chen Z and Sun M:
LncRNA HOTAIR influences cell growth, migration, invasion, and
apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer
Med. 7:842–855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Li H, Cao R, Sun L, Wang Y, Fan
S, Zhao Y, Kong D, Cui L, Lin L, et al: Suppression of Mir-708
promotes DKK3 to inhibit Wnt/β-catenin signaling pathway in adult
B-ALL. Blood. 128:50902016. View Article : Google Scholar
|
|
34
|
Katoh M and Katoh M: WNT signaling pathway
and stem cell signaling network. Clin Cancer Res. 13:4042–4045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ganesan K, Ivanova T, Wu Y, Rajasegaran V,
Wu J, Lee MH, Yu K, Rha SY, Chung HC, Ylstra B, et al: Inhibition
of gastric cancer invasion and metastasis by PLA2G2A, a novel
beta-catenin/TCF target gene. Cancer Res. 68:4277–4286. 2008.
View Article : Google Scholar : PubMed/NCBI
|