Introduction
Asthma, characterized by inflammation, the shedding
of airway epithelial cells (AECs) and airway remodeling, has become
one of the most prevalent chronic inflammatory airway disorders. It
promotes the contractility of surrounding smooth muscles and
aggravates pulmonary function, posing global economic and social
burdens (1,2). AECs can function as progenitors for
ciliated and goblet columnar cells in major airways, accounting for
~30% of the epithelium (3,4).
The airway epithelium can normally function as the frontline
defense against respiratory viruses through the mucociliary
apparatus and its immunological functions (5). It has also been found that barrier
damage and the dysfunction of AECs may be related to the onset of
asthma (6). Consequently, the
therapeutic methods used for the prevention of the damage and
dysfunction of AECs may be employed for the treatment of
asthma.
Inhaled glucocorticoids (GCs), also termed inhaled
corticosteroids (ICS), have been widely applied in the treatment
and prevention of asthma, with anti-allergy, anti-inflammatory and
immunosuppressive properties (7), which mainly function by exerting
suppressive effects on inflammation in the airways (8). However, GCs were also considered to
possibly adversely affect the repair process during which the
proliferation and migration of AECs are suppressed (9). At present, the underlying molecular
mechanisms of GCs in these processes remain unclear. Therefore,
further insight into the mechanisms of GCs may be conducive to
providing novel genetic strategies for the treatment of asthma.
The altered expression levels of microRNAs
(miRNAs/miRs) have also been found to be involved in the
development of asthma (10).
Zhang et al (11)
proposed that miR-221 was involved in AEC injury in asthma by
targeting sirtuin 1 (SIRT1), whilst Zhou et al (12) indicated that miR-155 could
function as a novel target in allergic asthma. In addition, miR-29c
has been found to play a vital role in children with asthma by
regulating Th2/Th17 cell differentiation (13). Lu et al (14) indicated that miR-375 was
predominately expressed in esophageal and bronchial epithelial
cells, and the upregulation of miR-375 was sufficient to modify
interleukin 13-associated immunoinflammatory pathways in epithelial
cells. However, the mechanisms of miR-375 as regards the
amelioration of GC-induced AEC dysfunction warrant further
investigation.
Dual specificity phosphatases (DUSPs) are considered
to be major modulators of signaling pathways affecting various
physiological processes (15).
DUSP6, as a member of the DUSPs, is a cytoplasmic enzyme, which is
perceived as a key cytoplasmic anchor of extracellular
signal-regulated kinases (ERKs) and a regulator of the ERK
signaling cascade (15).
Previous studies have suggested that DUSP6 is involved in the
progression of multiple diseases, such as cancer,
inflammation-related diseases and chronic obstructive pulmonary
disease (16-18). Recent evidence has suggested that
long non-coding RNA taurine-upregulated gene 1 promotes airway
remodeling by inhibiting the miR-145-5p/DUSP6 axis in
smoking-induced chronic obstructive pulmonary disease (18). Of note, DUSP6 is a known oncogene
regulating cellular differentiation and proliferation in thyroid
cancer, whose expression is inferred to correlate with that of
miR-375 (19). Therefore, it was
hypothesized that miR-375 may modulate AEC dysfunction by
regulating DUSP6 expression.
The present study mainly focused on the role and
function of miR-375 in ameliorating dexamethasone (Dex)-induced AEC
dysfunction, with the aim of assisting in the development of a
possible treatment for AEC dysfunction.
Materials and methods
Cells, cell culture and reagents
Human AECs (the 9HTE cell line), were obtained from
the Respiratory Research Laboratory (Key Laboratory of Child
Development and Disorders of Ministry of Education, Children's
Hospital, Chongqing, China). The cells were grown in Dulbecco's
modified Eagle's medium (DMEM; 01-057-1A, Biological Industries)
supplemented with 10% fetal bovine serum (FBS; 04-001-1A,
Biological Industries) at 37°C with 5% CO2.
Dex (D1756) was purchased from Sigma-Aldrich; Merck
KGaA. The 9HTE cells were divided into four groups as follows: i)
The control group, cells were incubated in DMEM without Dex
treatment; ii) Dex 6 group, cells were incubated in DMEM and then
treated with 10 µmol/l Dex for 6 h; iii) Dex 12 group, cells
were incubated in DMEM and then treated with 10 µmol/l Dex
for 12 h; and iv) Dex 24 group, cells were incubated in DMEM and
then treated with 10 µmol/l Dex for 24 h.
It was found that 10 µmol/l Dex treatment
exerted the optimal effects on the AECs after 24 h. Subsequently,
to determine the effects of Dex treatment, miR-375 and DUSP6 on
9HTE cells, the cells were transfected with miR-375 mimic (M) and
its control (MC), as well as with overexpression DUSP6 plasmid and
its negative control (NC), followed by treatment with or without 10
µmol/l Dex for 24 h. The transfection protocol is described
below.
Cell Counting Kit-8 (CCK-8) assay
The transfected 9HTE cells (1×104
cells/well) were seeded into a 96-well plate in DMEM containing 10%
FBS at 37°C with 5% CO2. Subsequently, 10 µl
CCK-8 reagent (GK10001; GLPBio) with serum-free DMEM was then added
into each well to detect cell viability at 12 and 24 h. The
absorbance at 450 nm was measured using an iMark™ Microplate
Absorbance Reader (168-1020; Bio-Rad Laboratories, Inc.).
Flow cytometry
9HTE cell apoptosis was detected using flow
cytometry with an Annexin V-FITC/propidium iodide (PI) apoptosis
kit (A211; GeneBio Systems, Inc.) as per the manufacturer's
instructions. Following transfection for 48 h, the 9HTE cells were
harvested and then washed with cold phosphate-buffered saline (PBS)
twice, followed by treatment with both Annexin V and PI (5
µl/well) for 20 min in the dark at room temperature. Cell
apoptosis was further analyzed using a Guava easyCyte Benchtop Flow
Cytometer (BR168323; Luminex Corporation) and Kaluza C Analysis
Software (version 1.1.1, Beckman Coulter, Inc.).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the 9HTE cells using
TRIzol® reagent (15596026; Invitrogen; Thermo Fisher
Scientific, Inc.) and cryopreserved at −80°C. The concentration of
total RNA was quantified using a NanoDrop Lite UV-Vis spectrometer
(ND-LITE, Thermo Fisher Scientific, Inc.). cDNA was synthesized
from 1 µg of total RNA with an Optimax First strand cDNA
Synthesis kit (K4201100, BioChain Institute, Inc.). In detail, the
reaction components were mixed, and the mixture was incubated at
42°C for 60 min; the reaction was then terminated by incubating the
tube at 70°C for 10 min. The qPCR experiment was conducted using a
QCell-Pro One-Step qRT-PCR SuperMix kit (K5055400, BioChain
Institute, Inc.) on a Touch real-time PCR Detection system (CFX384,
Bio-Rad Laboratories, Inc.) under the following conditions: 95°C
for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1
min. The primer sequences are presented in Table I. β-actin and U6 were used as
internal controls. The expression levels of relative genes were
quantified using the 2−ΔΔCq calculation method (20).
 | Table ISequences of primers used for reverse
transcription-quantitative PCR in the present study. |
Table I
Sequences of primers used for reverse
transcription-quantitative PCR in the present study.
| Gene ID | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) |
|---|
| miR-375 C |
TCGCGTGAGTCGTATCCAG |
GTATCCAGTGCGTGTCGTGG |
| miR-let-7 C |
AGCACTGAGGTAGTAGGTT |
CTGAGGCTCACTGACACAA |
| miR-21 |
GTGCAGGGTCCGAGGT |
GCCGCTAGCTTATCAGACTGATGT |
| miR-19 |
TGTGCAAATCTATGCAAA |
GTGCAGGGTCCGAGGTATTC |
| miR-455 |
ACACTCCAGCTGGGGCAGTCCATGGGCAT |
TGGTGTCGTGGAGTCG |
| DUSP6 |
GCTATACGAGTCGTCGCACA |
CGGGCTTCATCTTCCAGGTA |
| U6 C |
TCGCTTCGGCAGCACATATACT |
ACGCTTCACGAATTTGCGTGTC |
| β-actin |
ATTGGCAATGAGCGGTTC |
GGATGCCACAGGACTCCA |
Cell transfection
miR-375 mimic (B02003, sequence:
5′-UUUGUUCGUUCGGCUCGCGUGA-3′) and its control (B04001, sequence:
5′-UUGCCAUUUGGUAUGUGCGG UU-3′) were purchased from Shanghai
GenePharma Co., Ltd. The DUSP6 overexpression sequence was
structured by Thermo Fisher Scientific, Inc. and inserted into the
PcDNA3.1 plasmid (V79020; Thermo Fisher Scientific, Inc.) for
preparing the DUSP6 overexpression plasmid, and empty PcDNA3.1
plasmid was used as a negative control. The 9HTE cells were then
cultured in a 96-well plate at a density of 2×104
cells/well until reaching 80% confluence, and 0.2 µg mimic
and its control, as well as 50 nmol DUSP6 overexpression plasmid
were transfected into the cells using Lipofectamine®
3000 reagent (L3000-001; Thermo Fisher Scientific, Inc.) at 37°C.
The cells were harvested at 48 h post-transfection and were then
treated with 10 µmol/l Dex for 24 h. The expression levels
of miR-375 and DUSP6 in the treated cells were measured using
RT-qPCR or western blot analysis.
Wound healing assay
At 48 h post-transfection, the 9HTE cells
(1×105 cells/well) were cultured in a 24-well plate.
After the cells reached 80% confluency, a scratch was created in
the middle of each well using a sterile pipette tip. The cells were
then washed twice with PBS to smooth the scratch edge and remove
floating cells. Subsequently, the cells were cultured in serum-free
DMEM at 37°C with 5% CO2. Cell images at 0 and 24 h were
captured under an automated fluorescence microscope (BX63; Olympus
Corporation). Cell migration was detected at ×100 magnification and
quantified using Image-Pro Plus Analysis software 7.0 (Media
Cybernetics, Inc.).
Transwell invasion assay
Transwell chambers (8-µm-pore size; CLS3422,
Sigma-Aldrich; Merck KGaA) were placed in a 24-well plate, the
upper chamber of which was coated with 50 µl Matrigel
(356235, Corning, Inc.). The transfected 9HTE cells were
subsequently transferred onto the upper chamber at 37°C with 5%
CO2, and 500 µl DMEM containing 10% FBS were
added to the lower chamber as a chemoattractant. After 24 h, the
lower chamber was washed multiple times with PBS, and the
unmigrated cells in the upper chamber were gently removed using
cotton swabs. The lower Transwell chamber was first fixed in 4%
paraformaldehyde solution for 30 min and subsequently stained with
0.1% hematoxylin (H3136, Sigma-Aldrich; Merck KGaA) for 20 min at
room temperature. The number of cells in five randomly selected
fields was counted under an inverted optical microscope (DP27,
Olympus Corporation) and photographed at ×100 magnification.
Target gene prediction and
dual-luciferase reporter assay
The target gene and potential binding sites between
miR-375 and DUSP6 were predicted using TargetScan (http://www.targetscan.org/vert_72/) and confirmed
using dual-luciferase reporter assay.
Subsequently, a luciferase reporter with their
3′-untranslated regions (3′-UTRs) was constructed. PMIR-REPORT
Luciferase reporter (AM5795; Thermo Fisher Scientific, Inc.)
containing the wild-type (WT) or mutated (MUT) DUSP6 sequence was
cloned into the pMirGLO reporter vector (E1330; Promega
Corporation) to form DUSP6-WT (sequence, 5′-CCACU U CU UA A A ACAGA
ACA A A-3′) a nd DUSP6-MUT (sequence, 5′-CCACUUCUUAAAACAACC
AGUC-3′). The 9HTE cells were cultured in a 24-well plate at a
density of 3×104 cells/well and co-transfected or not
with DUSP6-WT (500 ng/well) and DUSP6-MUT (500 ng/well) with 500 ng
of miR-375 mimic (M; B02003; Gene Pharma, China) using
Lipofectamine® 3000 reagent at 37°C. After 48 h, the
Firefly luciferase activity was detected and normalized to
Renilla luciferase activity using the dual-luciferase
reporter assay system (E1910; Promega Corporation).
Western blot analysis
The protein expression levels of related genes were
measured by western blot analysis as previously described (3). After collecting the transfected
cells, proteins were lysed and extracted using RIPA buffer
(RIPA-50; FIVEphoton Biochemicals. The protein concentration was
measured using a Bicinchoninic Acid (BCA) Protein kit (SK3021, Bio
Basic Inc.). Sample protein lysates were electrophoresed by sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12%
gel; P0012A; Beyotime Institute of Biotechnology), and then
transferred onto a polyvinylidene fluoride (PVDF) membrane (FFP28;
Beyotime Institute of Biotechnology). After blocking with 5%
non-fat milk for 2 h at room temperature, the membrane was
incubated with primary antibodies including anti-Bcl-2 antibody
(rabbit, 1:1,000, cat. no. ab59348), anti-Bax antibody (rabbit,
1:10,000, cat. no. ab32503), anti-cleaved caspase-3 antibody
(rabbit, cat. no. ab2302, 1:1,000), anti-DUSP6 antibody (goat,
1:450, cat. no. ab166922) and anti-β-actin antibody (mouse,
1:1,000, cat. no. ab8226) (all from Abcam) at 4°C overnight.
β-actin was used as an internal reference. The membrane was then
incubated with the secondary horseradish peroxidase
(HRP)-conjugated antibodies, including goat anti-rabbit IgG H&L
(HRP; 1:10,000, cat. no. 65-6120), goat anti-mouse IgG H&L
(HRP; 1:10,000, cat. no. 62-6520) (both from Thermo Fisher
Scientific, Inc.) and donkey anti-goat IgG H&L (HRP; 1:20,000,
cat. no. ab6885, Abcam) at room temperature for 1 h and washed with
tris-buffered saline Tween-20 (TBST) three times. Protein bands
were collected from the samples and analyzed using an Enhanced
Chemiluminescence (ECL) kit (PCD-250, FIVEphoton Biochemicals).
Gray values of the bands were further analyzed and calculated using
ImageJ 5.0 software (Bio-Rad Laboratories, Inc.).
Statistical analysis
All experiments were conducted in triplicate
independently. Data are expressed as the mean ± standard deviation
(SD). Statistical analysis was performed using SPSS 17.0 software
(IBM Corp). Statistical significance was determined using one-way
analysis of variance (ANOVA) followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Dex treatment suppresses miR-375
expression and the viability of 9HTE cells, and promotes apoptosis
in a time-dependent manner
The 9HTE cells were pre-treated with Dex for 0, 6,
12 and 24 h and then incubated for a further 12 or 24 h to examine
the effects of Dex treatment on cell viability and apoptosis, and
the expression of miR-375. The results revealed that 9HTE cell
viability was decreased in a time-dependent manner (P<0.01,
Fig. 1A), suggesting that Dex
treatment suppressed 9HTE cell viability.
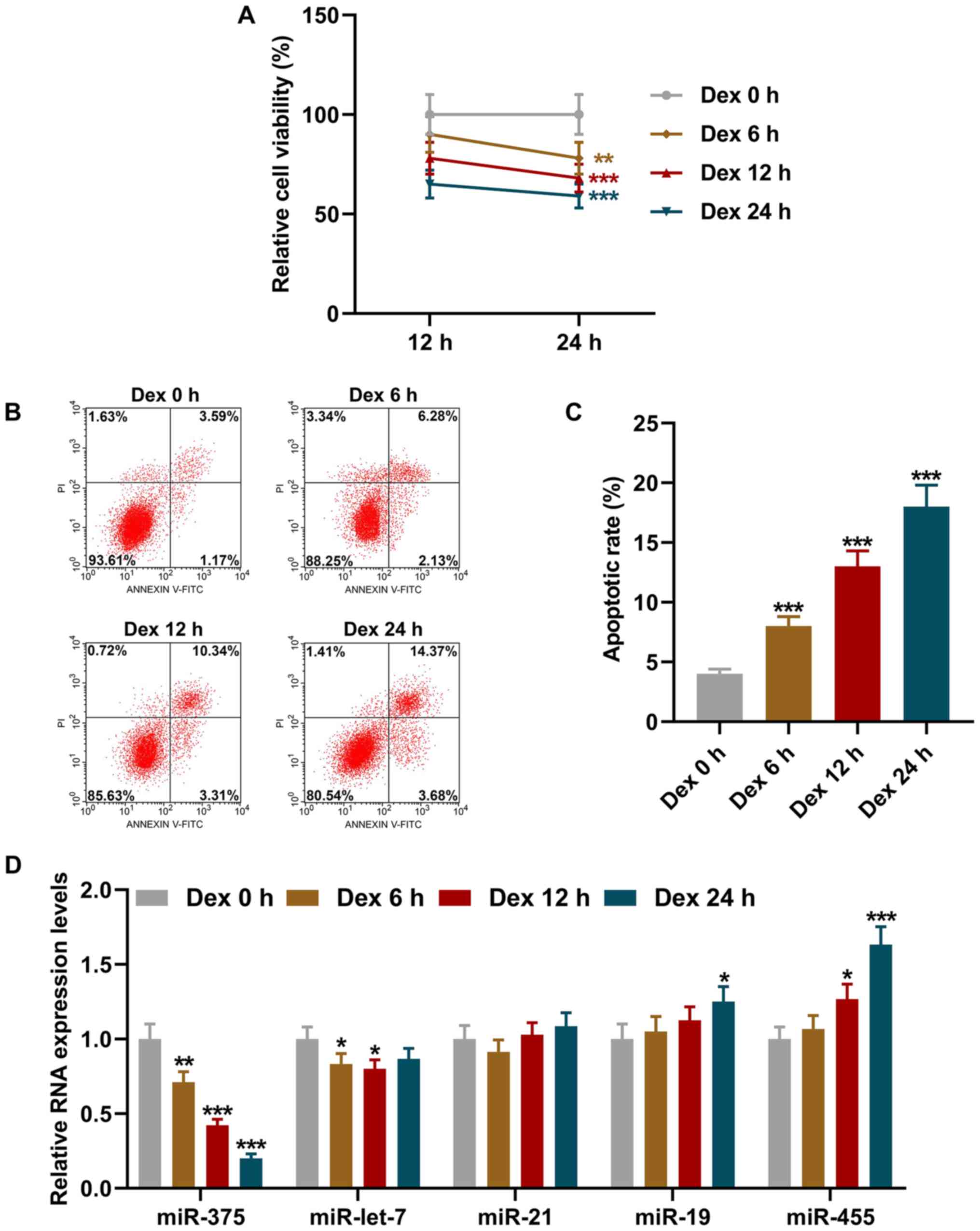 | Figure 1Dex treatment suppressed miR-375
expression and the viability of 9HTE cells, and promoted apoptosis
in a time-dependent manner. (A) Cell viability following Dex
pre-treatment for different periods of time (0, 6, 12 and 24 h)
followed by incubation for a further 12 or 24 h detected using Cell
Counting Kit-8 assay. (B and C) Cell apoptotic rate following Dex
treatment for different periods of time (0, 6, 12 and 24 h)
detected using flow cytometry. (D) Relative expression levels of
miR-375, miR-let-7, miR-21, miR-19 and miR-455 in cells following
Dex treatment for different periods of time (0, 6, 12 and 24 h)
measured using reverse transcription-quantitative PCR. U6 was
employed as an internal control. All experiments were performed in
triplicate and experimental data are expressed as the mean ±
standard deviation. *P<0.05, **P<0.01
and ***P<0.001 vs. Dex 0 h. Dex, dexamethasone. |
The effects of Dex treatment for different periods
of time (0, 6, 12 and 24 h) on 9HTE cell apoptosis were
subsequently detected using flow cytometry. As shown in Fig. 1B and C, the apoptotic rate of the
9HTE cells increased in a time-dependent manner following treatment
with Dex (P<0.001, Fig. 1B
and 1C), which indicated that
Dex treatment promoted 9HTE cell apoptosis.
Previous studies have reported that differentially
expressed miRNAs are involved in the progression of asthma, such as
miR-375, miR-let-7, miR-21, miR-19 and miR-455 (21,22). Subsequently, the effects of Dex
treatment for different periods of time (0, 6, 12 and 24 h) on the
expression levels of miR-375, miR-let-7, miR-21, miR-19 and miR-455
in the 9HTE cells were determined using RT-qPCR. It was found that
the expression of miR-375 in the 9HTE cells was downregulated
following treatment with Dex for different periods of time
(P<0.01, Fig. 1D), suggesting
that Dex treatment downregulated the expression of miR-375 in 9HTE
cells.
Overexpression of miR-375 reverses the
effects of Dex treatment on the expression of miR-375, and on the
viability, migration and invasion of 9HTE cells
Dex treatment for 24 h was proven to exert optimal
effects on the AECs. Thus, the present study then transfected the
9HTE cells with miR-375 mimic control or miR-375 mimic. It was
found that the level of miR-375 was increased by transfection with
miR-375 mimic (P<0.001, Fig.
2A). Subsequently, to further investigate the association
between miR-375 and Dex treatment, the 9HTE cells were treated with
Dex with or without transfection with miR-375 mimic control or
miR-375 mimic. As depicted in Fig.
2B, the expression of miR-375 was notably downregulated
following Dex treatment, whereas the overexpression of miR-375
reversed the effects of Dex treatment (P<0.001, Fig. 2B).
The viability of the 9HTE cells was assessed using
CCK-8 assay following Dex treatment and transfection with miR-375
mimic. It was noted that following treatment with Dex for 24 h,
9HTE cell viability was decreased, whereas the overexpression of
miR-375 reversed the effects of Dex treatment on 9HTE cell
viability (P<0.01, Fig.
2C).
Following Dex treatment and transfection with
miR-375 mimic, the migration and invasion of the 9HTE cells were
measured using wound healing assay and Transwell assay,
respectively. The results of the two assays demonstrated that the
cell migration and invasion rates decreased following treatment
with Dex for 24 h; these effects were reversed by the
overexpression of miR-375 (P<0.05, Fig. 2D-G).
Overexpression of miR-375 reverses the
effects of Dex treatment on the expression levels of
apoptosis-related proteins
Bcl-2, Bax and cleaved caspase-3 are considered as
apoptosis-related proteins (23). Thus, the present study then
measured their expression levels in 9HTE cells using western blot
analysis after the cells were treated with Dex and transfected with
miR-375 mimic. As illustrated in Fig. 3, the expression of Bcl-2 was
decreased and that of Bax and cleaved caspase-3 were increased
following treatment with Dex (P<0.001, Fig. 3A and B). However, the effects of
Dex treatment on the levels of these apoptosis-related proteins
were reversed by the overexpression of miR-375 (P<0.05, Fig. 3).
Expression of DUSP6, the target of
miR-375, is increased in a time-dependent manner following
treatment with Dex
DUSP6 was predicted and recognized as the target of
miR-375 using TargetScan, and the conserved binding sites between
DUSP6 and miR-375 are illustrated in Fig. 4A. To verify these results, a
luciferase reporter with their 3′-UTRs was constructed. The results
from dual-luciferase reporter assay demonstrated that the
luciferase activity of the DUSP6-WT-mimic (M) group was decreased
in comparison with that of the DUSP6-WT-Blank group (P<0.001,
Fig. 4B). However, no
significant difference was found in the luciferase activity of the
DUSP6-MUT-M group as compared to that of the DUSP6-MUT-Blank group.
Therefore, it was suggested that DUSP6 was the target gene of
miR-375.
 | Figure 4The expression of DUSP6, the target
gene of miR-375, is upregulated in a time-dependent manner
following Dex treatment. (A) Sequences of DUSP6 WT (top row),
miR-375 (middle row) and DUSP6 MUT (bottom row) are listed. (B)
Dual-luciferase reporter assay revealed that DUSP6 was the target
gene of miR-375. (C) Relative mRNA expression of DUSP6 following
Dex treatment for different periods of time (0, 6, 12 and 24 h)
measured using reverse transcription-quantitative PCR. β-actin was
used as an internal control. (D and E) Relative protein/β-actin
expression of DUSP6 following Dex treatment for different periods
of time (0, 6, 12 and 24 h) measured using western blot analysis.
β-actin was used as an internal control. All experiments were
performed in triplicate and experimental data are expressed as the
mean ± standard deviation. ***P<0.001 vs.
blank; ^^^P<0.001 vs. Dex 0 h. DUSP6, dual
specificity phosphatase 6; M, miR-375 mimic; Dex, dexamethasone;
WT, wild-type; MUT, mutant-type. |
To determine the role of DUSP6 in the 9HTE cells,
its expression was measured following treatment with Dex for
different periods of time (0, 6, 12 and 24 h). The results revealed
that the mRNA and protein expression levels of DUSP6 were increased
in a time-dependent manner following Dex treatment (P<0.001,
Fig. 4C-E).
Overexpression of DUSP6 reverses the
effects of the overexpression of miR-375 on the expression of DUSP6
and the viability of Dex-treated 9HTE cells
Subsequently, the 9HTE cells were transfected with
DUSP6 overexpression plasmid, and it was found that DUSP6
expression was increased following transfection with DUSP6
overexpression plasmid (P<0.001, Fig. 5A). To further examine the effects
of DUSP6 and miR-375 on Dex-treated 9HTE cells, miR-375 mimic and
DUSP6 overexpression plasmid were transfected into the cells
followed by treatment with Dex. The results revealed that the
protein and mRNA expression levels of DUSP6 in the 9HTE cells were
notably increased following Dex treatment, and these effects were
reversed by miR-375 overexpression (P<0.001, Fig. 5B-D). Furthermore, the
overexpression of DUSP6 abrogated the effects of miR-375
overexpression on the protein and mRNA expressions of DUSP6
(P<0.05, Fig. 5B-D).
The viability of the Dex-treated 9HTE cells was then
detected using CCK-8 assay following transfection with miR-375
mimic and DUSP6 overexpression plasmid. The results indicated that
9HTE cell viability was decreased following Dex treatment, which
was reversed by miR-375 overexpression. Moreover, the
overexpression of DUSP6 reversed the effects of miR-375
overexpression on the viability of Dex-treated 9HTE cells
(P<0.01, Fig. 5E).
Overexpression of DUSP6 reverses the
effects of the overexpression of miR-375 on the migration and
invasion of Dex-treated 9HTE cells
The migration of Dex-treated 9HTE cells was assessed
using wound healing assay following transfection with miR-375 mimic
and DUSP6 overexpression plasmid. The results revealed that the
cell migration rate following Dex treatment was suppressed, and
this effect was reversed by the overexpression of miR-375
(P<0.001, Fig. 6A and C). In
addition, DUSP6 overexpression reversed the effects of the
overexpression of miR-375 on the migration of Dex-treated 9HTE
cells (P<0.001, Fig. 6A and
C).
The invasion of Dex-treated 9HTE cells was examined
using Transwell assay following transfection of miR-375 mimic and
DUSP6 overexpression plasmid. The cell invasion rate following Dex
treatment was reduced, which was counteracted by the overexpression
of miR-375 (P<0.05, Fig. 6B and
D). The overexpression of DUSP6 abrogated the effects of the
overexpression of miR-375 on the invasion of Dex-treated 9HTE cells
(P<0.05, Fig. 6B and D).
Overexpression of DUSP6 reverses the
effects of the overexpression of miR-375 on the expression levels
of cell apoptosis-related proteins in Dex-treated 9HTE cells
Following transfection with miR-375 mimic and DUSP6
overexpression plasmid, the expression levels of cell
apoptosis-related proteins (Bcl-2, Bax and cleaved caspase-3) in
the Dex-treated 9HTE cells were measured using western blot
analysis. The results confirmed that following Dex treatment, the
expression of Bcl-2 was decreased, while that of Bax and cleaved
caspase-3 was increased (P<0.001, Fig. 7). The overexpression of miR-375
led to an opposite result (P<0.01, Fig. 7). In addition, the overexpression
of DUSP6 was found to reverse the effects of miR-375 on the protein
expression levels of Bcl-2, Bax and cleaved caspase-3 in the
Dex-treated 9HTE cells (P<0.01, Fig. 7).
Discussion
The human airway epithelium forms the mucosal
interface between inhaled environment and the lung (24). Increasing evidence has indicated
that in patients with asthma, the airway epithelium is structurally
and functionally abnormal, with increased mucus production, higher
permeability, enhanced oxidant sensitivity, and deficient innate
immune response to viruses (25). Nowadays, ICS combined with second
controller medications have been widely adopted for the management
of mild or moderate asthma in adults and children (26), although the results may vary in
patients.
Dex is a synthetic GC compound with
anti-inflammatory, immunosuppressive and decongestant effects
(27,28). A number of studies have proposed
that Dex exerts promising effects in the treatment of several
diseases, including acute myeloid leukemia (29), macular edema (30) and noninfectious uveitis (31). Furthermore, Dex has also been
found useful in preventing respiratory distress, particularly
asthma (32,33). The present study concentrated on
human AECs, namely the 9HTE cell line. It was found that following
Dex treatment, cell viability was suppressed, whereas cell
apoptosis was promoted in a time-dependent manner. Further
investigations also confirmed that Dex treatment led to the
downregulation of miR-375, a novel potential therapeutic target for
asthma treatment (34).
miRNAs have been found to affect cell proliferation,
metastasis and apoptosis (35).
Previous studies have suggested that miR-375 can promote the
progression of inflammatory bowel disease by upregulating Toll-like
receptor 4 (36). In addition,
miR-375 has been shown to prevent nasal mucosa cells from apoptosis
and to ameliorate allergic rhinitis by suppressing the JAK2/STAT3
pathway (37). Furthermore,
miR-375 has been verified to promote cell growth in small cell lung
carcinoma (38). In the present
study, the overexpression of miR-375 reversed the suppressive
effects of Dex treatment on AEC viability, migration and invasion.
Bcl-2, Bax and cleaved caspase-3 have been found to exert effects
on apoptosis, an important process where the function of normal
epithelial tissue is maintained (23,39). It has been reported that the
upregulation of Bcl-2 suppresses apoptosis, while the upregulation
of Bax and cleaved caspase-3 promotes apoptosis (40,41). Moreover, the present study also
proved that the expression of Bcl-2 was downregulated, while that
of Bax and cleaved caspase-3 was upregulated following Dex
treatment; these effects were reversed by the overexpression of
miR-375, indicating that the overexpression miR-375 abrogated the
promoting effects of Dex on cell apoptosis. However, the detailed
molecular mechanisms remain to be further addressed.
DUSP6 belongs to the mitogen-activated protein
kinase (MAPK) family that can serve as a feedback regulator of MAPK
cascades (18). Accumulating
evidence has indicated that DUSP6 functions as a critical mediator
in inflammatory responses. Hsu et al (42) proposed that DUSP6 promoted
endothelial inflammation via intercellular adhesion molecule-1. It
has also been demonstrated that DUSP6 deletion can enhance the
regulation of colonic inflammatory responses and the protection of
the intestinal epithelium against oncogenic stress by controlling
the activation of ERK1/2 (43).
In addition, chemokine (C-C motif) ligand 2-enhanced macrophage
inflammation responses may be related to the suppression of ERK
phosphatase DUSP6 (44). A
previous study verified that DUSP6 played a tumor-suppressive role
by inhibiting apoptosis (45).
In the present study, it was confirmed that the expression of
DUSP6, the target of miR-375, was upregulated in a time-dependent
manner following Dex treatment. Furthermore, the overexpression of
DUSP6 was proven to reverse the effects of the overexpression of
miR-375 on AEC viability, migration and invasion, as well as the
expression of apoptosis-related proteins (Bcl-2, Bax and cleaved
caspase-3) in Dex-treated cells. Therefore, it could be summarized
that the overexpression of miR-375 reverses the effects of Dex
treatment on AEC viability, migration, invasion and apoptosis by
targeting DUSP6.
However, since the in vitro exploration of the roles
of Dex, miR-375 and DUSP6 in AECs was performed in the present
study, the authors aim to validate these results through in vivo
investigations in the future.
In conclusion, the present study demonstrated study
that the overexpression of miR-375 reversed the effects of Dex
treatment on human AEC viability, migration, invasion and apoptosis
in vitro by targeting DUSP6. It is hoped that these findings may
provide novel roles and evidence of Dex and miR-375 in AEC
dysfunction, thereby providing a potential therapeutic strategy for
AEC dysfunction.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
XZ made substantial contributions to the conception
and design of the study. CL and XG were involved in data
acquisition, data analysis and interpretation. XZ was involved in
the drafting of the article or critically revising it for important
intellectual content. All authors confirm the authenticity of all
the raw data. All authors gave the final approval of the final
version of the manuscript to be published. All authors agree to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Human AECs, the 9HTE cell line, were obtained from
the Respiratory Research Laboratory (Key Laboratory of Child
Development and Disorders of Ministry of Education, Children's
Hospital, Chongqing, China). The authors are sincerely grateful to
Ying Huang (chief physician), for providing the 9HTE cells for this
study.
Funding
No funding was received.
References
|
1
|
Shine S, Muhamud S and Demelash A:
Prevalence and associated factors of bronchial asthma among adult
patients in Debre Berhan Referral Hospital, Ethiopia 2018: A
cross-sectional study. BMC Res Notes. 12:6082019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ye C, Huang C, Zou M, Hu Y, Luo L, Wei Y,
Wan X, Zhao H, Li W, Cai S, et al: The role of secreted Hsp90α in
HDM-induced asthmatic airway epithelial barrier dysfunction. BMC
Pulm Med. 19:2182019. View Article : Google Scholar
|
|
3
|
Liu J, Zhang M, Niu C, Luo Z, Dai J, Wang
L, Liu E and Fu Z: Dexamethasone inhibits repair of human airway
epithelial cells mediated by glucocorticoid-induced leucine zipper
(GILZ). PLoS One. 8:e607052013. View Article : Google Scholar :
|
|
4
|
Xie B, Laxman B, Hashemifar S, Stern R,
Gilliam TC, Maltsev N and White SR: Chemokine expression in the
early response to injury in human airway epithelial cells. PLoS
One. 13:e01933342018. View Article : Google Scholar :
|
|
5
|
Vareille M, Kieninger E, Edwards MR and
Regamey N: The airway epithelium: Soldier in the fight against
respiratory viruses. Clin Microbiol Rev. 24:210–229. 2011.
View Article : Google Scholar :
|
|
6
|
Gon Y and Hashimoto S: Role of airway
epithelial barrier dysfunction in pathogenesis of asthma. Allergol
Int. 67:12–17. 2018. View Article : Google Scholar
|
|
7
|
He Y, Shi J, Nguyen QT, You E, Liu H, Ren
X, Wu Z, Li J, Qiu W, Khoo SK, et al: Development of highly potent
glucocorticoids for steroid-resistant severe asthma. Proc Natl Acad
Sci USA. 116:6932–6937. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adcock IM and Mumby S: Glucocorticoids.
Handb Exp Pharmacol. 237:171–196. 2017. View Article : Google Scholar
|
|
9
|
Jia S, Guo P, Ge X, Wu H, Lu J and Fan X:
Overexpression of indoleamine 2, 3-dioxygenase contributes to the
repair of human airway epithelial cells inhibited by dexamethasone
via affecting the MAPK/ERK signaling pathway. Exp Ther Med.
16:282–290. 2018.PubMed/NCBI
|
|
10
|
Svitich OA, Sobolev VV, Gankovskaya LV,
Zhigalkina PV and Zverev VV: The role of regulatory RNAs (miRNAs)
in asthma. Allergol Immunopathol (Madr). 46:201–205. 2018.
View Article : Google Scholar
|
|
11
|
Zhang H, Sun Y, Rong W, Fan L, Cai Y, Qu
Q, Gao Y and Zhao H: miR-221 participates in the airway epithelial
cells injury in asthma via targeting SIRT1. Exp Lung Res.
44:272–279. 2018. View Article : Google Scholar
|
|
12
|
Zhou H, Li J, Gao P, Wang Q and Zhang J:
miR-155: A Novel Target in Allergic Asthma. Int J Mol Sci.
17:17732016. View Article : Google Scholar :
|
|
13
|
Zhang X, Zhao X, Sun H, Yan Y, Huang L, Gu
W, Jiang W, Wang Y, Zhu C, Ji W, et al: The role of miR-29c/B7-H3
axis in children with allergic asthma. J Transl Med. 16:2182018.
View Article : Google Scholar :
|
|
14
|
Lu TX, Lim EJ, Wen T, Plassard AJ, Hogan
SP, Martin LJ, Aronow BJ and Rothenberg ME: MiR-375 is
downregulated in epithelial cells after IL-13 stimulation and
regulates an IL-13-induced epithelial transcriptome. Mucosal
Immunol. 5:388–396. 2012. View Article : Google Scholar
|
|
15
|
Ma R, Ma L, Weng W, Wang Y, Liu H, Guo R,
Gao Y, Tu J, Xu TL, Cheng J, et al: DUSP6 SUMOylation protects
cells from oxidative damage via direct regulation of Drp1
dephosphorylation. Sci Adv. 6:eaaz03612020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng Y, Zhu Y, Xu J, Yang M, Chen P, Xu
W, Zhao J, Geng L and Gong S: PKN2 in colon cancer cells inhibits
M2 phenotype polarization of tumor-associated macrophages via
regulating DUSP6-Erk1/2 pathway. Mol Cancer. 17:132018. View Article : Google Scholar :
|
|
17
|
Chen L, Wang Y, Luan H, Ma G, Zhang H and
Chen G: DUSP6 protects murine podocytes from high glucose-induced
inflammation and apoptosis. Mol Med Rep. 22:2273–2282. 2020.
View Article : Google Scholar
|
|
18
|
Gu W, Yuan Y, Wang L, Yang H, Li S, Tang Z
and Li Q: Long non-coding RNA TUG1 promotes airway remodelling by
suppressing the miR-145-5p/DUSP6 axis in cigarette smoke-induced
COPD. J Cell Mol Med. 23:7200–7209. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu M, Xu X, Xi B, Dai Q, Li C, Su L, Zhou
X, Tang M, Yao Y and Yang J: Molecular network-based identification
of competing endogenous RNAs in thyroid carcinoma. Genes (Basel).
9:442018. View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
van den Berge M and Tasena H: Role of
microRNAs and exosomes in asthma. Curr Opin Pulm Med. 25:87–93.
2019. View Article : Google Scholar
|
|
22
|
Zhao L, Shi X, Wang N, Liu C and Wang J:
YAP1, targeted by miR-375, enhanced the pro-angiogenesis of airway
smooth muscle cells in asthma via STAT3 activation. Cell Cycle.
19:1275–1284. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dolka I, Król M and Sapierzyński R:
Evaluation of apoptosis-associated protein (Bcl-2, Bax, cleaved
caspase-3 and p53) expression in canine mammary tumors: An
immunohistochemical and prognostic study. Res Vet Sci. 105:124–133.
2016. View Article : Google Scholar
|
|
24
|
Clifford RL, Patel J, MacIsaac JL, McEwen
LM, Johnson SR, Shaw D, Knox AJ, Hackett TL and Kobor MS: Airway
epithelial cell isolation techniques affect DNA methylation
profiles with consequences for analysis of asthma related
perturbations to DNA methylation. Sci Rep. 9:144092019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grainge C, Dennison P, Lau L, Davies D and
Howarth P: Asthmatic and normal respiratory epithelial cells
respond differently to mechanical apical stress. Am J Respir Crit
Care Med. 190:477–480. 2014. View Article : Google Scholar :
|
|
26
|
Wang L, Liu XH, Chen H, Chen ZY, Weng XD,
Qiu T and Liu L: Picroside II protects rat kidney against
ischemia/reperfusion-induced oxidative stress and inflammation by
the TLR4/NF-κB pathway. Exp Ther Med. 9:1253–1258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bordag N, Klie S, Jürchott K, Vierheller
J, Schiewe H, Albrecht V, Tonn JC, Schwartz C, Schichor C and
Selbig J: Glucocorticoid (dexamethasone)-induced metabolome changes
in healthy males suggest prediction of response and side effects.
Sci Rep. 5:159542015. View Article : Google Scholar :
|
|
28
|
Giles AJ, Hutchinson MND, Sonnemann HM,
Jung J, Fecci PE, Ratnam NM, Zhang W, Song H, Bailey R, Davis D, et
al: Dexamethasone-induced immunosuppression: Mechanisms and
implications for immunotherapy. J Immunother Cancer. 6:512018.
View Article : Google Scholar :
|
|
29
|
Bertoli S, Picard M, Bérard E, Griessinger
E, Larrue C, Mouchel PL, Vergez F, Tavitian S, Yon E, Ruiz J, et
al: Dexamethasone in hyperleukocytic acute myeloid leukemia.
Haematologica. 103:988–998. 2018. View Article : Google Scholar :
|
|
30
|
Bonfiglio V, Reibaldi M, Fallico M, Russo
A, Pizzo A, Fichera S, Rapisarda C, Macchi I, Avitabile T and Longo
A: Widening use of dexamethasone implant for the treatment of
macular edema. Drug Des Devel Ther. 11:2359–2372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pohlmann D, Vom Brocke GA, Winterhalter S,
Steurer T, Thees S and Pleyer U: Dexamethasone inserts in
noninfectious uveitis: a single-center experience. Ophthalmology.
125:1088–1099. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Piastra M, Pizza A, Gaddi S, Luca E,
Genovese O, Picconi E, De Luca D and Conti G: Dexmedetomidine is
effective and safe during NIV in infants and young children with
acute respiratory failure. BMC Pediatr. 18:2822018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abaya R, Jones L and Zorc JJ:
Dexamethasone compared to prednisone for the treatment of children
with acute asthma exacerbations. Pediatr Emerg Care. 34:53–58.
2018. View Article : Google Scholar
|
|
34
|
Ji Y, Yang X and Su H: Overexpression of
microRNA-375 impedes platelet-derived growth factor-induced
proliferation and migration of human fetal airway smooth muscle
cells by targeting Janus kinase 2. Biomed Pharmacother. 98:69–75.
2018. View Article : Google Scholar
|
|
35
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu CP, Bi YJ, Liu DM and Wang LY:
Hsa-miR-375 promotes the progression of inflammatory bowel disease
by upregulating TLR4. Eur Rev Med Pharmacol Sci. 23:7543–7549.
2019.
|
|
37
|
Wang T, Chen D, Wang P, Xu Z and Li Y:
miR-375 prevents nasal mucosa cells from apoptosis and ameliorates
allergic rhinitis via inhibiting JAK2/STAT3 pathway. Biomed
Pharmacother. 103:621–627. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jin Y, Liu Y, Zhang J, Huang W, Jiang H,
Hou Y, Xu C, Zhai C, Gao X, Wang S, et al: The expression of
miR-375 is associated with carcinogenesis in three subtypes of lung
cancer. PLoS One. 10:e01441872015. View Article : Google Scholar :
|
|
39
|
Novaleski CK, Carter BD, Sivasankar MP,
Ridner SH, Dietrich MS and Rousseau B: Apoptosis and vocal fold
disease: clinically relevant implications of epithelial cell death.
J Speech Lang Hear Res. 60:1264–1272. 2017. View Article : Google Scholar
|
|
40
|
Hassan M, Watari H, AbuAlmaaty A, Ohba Y
and Sakuragi N: Apoptosis and molecular targeting therapy in
cancer. BioMed Res Int. 2014:1508452014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu G, Kuang G, Jiang W, Jiang R and Jiang
D: Polydatin promotes apoptosis through upregulation the ratio of
Bax/Bcl-2 and inhibits proliferation by attenuating the β-catenin
signaling in human osteosarcoma cells. Am J Transl Res. 8:922–931.
2016.
|
|
42
|
Hsu SF, Lee YB, Lee YC, Chung AL, Apaya
MK, Shyur LF, Cheng CF, Ho FM and Meng TC: Dual specificity
phosphatase DUSP6 promotes endothelial inflammation through
inducible expression of ICAM-1. FEBS J. 285:1593–1610. 2018.
View Article : Google Scholar
|
|
43
|
Beaudry K, Langlois MJ, Montagne A, Cagnol
S, Carrier JC and Rivard N: Dual-specificity phosphatase 6 deletion
protects the colonic epithelium against inflammation and promotes
both proliferation and tumorigenesis. J Cell Physiol.
234:6731–6745. 2019. View Article : Google Scholar
|
|
44
|
Carson WF IV, Salter-Green SE, Scola MM,
Joshi A, Gallagher KA and Kunkel SL: Enhancement of macrophage
inflammatory responses by CCL2 is correlated with increased miR-9
expression and downregulation of the ERK1/2 phosphatase Dusp6. Cell
Immunol. 314:63–72. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Z, Kobayashi S, Borczuk AC, Leidner
RS, Laframboise T, Levine AD and Halmos B: Dual specificity
phosphatase 6 (DUSP6) is an ETS-regulated negative feedback
mediator of oncogenic ERK signaling in lung cancer cells.
Carcinogenesis. 31:577–586. 2010. View Article : Google Scholar :
|





















