Introduction
Lung cancer was the most commonly diagnosed cancer
(11.6% of total cancer cases) and the leading cause of
cancer-related death (18.4% of total cancer-related deaths)
worldwide according to the global cancer statistics from 2018
(1). Among all lung cancer
cases, at least 80% are non-small cell lung cancer (NSCLC)
(2) and among these, lung
adenocarcinoma (LUAD) accounts for 50% of all the cases (3). The high prevalence rate of LUAD
requires further investigation to elucidate the mechanisms that
drive its pathogenesis and to determine potential targeted
therapies.
The endoplasmic reticulum is a complex organelle
that functions to orchestrate protein folding, Ca2+
storage, and lipid and carbohydrate metabolism. When cells are
exposed to intracellular or extracellular stimuli that cause
unfolded or misfolded proteins to accumulate in the endoplasmic
reticulum lumen, a condition known as endoplasmic reticulum stress
(ERS) is initiated to restore homeostasis. There are three
endoplasmic reticulum membrane-embedded sensors which transduce
signals during ERS, including inositol-requiring enzyme 1 (IRE1),
double-stranded RNA-activated protein kinas-like ER kinase (PERK)
and activating transcription factor 6 (ATF6) (4). In rapidly growing solid tumors, low
levels of oxygen and glucose often trigger ERS and lead to the
activation of spliced X-box binding protein (XBP1s) (5).
XBP1s is a key transcription factor during ERS. Upon
ERS, IRE1α splices a 26-base intron from the XBP1 mRNA,
transforming it from its unspliced form (XBP1u) to its spliced form
(XBP1s) (6). XBP1s was reported
to participate in the development of numerous types of cancer
(7). IRE1α-XBP1 was reported to
promote prostate cancer by activating the c-MYC pathway (8) and control T cell function in
ovarian cancer by regulating mitochondrial activity (9). XBP1 was also reported to promote
triple-negative breast cancer by regulating the hypoxia inducible
factor α (HIF1α) pathway (10).
Several studies have revealed that XBP1s plays a significant role
in tumorigenesis and cancer progression; however, to the best of
our knowledge, research on the role of XBP1s in LUAD is limited. A
recent study revealed that XBP1s was overexpressed in NSCLC tissues
and associated with Tumor Node Metastasis (TNM) stages, lymph node
metastasis and poor prognosis (11). However, the mechanisms involved
requires further investigation.
There are three main branches of MAPKs, including
JNK, ERK and p38 MAPK. These enzymes regulate various cellular
activities, including proliferation, differentiation, apoptosis,
survival, inflammation and innate immunity. These proteins
contribute to the pathology of diverse human diseases, including
cancer (12). Numerous studies
have proven MAPK to be associated with the initiation and
progression of LUAD (13-20).
However, whether there is an association between XBP1s and MAPK in
LUAD has not yet been investigated.
The present study was designed to investigate
whether and how XBP1s participates in the development of LUAD and
whether MAPK was involved in this process.
Materials and methods
Datasets
Both mRNA and protein expression of XBP1 were
analyzed using bioinformatics analysis. The mRNA expression level
of XBP1 in various types of cancer was analyzed using the ONCOMINE
(https://www.oncomine.org/resource/login.html)
(21) and the TIMER2.0 databases
(https://cistrome.shinyapps.io/timer)
(22). The thresholds in the
ONCOMINE database were set as follows: P<0.05, fold change of
all and gene rank of all. The protein expression level of XBP1 in
patients with LUAD and normal tissue from the Clinical Proteomic
Tumor Analysis Consortium, and the association between XBP1 and the
clinical characteristics was analyzed using the UALCAN database
(https://ualcan.path.uab.edu) (23). The proteins associated with XBP1,
and Gene Ontology (biological processes) analysis, and pathway
enrichment were analyzed (false discovery rate <0.01) using the
LinkedOmics database (https://www.linkedomics.org/login.php) (24). RNA sequencing data in patients
with LUAD from TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
were analyzed. The pathways involved in LUAD were analyzed using
the Kyoto Encyclopedia of Genes and Genomes (KEGG) database
(https://kegg.jp) (25). The association between XBP1 and
the MAPK pathway was analyzed using the LinkedOmics and GEPIA
databases (https://gepia.cancer-pku.cn) (26).
Cell culture
The human LUAD cell lines, A549, H1299 and PC9, and
the large cell lung cancer cell line, H460 were purchased from
American Type Culture Collection. Human bronchial epithelial (HBE)
cell line was purchased from NTCC Preservation Center. RPMI-1640
(Nanjing KeyGen Biotech Co., Ltd.), supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) was used to culture the
A549, H1299 and H460 cell lines. DMEM/High Glucose (HyClone;
Cytiva), supplemented with 10% FBS was used to culture the HBE and
PC9 cell lines. The cells were routinely cultured at 37°C in a
humidified incubator (Thermo Fisher Scientific, Inc.) with 21%
O2, 5% CO2 and 74% N2. For the
hypoxic culture environment, the A549 cell line was cultured at
37°C in a humidified incubator (Thermo Fisher Scientific, Inc.) for
0, 12, 24, 36, 48 and 60 h with 2% O2, 5% CO2
and 93% N2. SP600125 (cat. no. HY-12041; MedChemExpress)
was used to inhibit the JNK MAPK pathway in rescue experiments.
Small interfering (si)RNA and plasmid
transfection
siRNAs targeting XBP1s were designed by Guangzhou
RiboBio Co. Ltd. The following sequences were used: siNC
(non-targeting), 5′-TTC TCC GAA CGT GTC ACG TdTdT-3′; siXBP1s-1,
5′-GCA AGT GGT AGA TTT AGA A-3′; siXBP1s-2, 5′-GAT CGA AAG AAG GCT
CGA A-3′; and siXBP1s-3, 5′-TGA GAA CCA GGA GTT AAG A-3′. siRNA (50
µM) was transfected into the A549 cell line seeded
(2.0×105 cells/well) in the 6-well plate. The cells were
incubated with transfection reagent in the incubator for 24 h at
37°C with 21% O2. Then, the medium was replaced with
fresh culture medium and prepared for subsequent experiments. siRNA
transfection was conducted using Lipofectamine™ 3000 reagent (cat.
no. 2241260; Invitrogen; Thermo Fisher Scientific, Inc.).
The A549 and HBE cell lines were transfected with
either blank (no vector), liposome, siNC and siXBP1s-1/2/3 or
control and XBP1s overexpression vector to knockdown the expression
level of XBP1s or increase the expression level of XBP1s,
respectively.
The XBP1s overexpression plasmid was designed by
Shanghai GeneChem Co., Ltd., using Pcdna3.1-flag plasmid. Plasmid
transfection was conducted using Lipofectamine™ 3000 and P3000™
reagent (cat. no. 2241260; Invitrogen; Thermo Fisher Scientific,
Inc.), and the cells were transfected with 800 ng control or XBP1s
overexpression vector. Fresh culture medium was replaced 8 h
following transfection and the cells were cultured for another 16 h
at 37°C, then used for subsequent experiments.
Western blot analysis
Protein from the cells was extracted with protein
extraction buffer (RIPA, protease inhibitor cocktail, PMSF,
phosphorylation protease inhibitor A and B; ratio, 100:2:1:1:1.
Protein concentration was determined using a BCA protein
Concentration kit (cat. no. 16F17B97; Boster Biological
Technology). Protein (30 µg) was loaded into each lane and
separated using 10% SDS-PAGE, then transferred onto a PVDF
membrane. The membrane was blocked with 5% skimmed milk for 1 h at
room temperature, washed with TBS with 0.05% Tween-20 (TBST) and
incubated with the primary antibodies overnight at 4°C. The
following primary antibodies were used: XBP1s (cat. no. 40435s;
1:1,000; Cell Signaling Technology, Inc.), phosphorylated (p)-c-JNK
(cat. no. 4668; 1:1,000; Cell Signaling Technology, Inc.), JNK
(cat. no. 9252; 1:1,000; Cell Signaling Technology, Inc.), p-ERK
(cat. no. 4370; 1:1,000; Cell Signaling Technology, Inc.), ERK
(cat. no. 4695; 1:1,000; Cell Signaling Technology, Inc.), p-p38
(cat. no. 4511; 1:1,000; Cell Signaling Technology, Inc.), p38
(cat. no. 8690; 1:1,000; Cell Signaling Technology, Inc.) and actin
(cat. no. 66009-1-Ig; 1:4,000; ProteinTech Group, Inc.). Then, the
membrane was washed with TBST, incubated with HRP-conjugated
secondary antibodies (goat anti-rabbit, cat. no. AS1107; goat
anti-mouse, cat. no. AS1106) (both 1:4,000 and purchased from Wuhan
Aspen Biotechnology, Co., Ltd.) for 1 h at room temperature. The
membranes were washed with TBST again and the protein bands were
detected using a detection kit (cat. no. 201005-79; Advansta,
Inc.). ImageJ software (v1.46r; National Institutes of Health) was
used to semi-quantify protein expression.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). Then, RT using
PrimeScript RT Master Mix (cat. no. RR036A; Takara Biotechnology
Co., Ltd.) was performed with 500 ng RNA and a total volume of 10
µl. The following temperature conditions were used: 37°C for
15 min, 85°C for 5 sec and held at 4°C until further
experimentation. qPCR was performed using TB Green®
Premix Ex Taq (cat. no. RR420A; Takara Biotechnology Co., Ltd.) and
the following thermocycling conditions: Initial denaturation at
95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C
for 30 sec, and a final extension at 65-95°C, in 0.5°C increments
for 5 sec. Relative quantification analysis was performed using the
2−ΔΔCq method (27).
The following primers were used to amplify the genes of interest:
XBP1s, forward, 5′-GCT GAG TCC GCA GCA GG-3′ and reverse, 5′-CTC
TGG GGA AGG GCA TTT GA-3′; actin forward, 5′-AGC GAG CAT CCC CCA
AAG TT-3′ and reverse, 5′-GGG CAC GAA GGC TCA TCA TT-3′.
EdU staining assay and cytotoxicity
assay
The cells were seeded into 96-well plates,
transfected with siXBP1s or siNC (A549 cells; 6,000 cells/well), or
XBP1s overexpression vector or control vector (A549 and HBE cells)
(both 8,000 cells/well). The cells were then cultured under 21%
O2 normoxic or 2% O2 hypoxic conditions, 24 h
following transfection. Subsequently, EdU staining (cells were
incubated with the EdU reagent A for 2 h in the incubator at 37°C,
and subsequent procedures were performed according to the
manufacturer's instructions; Cell-Light™ EdU Apollo567 In
Vitro kit; Guangzhou RiboBio, Co., Ltd.) or cell viability
(cells were incubated with the Cell Counting Kit (CCK)-8 reagent
for 30 mins and OD was measured every 15 mins) (Cell Counting Kit-8
assay; cat. no. HY-K0301; MedChem Express) was performed. The
detailed procedures were conducted according to the manufacturers'
instructions. Images were captured at ×200 magnification.
Colony formation assay
The cells were cultured with siXBP1s or siNC
transfection (A549 cells), or XBP1s overexpression vector or
control vector transfection (A549 and HBE cells) for 24 h, then 300
cells/well were seeded into 6-well plates. After ~2 weeks, colony
formation was assessed, and the cells were fixed with 4%
paraformaldehyde for 15 mins, then stained with crystal violet for
15 mins, both at room temperature. Then, images of the colonies
(>50 cells) were captured and manually calculated.
Transwell assay
Migration ability was evaluated using Transwell
chambers with 8-µm pores (Corning, Inc.). siXBP1s or siNC
was transfected into the A549 cell line (2.0×105
cells/well) or XBP1s overexpression vector or control vector was
transfected into the HBE (3.0×105 cells/well) or A549
cell lines (3.0×105 cells/well). Subsequently, the cells
were digested and a total of 1.0×104 cells suspended in
200 µl culture medium (10% FBS) were added to the upper
chamber, and 600 µl culture medium was added to the lower
chamber, replacing the cell culture medium with fresh culture
medium 24 h after suspension. The cells were cultured at 37°C in
the incubator for 24 h with 21% O2 (normoxic conditions)
in the A549 and HBE cell lines, or 36 h with 21% O2 or
2% O2 (hypoxic conditions) in rescue experiments with
the A549 cell line, until they were harvested. Then, the cells were
fixed with 4% paraformaldehyde for 15 mins, then stained with
crystal violet for 15 mins, both at room temperature. The cells
were washed with PBS, then images of the migrated cells were
captured (TE2000 U; light; Nikon Corporation) at ×200
magnification.
Wound healing assay
The A549 (2.0×105 or 3.0×105
cells/well) or HBE cell lines (3.0×105 cells/well) were
seeded in a 6-well plate and transfected 24 h later with siXBP1s or
siNC, or XBP1s overexpression or control vector, respectively. When
the cells reached 80-90% confluence, a wound was generated using a
pipette tip. Then, the cells were cultured for 24 h with serum-free
medium. The images of the cells were captured at 0 and 24 h, at
×200 magnification (TE2000 U; Nikon Corporation). The widths of the
wound were recorded, and wound closure was calculated as [wound
width (0-24 h)/wound width (0 h)] ×100%.
Flow cytometry
The A549 (2.0×105 or
3.0×105cells/well) or HBE cell lines (3.0×105
cells/well) were seeded in a 6-well plate and transfected 24 h
later with siXBP1s or siNC, or XBP1s overexpression or control
vector, respectively. The cells were cultured for 24 h with 21%
O2 (normoxic conditions) in the A549 and HBE cell lines,
or for 36 h with 21% O2 or 2% O2 (hypoxic
conditions) in rescue experiments with the A549 cell line. Then,
the cells were collected, washed three times with cold PBS and
suspended in 300 µl binding buffer. Next, the cells were
stained with 3 µl Annexin V-FITC and 3 µl PI at room
temperature for 30 mins with an apoptosis detection kit (cat. no.
556547; BD Pharmingen; BD Biosciences).
Statistical analysis
All the quantitative data are presented as the mean
± SD. GraphPad Prism (V8.0) was used for statistical analysis.
Statistical significance was determined using an unpaired Student's
t-test for comparisons between two groups, while one-way ANOVA
followed by Bonferroni post hoc test was used for comparisons among
more than two groups. All the experiments were performed
independently at least three times. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of XBP1s is increased in LUAD
and A549 cell line
Analysis of data from the TIMER2.0, ONCOMINE and
UALCAN databases revealed that XBP1 mRNA and protein expression
level was highly expressed in various types of cancer compared with
that in normal tissues, including LUAD (Fig. 1A, B and D, respectively). The
mRNA expression level of XBP1 was significantly different in
patients with different clinical features, including age, smoking
habit, individual cancer stage and nodal metastasis status
(Fig. 1C). Since XBP1s is a
transcription factor, that is more active than XBP1u (28), the mRNA and protein expression
level of XBP1s in the normal HBE, the A549, H1299 and PC9 LUAD, and
the H460 large cell lung cancer cell lines was analyzed. XBP1s
protein expression level was significantly elevated in the A549,
H1299, PC9 and H460 cell lines compared with that in the HBE cell
line. In addition, XBP1s mRNA expression level was significantly
elevated in the A549 cell line compared with that in the HBE cell
line (Fig. 2A). Since tumor
cells survive in a hypoxic microenvironment (29), the A549 cell line was cultured
under 2% O2 hypoxia to further investigate the
expression level of XBP1s. The protein expression level of XBP1s in
the A549 cell line was increased under hypoxic conditions. The
protein expression level reached its peak at 36 h, then declined,
followed by another elevation at 60 h (Fig. 2B). To further investigate the
role of XBP1s in LUAD, siRNA was designed to knockdown the
expression level of XBP1s in the A549 cell line, which had the
highest protein and mRNA expression level compared with that in the
HBE cell line. In addition, XBP1s overexpression vector was
designed to overexpress XBP1s in the normal HBE cell line, which
had the lowest mRNA and protein expression level. Western blot and
RT-qPCR was performed to verify the knockdown and overexpression of
XBP1s at the protein and mRNA level in the A549 (Fig. 2C) and HBE (Fig. 2D) cell lines, respectively.
siXBP1s-1 significantly knocked down the expression level of XBP1s
compared with that for siXBP1s-2 and -3. These results indicated
that the expression level of XBP1s was increased in LUAD and in the
A549 cell line.
 | Figure 2High expression level of XBP1s in the
A549 cell line. (A) XBP1s protein and mRNA expression level was
analyzed in the normal HBE and the A549, H1299, PC9 and H460 lung
cancer cell lines. The protein expression level was subsequently
analyzed using densitometry. (B) XBP1s protein expression level was
measured using western blot analysis in the A549 cell line cultured
under hypoxic conditions for the indicated times, and subsequently
analyzed using densitometry. (C) Protein and mRNA expression levels
of XBP1s was measured in the A549 cell line following no
transfection (blank), transfection with liposome, siNC and
siXBP1s-1/2/3. (D) Protein and mRNA expression levels of XBP1s was
measured in the HBE cell line following no transfection (blank),
transfection with liposome, control vector or XBP1 overexpression
vector. n=3. The data are presented as the mean ± SD.
**P<0.01, ***P<0.001,
****P<0.0001. XBP1s, spliced X-box binding protein 1;
LUAD, lung adenocarcinoma; NC, negative control; si, small
interfering. |
XBP1s is positively associated with
proliferation, colony formation, and cell viability in both the
A549 and HBE cell lines
The aforementioned results confirmed the high
protein and mRNA expression level of XBP1s in the A549 cells;
therefore, it was investigated whether XBP1s was associated with
the proliferation, colony formation, and cell viability of the A549
and HBE cell lines. Proliferation was detected using EdU staining.
The cells positive for EdU staining were stained red and were in a
mitotic state. The proliferation rate of the A549 cells was
significantly decreased when the expression level of XBP1s was
knocked down. By contrast, it was significantly increased when
XBP1s was overexpressed in the HBE cell line (Fig. 3A). Similarly, colony formation
ability was significantly reduced in the A549 cell line following
knock down of XBP1s expression, but was significantly increased in
the HBE cells following overexpression of XBP1s (Fig. 3B). Similar results were also
obtained when cell viability was measured (Fig. 3C). In conclusion, XBP1s was
associated with proliferation, colony formation and cell viability
in both the A549 and HBE cell lines.
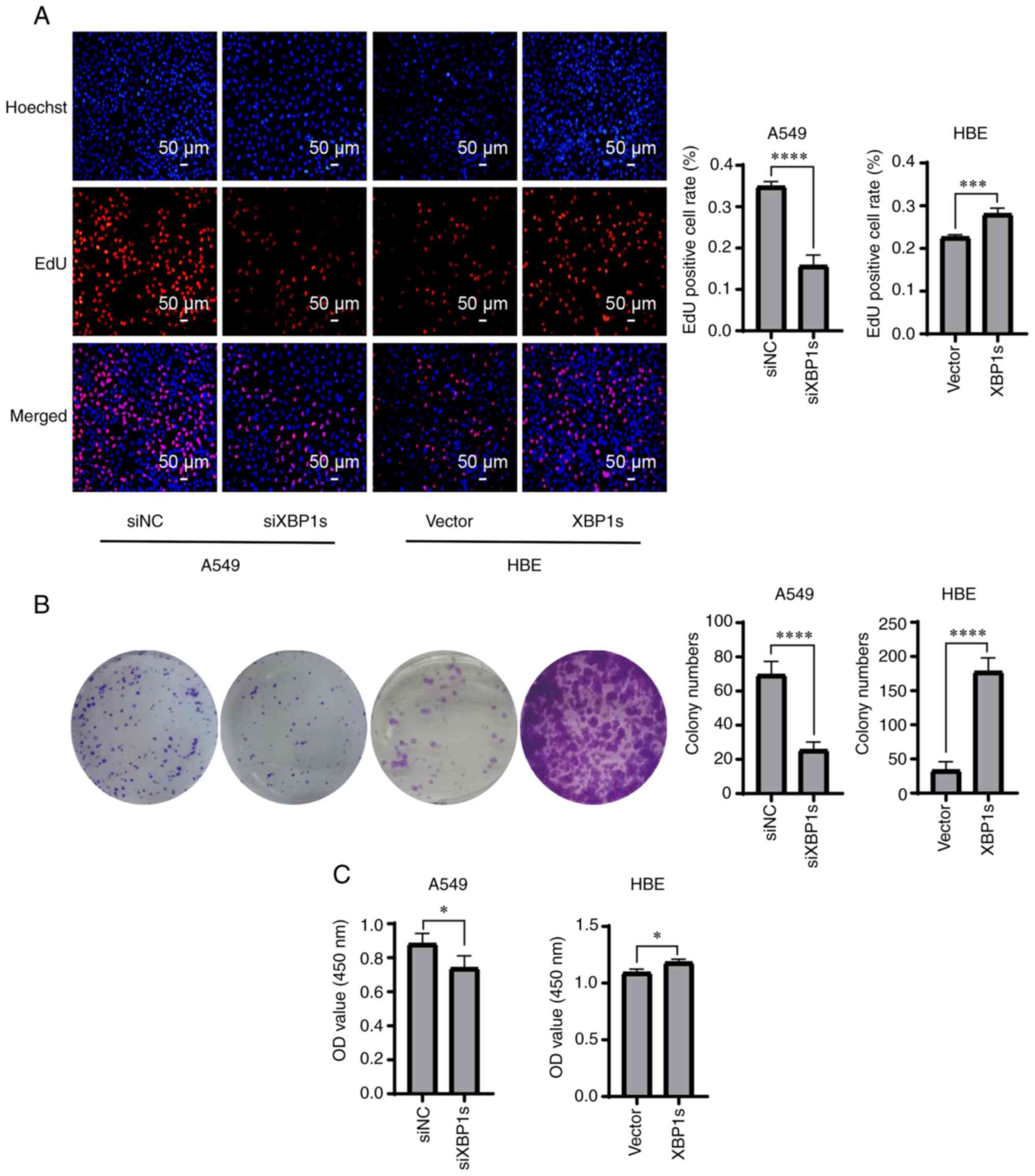 | Figure 3Knockdown of XBP1s expression in the
A549 cell line decreases proliferation, colony formation and cell
viability, while overexpressing XBP1s in the HBE cell line
increases proliferation, colony formation and cell viability. The
A549 cell line was transfected with siNC or siXBP1s, while the HBE
cell line was transfected with control or XBP1s overexpression
vector. Then, (A) EdU staining, (B) colony formation assay and (C)
cytotoxicity assay was performed to determine the number of
EdU-positive cells, number of colonies and cell viability,
respectively. n=3. The data are presented as the mean ± SD.
*P<0.05, ***P<0.001,
****P<0.0001. XBP1s, spliced X-box binding protein 1;
NC, negative control; si, small interfering; OD, optical
density. |
XBP1s is associated with migration,
invasion and apoptosis in the A549 and HBE cell lines
In addition to proliferation, abnormally increased
migration and invasion abilities and decreased apoptosis are also
crucial factors that drive the worsening of tumor development
(30,31). Therefore, it was investigated
whether XBP1s was associated with migration, invasion and apoptosis
in the LUAD cell line, and in the normal HBE cell line as a
control. Transwell assay demonstrated that knock down of XBP1s
expression decreased the numbers of migrating A549 cells, while
overexpressing XBP1s resulted in increased numbers of migrating HBE
cells (Fig. 4A). A wound healing
assay demonstrated that the wound healing ability of the cells was
decreased following knock down of XBP1s expression in the A549 cell
line and was increased following overexpression of XBP1s in the HBE
cell line (Fig. 4B). Annexin-V
FITC/PI double staining and flow cytometry was performed to detect
apoptosis in the cell lines. The apoptosis rate was increased in
the A549 cell line following knock down of XBP1s expression and
decreased in the HBE cell line following overexpression of XBP1s
(Fig. 4C). These results showed
that XBP1s was associated with migration, invasion and apoptosis in
the A549 and HBE cell lines.
 | Figure 4Knockdown of XBP1s expression in the
A549 cell line decreases migration, wound healing rate, and
increases apoptosis, while overexpressing XBP1s in the HBE cell
line led to the opposite biological effects. The A549 cell line was
transfected with siNC or siXBP1s, while the HBE cell line was
transfected with control or XBP1s overexpression vector. Then, (A)
Transwell assay, (B) wound healing assay and (C) flow cytometry was
performed to analyze the migratory and wound healing ability, and
apoptosis, respectively. The data are presented as the mean ± SD.
n=3. **P<0.01, ***P<0.001,
****P<0.0001. XBP1s, spliced X-box binding protein 1;
NC, negative control; si, small interfering. |
p-JNK is the downstream target of
XBP1s
To investigate the possible pathways that mediate
the biological effects of XBP1s, GO and pathway enrichment analysis
of the genes associated with XBP1 was analyzed using the
LinkedOmics database. A total of 515 patients with LUAD from TCGA
were analyzed. Genes that were associated with high XBP1 expression
were found using the Volcano plot (Fig. 5A). The GO terms from the genes
associated with XBP1, based on the LUAD samples revealed that these
genes were most frequently involved in protein processing in the
endoplasmic reticulum (Fig. 5B).
GSEA pathway enrichment revealed that both endoplasmic reticulum
stress (Fig. 5C) and protein
activation cascade (Fig. 5D)
were enriched with high expression of XBP1. As aforementioned, XBP1
is a transcription factor during ERS; therefore, it would be
important to identify genes enriched in ERS. With respect to the
enrichment of protein activation cascade, the MAPK pathway could be
a possible candidate. In addition, activation of the MAPK pathway
involves protein phosphorylation cascade and the MAPK pathway is a
classic tumor-related pathway (32). Furthermore, when the pathways
enriched in NSCLC were analyzed, the MAPK pathway was associated
(Fig. S1A). To further identify
whether the MAPK pathway was associated with XBP1, correlation
analysis of XBP1 and the three main genes in the MAPK pathway was
analyzed using a Pearson's correlation test. p-JNK (MAPK8) was
found to be the only highly positively correlated gene both via
LinkedOmics (Pearson's correlation, 0.09959;
P=2.381×10−2) (Fig.
6A-a) and GEPIA database (Pearson's correlation, 0.18;
P=1.7×10−5) (Fig.
6A-b). p-ERK (MAPK1) was not correlated with XBP1s using the
LinkedOmics database (Fig. 6B).
p-p38 (MAPK14) was not positively correlated with XBP1s using the
LinkedOmics database (Fig. 6C).
Correlation analysis between XBP1 and p-ERK or p-p38 was also
performed using the GEPIA database (Fig. S1B), and neither were correlated.
To further verify the association between XBP1s and the MAPK
pathway, the protein expression level of proteins in the MAPK
pathway was analyzed following knockdown of XBP1s expression. Since
tumor cells often grow in a hypoxic microenvironment and it was
aforementioned that XBP1s reached its peak expression level at 36 h
when cultured in 2% O2, the cells transfected with siNC
or siXBP1s were cultured under normoxic (21% O2, 36 h)
or hypoxic (2% O2, 36 h) conditions and the protein
expression level of proteins in the MAPK pathway was analyzed.
p-JNK expression was highly consistent with the expression level of
XBP1s. The protein expression level of both p-JNK and XBP1s were
increased under hypoxic conditions compared with that in normoxic
conditions. When XBP1s expression was knocked down in cells
cultured under normoxic or hypoxic conditions, p-JNK expression
level was also decreased under both normoxic and hypoxic
conditions. The expression levels of p-ERK and p-p38 were both
increased under hypoxic conditions compared with that in normoxic
conditions, but showed no change when XBP1s expression was knocked
down under both normoxic and hypoxic conditions (Fig. 6D). Taken together, p-JNK rather
than p-ERK or p-p38 was found to be the downstream target of
XBP1s.
Inhibition of p-JNK mitigates the
biological effects caused by overexpression of XBP1s
Since p-JNK was hypothesized to be the downstream
target of XBP1s, it was investigated whether inhibition of p-JNK
could mitigate the effects caused by XBP1s overexpression. The A549
cells were cultured under normoxia, hypoxia, hypoxia with XBP1s
overexpression, hypoxia with XBP1s overexpression and inhibition of
p-JNK with SP600125. Hypoxia increased the proliferation rate
(Fig. 7A and B), cell viability
(Fig. 7C), colony formation
ability (Fig. 7D and E), numbers
of migrated cells (Fig. 8A),
invasion ability (Fig. 8B), and
decreased the apoptosis rate (Fig.
8C) of the A549 cells. XBP1s overexpression under hypoxia
further enhanced these effects. However, cells treated with the
p-JNK inhibitor exhibited a notable decrease in these effects
(Figs. 7A-E and 8A-C). Taken together, the results
indicated that inhibiting p-JNK with SP600125 alleviated the
biological effects of the overexpression of XBP1s.
Discussion
In brief, the results from the present study
demonstrated that XBP1s promoted the development of LUAD via the
p-JNK MAPK pathway. XBP1s was expressed at notably high levels in
lung cancer tissues compared with that in normal tissues, and in
the A549 cell line compared with that in the HBE, H1299, PC9 and
H460 cell lines. High expression of XBP1s promoted cell
proliferation, colony formation, cell viability, migration,
invasion, wound healing rate and reduction in apoptosis. Knockdown
of XBP1s in the A549 cells resulted in the impairment of these
prosurvival effects, while overexpression of XBP1s in the HBE cell
line resulted in enhancement of these effects. Further
investigation revealed that p-JNK was the downstream effector of
XBP1s. Inhibition of p-JNK with SP600125 abolished the increased
prosurvival effects caused by overexpressing XBP1s.
Targeting XBP1s for cancer therapy has been studied
in numerous malignancies. In the tumor microenvironment, XBP1s
cooperated with HIF1α to promote cell survival and thus enabled
some tumor subtypes to grow under hypoxic conditions (33). In triple-negative breast cancer,
XBP1s was activated and drove tumorigenicity by forming a
transcriptional complex with HIF1α and regulating its expression
(10). In ovarian cancer, XBP1
was highly expressed in T cells and tumor-associated dendritic
cells, and upregulation of XBP1s expression impaired mitochondrial
respiration and antitumor function (9,34). In pancreatic ductal
adenocarcinoma, XBP1s was associated with immune escape and
metastasis (35). In prostate
cancer, XBP1s expression was notably increased, promoted cancer
development and was associated with prognosis (8,36). In intestinal tumors, XBP1
determined the propensity of the epithelium to develop tumors by
instructing a multilayered regenerative response in the intestinal
epithelium (37). In
glioblastoma multiform, IRE1-XBP1 was associated with tumor
progression (38). The protein
expression level of XBP1s is usually increased in these tumors and
associated with tumorigenicity, metastasis and poor prognosis. In
some malignancies, such as breast cancer, knockdown of XBP1s
expression selectively blocked the growth of tumor cells (39). All these studies demonstrate that
XBP1s promoted tumorigenesis, progression and prognosis. However,
studies investigating how XBP1s functions in lung cancer are
relatively limited. In the present study it was demonstrated that
XBP1s was associated with the development of LUAD via the p-JNK
MAPK pathway, providing further support for targeting XBP1s in LUAD
treatment.
There are already drugs or inhibitors that target
ERS for cancer therapy. Activation of ERS has been described in
different human tumors, such as breast cancer, prostate cancer and
kidney tumor, and in multiple cellular and animal models of cancer,
such as murine cancer and prostate cancer cells (40). Tumor cells survive in a more
hostile microenvironment, facing nutrient deprivation, oxygen
limitation, high metabolic demand, and oxidative stress, and all
these stimuli induce ERS in tumor cells. Sustained ERS endows
malignant cells with greater tumorigenic, metastatic and
drug-resistant ability (41). A
large number of drugs or inhibitors that target IRE1α, PERK and
ATF6 have been developed for cancer therapy. Among these IRE1α
inhibitors, STF-083010 and MKC-3946 are already at the preclinical
stage. Other IRE1α inhibitors, including MKC3946, B-I09 and
MKC8866, were proven to induce apoptosis, and inhibit tumor growth
(42,43). These drugs and inhibitors provide
exciting clinical perspectives for the treatment of various tumor
subtypes, such as multiple myeloma xenografts and triple-negative
breast cancer (39,43,44). Based on the results from the
current study, XBP1s played an important role in promoting survival
in the A549 cell line. It is highly promising that IRE1α inhibitors
would be applicable for patients with LUAD.
The role of IRE1α/XBP1s in lung cancer has not been
thoroughly studied; however, the role of MAPK in lung cancer has
been investigated by numerous researchers, although the results are
controversial. Some studies indicated that MAPK promoted the
initiation and progression of tumors (13,45), mediated proliferation and the
antiapoptotic effects in LUAD (17,19), and was associated with poor
survival and treatment resistance in lung cancer (46). However, other studies indicated
that MAPK mediated migration inhibition (47) and the induction of cell apoptosis
in tumors (16,18,48). The results from the present study
were consistent with most studies (13,17,19,45,46). It was found that the MAPK pathway
promoted tumor growth rather than suppressing it. In addition, it
was found that p-JNK MAPK mediated the survival-promoting effects
of XBP1s in the A549 cell line; thus, contributing to the
development of LUAD.
There are also some limitations to the present
study. It was found that p-JNK was the downstream effector that
mediated the pro-survival effects of XBP1s; however, whether it
functioned via direct or indirect interaction requires further
investigation. As a transcription factor, XBP1 is able to
positively regulate protein phosphorylation (49), but how it promoted the
phosphorylation of JNK remains unknown. The upstream activator of
XBP1s, IRE1 was reported to be able to activate JNK during ERS
(50). Whether there is an
interplay between IRE1/JNK and XBP1s/p-JNK remains to be
elucidated.
To conclude, it was found that XBP1s promoted the
development of LUAD via the p-JNK MAPK pathway. Targeting
XBP1s/p-JNK could be a potentially effective strategy for the
treatment of LUAD.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HJ was involved in conceptualization of the study,
analysis, performed the experiments and writing the original draft
of the manuscript. QJ was involved in the conceptualization of the
study and methodology. YH and XLi were involved in the
conceptualization of the study and revising and editing the
manuscript. YX and XLiu contributed to the design of the study and
critically reviewed the manuscript, revised and edited the
manuscript. HJ and XLiu confirmed the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This study was funded by the National Natural Science Foundation
of China (grant nos. 81973987, 81700051 and 81700052).
Abbreviations:
|
XBP1s
|
spliced X-box binding protein 1
|
|
ERS
|
endoplasmic reticulum stress
|
|
IRE1
|
inositol-requiring enzyme 1
|
|
PERK
|
double-stranded RNA-activated protein
kinase-like ER kinase
|
|
ATF6
|
activating transcription factor 6
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
HIF1α
|
hypoxia inducible factor α
|
|
TNM
|
Tumor Node Metastasis
|
|
LUAD
|
lung adenocarcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
HBE
|
human bronchial epithelial
|
|
TCGA
|
The Cancer Genome Atlas
|
|
FDR
|
False Discovery Rate
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Devesa SS, Bray F, Vizcaino AP and Parkin
DM: International lung cancer trends by histologic type:
Male:Female differences diminishing and adenocarcinoma rates
rising. Int J Cancer. 117:294–299. 2005. View Article : Google Scholar
|
|
3
|
Chan BA and Hughes BG: Targeted therapy
for non-small cell lung cancer: Current standards and the promise
of the future. Transl Lung Cancer Res. 4:36–54. 2015.PubMed/NCBI
|
|
4
|
Senft D and Ronai ZA: UPR, autophagy, and
mitochondria crosstalk underlies the ER stress response. Trends
Biochem Sci. 40:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brenning G, Simonsson B, Kallander C and
Ahre A: Pretreatment serum beta 2-microglobulin in multiple
myeloma. Br J Haematol. 62:85–93. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shinya S, Kadokura H, Imagawa Y, Inoue M,
Yanagitani K and Kohno K: Reconstitution and characterization of
the unconventional splicing of XBP1u mRNA in vitro. Nucleic Acids
Res. 39:5245–5254. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen S, Chen J, Hua X, Sun Y, Cui R, Sha J
and Zhu X: The emerging role of XBP1 in cancer. Biomed
Pharmacother. 127:1100692020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sheng X, Nenseth HZ, Qu S, Kuzu OF,
Frahnow T, Simon L, Greene S, Zeng Q, Fazli L, Rennie PS, et al:
IRE1α-XBP1s pathway promotes prostate cancer by activating c-MYC
signaling. Nat Commun. 10:3232019. View Article : Google Scholar
|
|
9
|
Song M, Sandoval TA, Chae CS, Chopra S,
Tan C, Rutkowski MR, Raundhal M, Chaurio RA, Payne KK, Konrad C, et
al: IRE1α-XBP1 controls T cell function in ovarian cancer by
regulating mitochondrial activity. Nature. 562:423–428. 2018.
View Article : Google Scholar :
|
|
10
|
Chen X, Iliopoulos D, Zhang Q, Tang Q,
Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, et
al: XBP1 promotes triple-negative breast cancer by controlling the
HIF1α pathway. Nature. 508:103–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo Q, Shi W, Dou B, Wang J, Peng W, Liu
X, Zhao D, Tang F, Wu Y, Li X, et al: XBP1-IGFBP3 signaling pathway
promotes NSCLC invasion and metastasis. Front Oncol. 11:6549952021.
View Article : Google Scholar
|
|
12
|
Kim EK and Choi EJ: Compromised MAPK
signaling in human diseases: An update. Arch Toxicol. 89:867–882.
2015. View Article : Google Scholar
|
|
13
|
Cicchini M, Buza EL, Sagal KM, Gudiel AA,
Durham AC and Feldser DM: Context-dependent effects of amplified
MAPK signaling during lung adenocarcinoma initiation and
progression. Cell Rep. 18:1958–1969. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stutvoet TS, Kol A, de Vries EG, de Bruyn
M, Fehrmann RS, Terwisscha van Scheltinga AG and de Jong S: MAPK
pathway activity plays a key role in PD-L1 expression of lung
adenocarcinoma cells. J Pathol. 249:52–64. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao X, Fang X, Malik WS, He Y, Li X, Xie
M, Sun W, Xu Y and Liu X: TRB3 interacts with ERK and JNK and
contributes to the proliferation, apoptosis, and migration of lung
adenocarcinoma cells. J Cell Physiol. 235:538–547. 2020. View Article : Google Scholar
|
|
16
|
Shen H, Liu J, Wang Y, Lian H, Wang J,
Xing L, Yan X, Wang J and Zhang X: Aflatoxin G1-induced oxidative
stress causes DNA damage and triggers apoptosis through MAPK
signaling pathway in A549 cells. Food Chem Toxicol. 62:661–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du X, Wang S, Liu X, He T, Lin X, Wu S,
Wang D, Li J, Huang W and Yang H: MiR-1307-5p targeting TRAF3
upregulates the MAPK/NF-κB pathway and promotes lung adenocarcinoma
proliferation. Cancer Cell Int. 20:5022020. View Article : Google Scholar
|
|
18
|
Ong JY, Yong PV, Lim YM and Ho AS:
2-Methoxy-1,4-naphthoquinone (MNQ) induces apoptosis of A549 lung
adenocarcinoma cells via oxidation-triggered JNK and p38 MAPK
signaling pathways. Life Sci. 135:158–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou XM, Zhang T, Da Z and Wu XA: CHPF
promotes lung adenocarcinoma proliferation and anti-apoptosis via
the MAPK pathway. Pathol Res Pract. 215:988–994. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen YY, Liu FC, Chou PY, Chien YC, Chang
WS, Huang GJ, Wu CH and Sheu MJ: Ethanol extracts of fruiting
bodies of Antrodia cinnamomea suppress CL1-5 human lung
adenocarcinoma cells migration by inhibiting matrix
metalloproteinase-2/9 through ERK, JNK, p38, and PI3K/Akt signaling
pathways. Evid Based Complement Alternat Med. 2012:3784152012.
|
|
21
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar :
|
|
22
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar :
|
|
25
|
Kanehisa M, Sato Y and Kawashima M: KEGG
mapping tools for uncovering hidden features in biological data.
Protein Sci. 31:47–53. 2022. View Article : Google Scholar
|
|
26
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Yoshida H, Matsui T, Yamamoto A, Okada T
and Mori K: XBP1 mRNA is induced by ATF6 and spliced by IRE1 in
response to ER stress to produce a highly active transcription
factor. Cell. 107:881–891. 2001. View Article : Google Scholar
|
|
29
|
Huang CH, Chong KY and Lei KF: Analysis of
the internal hypoxic environment in solid tumor tissue using a
folding paper system. ACS Appl Mater Interfaces. 13:33885–33893.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jin X, Zhai B, Fang T, Guo X and Xu L:
FXR1 is elevated in colorectal cancer and acts as an oncogene.
Tumour Biol. 37:2683–2690. 2016. View Article : Google Scholar
|
|
31
|
Goldar S, Khaniani MS, Derakhshan SM and
Baradaran B: Molecular mechanisms of apoptosis and roles in cancer
development and treatment. Asian Pac J Cancer Prev. 16:2129–2144.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Drosten M and Barbacid M: Targeting the
MAPK pathway in KRAS-Driven tumors. Cancer Cell. 37:543–550. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xia Z, Wu S, Wei X, Liao Y, Yi P, Liu Y
and Liu J and Liu J: Hypoxic ER stress suppresses β-catenin
expression and promotes cooperation between the transcription
factors XBP1 and HIF1α for cell survival. J Biol Chem.
294:13811–13821. 2019. View Article : Google Scholar :
|
|
34
|
Cubillos-Ruiz JR, Silberman PC, Rutkowski
MR, Chopra S, Perales-Puchalt A, Song M, Zhang S, Bettigole SE,
Gupta D, Holcomb K, et al: ER stress sensor XBP1 controls
anti-tumor immunity by disrupting dendritic cell homeostasis. Cell.
161:1527–1538. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pommier A, Anaparthy N, Memos N, Kelley
ZL, Gouronnec A, Yan R, Auffray C, Albrengues J, Egeblad M,
Iacobuzio-Donahue CA, et al: Unresolved endoplasmic reticulum
stress engenders immune-resistant, latent pancreatic cancer
metastases. Science. 360:eaao49082018. View Article : Google Scholar
|
|
36
|
Sheng X, Arnoldussen YJ, Storm M, Tesikova
M, Nenseth HZ, Zhao S, Fazli L, Rennie P, Risberg B, Wæhre H, et
al: Divergent androgen regulation of unfolded protein response
pathways drives prostate cancer. EMBO Mol Med. 7:788–801. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Niederreiter L, Fritz TM, Adolph TE,
Krismer AM, Offner FA, Tschurtschenthaler M, Flak MB, Hosomi S,
Tomczak MF, Kaneider NC, et al: ER stress transcription factor Xbp1
suppresses intestinal tumorigenesis and directs intestinal stem
cells. J Exp Med. 210:2041–2056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lhomond S, Avril T, Dejeans N, Voutetakis
K, Doultsinos D, McMahon M, Pineau R, Obacz J, Papadodima O, Jouan
F, et al: Dual IRE1 RNase functions dictate glioblastoma
development. EMBO Mol Med. 10:e79292018. View Article : Google Scholar :
|
|
39
|
Zhao N, Cao J, Xu L, Tang Q, Dobrolecki
LE, Lv X, Talukdar M, Lu Y, Wang X, Hu DZ, et al: Pharmacological
targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven
breast cancer. J Clin Invest. 128:1283–1299. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Clarke HJ, Chambers JE, Liniker E and
Marciniak SJ: Endoplasmic reticulum stress in malignancy. Cancer
Cell. 25:563–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cubillos-Ruiz JR, Bettigole SE and
Glimcher LH: Tumorigenic and immunosuppressive effects of
endoplasmic reticulum stress in cancer. Cell. 168:692–706. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang M and Kaufman RJ: The impact of the
endoplasmic reticulum protein-folding environment on cancer
development. Nat Rev Cancer. 14:581–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hetz C, Axten JM and Patterson JB:
Pharmacological targeting of the unfolded protein response for
disease intervention. Nat Chem Biol. 15:764–775. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mimura N, Fulciniti M, Gorgun G, Tai YT,
Cirstea D, Santo L, Hu Y, Fabre C, Minami J, Ohguchi H, et al:
Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising
therapeutic option in multiple myeloma. Blood. 119:5772–5781. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu P, Wang H, Liang Y, Hu A, Xing R,
Jiang L, Yi L and Dong J: LINC00852 promotes lung adenocarcinoma
spinal metastasis by targeting S100A9. J Cancer. 9:4139–4149. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sato H, Schoenfeld AJ, Siau E, Lu YC, Tai
H, Suzawa K, Kubota D, Lui AJW, Qeriqi B, Mattar M, et al: mapk
pathway alterations correlate with poor survival and drive
resistance to therapy in patients with lung cancers driven by ROS1
fusions. Clin Cancer Res. 26:2932–2945. 2020. View Article : Google Scholar
|
|
47
|
Zhou Q, Gui S, Zhou Q and Wang Y:
Melatonin inhibits the migration of human lung adenocarcinoma A549
cell lines involving JNK/MAPK pathway. PLoS One. 9:e1011322014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu T, Wu L, Wang D, Wang H, Chen J, Yang
C, Bao J and Wu C: Role of reactive oxygen species-mediated MAPK
and NF-κB activation in polygonatum cyrtonema lectin-induced
apoptosis and autophagy in human lung adenocarcinoma A549 cells. J
Biochem. 160:315–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
QuickGO: Term GO:0001934. https://www.ebi.ac.uk/QuickGO/term/GO:0001934.
Accessed July 20, 2021.
|
|
50
|
Urano F, Wang X, Bertolotti A, Zhang Y,
Chung P, Harding HP and Ron D: Coupling of stress in the ER to
activation of JNK protein kinases by transmembrane protein kinase
IRE1. Science. 287:664–666. 2000. View Article : Google Scholar : PubMed/NCBI
|






















