Introduction
Cutaneous melanoma is a lethal form of skin cancer
with an incidence that has been rapidly increasing in the past
decades. Melanoma has high metastatic potential and shows therapy
resistance, resulting in extremely poor prognosis for this disease
(1,2). The development of checkpoint
inhibitors (anti-programmed death protein 1, anti-programmed death
ligand 1 and anti-cytotoxic T-lymphocyte antigen-4) and targeted
therapy with B-Raf (BRAF) and mitogen-activated protein kinase
kinase (MEK) nhibitors have revolutionized the treatment of
metastatic melanoma, but a number of patients eventually develop
progressive disease (3). The
understanding of melanoma pathogenesis is crucial for the
development of new therapeutic strategies.
Numerous studies have demonstrated that tumor
progression and resistance to therapies are driven by the close
interplay among tumor microenvironment (TME) cells and genetic
lesions and plasticity of cancer cells (4-6).
In particular, the communication between tumor cells and the
bystander endothelial cells (ECs) is essential in regulating
angiogenesis and instrumental for the spread of metastasis
(7). The interaction between
tumor cells and cells of TME can be direct or indirect through the
secretion of soluble factors, cytokines and chemokines and
extracellular cell-matrix remodeling (8). IL-8 is a well-known
pro-inflammatory and pro-angiogenic chemokine that is prominently
expressed in immune, endothelial and tumor cells (9). The effect of IL-8 signaling has
received considerable attention as a key modulator in the context
of TME (10).
During tumor progression, cancer cells must adapt to
changes in TME conditions, such as mechanical stress, altered
oxygen tension and nutrient availability (10,11). Reactive oxygen species (ROS)
overproduction in TME causes oxidation of polyunsaturated fatty
acids in the cellular membrane of cancer cells through free radical
chain reactions with the formation of aldehydes as final products,
which serve a crucial role in the pathogenesis and progression of
cancer (12). Aldehyde
dehydrogenases (ALDHs), a family of NADP-dependent enzymes involved
in the detoxification of endogenous/exogenous aldehydes, are
proposed as a marker of cancer stem cells in several types of
cancer, including melanoma, non-small cell lung cancer (NSCLC),
gastric and breast tumors (13-15). Furthermore, some ALDHs,
particularly aldehyde dehydrogenase 1A1 (ALDH1A1), 1A3 (ALDH1A3)
and 3A1 (ALDH3A1) are associated with cell self-protection,
differentiation, expansion, tumor progression and therapy
resistance (16). Building on
this evidence, the authors have previously demonstrated the
contribution of ALDH1A1 in tumor angiogenesis in pre-clinical
breast cancer models (17) and
the role of ALDH3A1 in promoting stem cell development,
epithelial-mesenchymal transition and immune evasion in melanoma
and NSCLC cell lines (18).
In light of the emerging complexities related to
ALDHs influence on cancer phenotype and shaping of TME, the present
study investigated the link between ALDH1A1 expression in melanoma
cells and the acquisition of pro-angiogenic phenotype of normal
endothelium in in vivo xenografts and in 2D and 3D
co-culture models, focusing on tumor and endothelial components of
the TME.
The present study demonstrated that ALDH1A1 activity
and expression in melanoma cells regulated angiogenesis features in
endothelium via the production and release of IL-8 and, in turn,
the modulation of the gene expression profile of the Notch
signaling pathway.
Materials and methods
Chemicals and reagents
The ALDH1A1 inhibitor CM037 was from ChemDiv Inc.
CM037 was dissolved in DMSO (10 mM). CelLytic MT Cell Lysis
Reagent, goat serum and Eukitt quick-hardening mounting medium for
microscopy, 3 kDa FITC-Dextran, ARA-C and DAPI were from Merck
KGaA. Fluoromount aqueous mounting medium was from Thermo Fisher
Scientific, Inc. Lentiviral particles were from OriGene
Technologies, Inc. Matrix Matrigel (growth factors and phenol
red-free) was from Becton Dickinson. Anti-IL-8 antibody from
R&D Systems (cat. no. MAB208). Tissue-Tek O.C.T. was from
Sakura. Retinoic acid and pan-RAR antagonist (cat. no. AGN 193109)
were from Tocris Bioscience.
Cell culture
Melanoma cells A375, metastatic human melanoma cells
WM-266-4 (passages 5-20; ATCC), immortalized human keratinocytes
HaCaT (Voden Medical, SpA) and normal human dermal fibroblasts NHDF
(Lonza Group, Ltd.) were cultured in DMEM 4500 high glucose
(Euroclone, SpA) supplemented with 10% fetal bovine serum (FBS;
HyClone; Cytiva) and 2 mM glutamine, 100 units penicillin and 0.1
mg/l streptomycin (Merck KGaA). Cells were propagated by splitting
1:6 twice a week for A375, WM-266-4 and HaCaT and 1:3 twice a week
for NHDF.
Human umbilical vein endothelial cells (HUVECs) were
purchased from PromoCell GmbH. They were grown in endothelial
growth medium (EGM-2), containing vascular endothelial growth
factor (VEGF), recombinant human long R3 insulin like growth factor
1 (R3-IGF-1), human epidermal growth factor (hEGF), human
fibroblastic growth factor (hFGF), hydrocortisone, ascorbic acid,
heparin and GA-1000 (Lonza Group, Ltd.), 10% FBS and 2 mM
glutamine, 100 units/ml penicillin and 0.1 mg/ml streptomycin
(Merck KGaA). HaCaT cells authentication was by STR profiling.
To create GFP-HUVECS, the third generation of
lentiviral particles was used. GFP-HUVECS were kindly provided by
Professor Ambra Grolla, University of Piemonte Orientale A.
Avogadro, Novara, Italy. Cells were cultured at 37°C in 5%
CO2.
To achieve a stable knockdown, 1.5×105
melanoma cells were seeded on 6-multiplates and transduced at 70%
confluence with lentiviral particles (Merck KGaA) carrying a
scrambled (SC; pLKO.1-puro Empty Vector Control Transduction
Particles also from Merck KGaA) or two ALDH1A1 short hairpin
(sh)RNA sequences (TRC N 0000276459 and TRC N 0000276397) and
expressing the puromycin-resistant gene (ALDH1A1KD). A MOI
(Multiplicity of Infection) of 10 was used. The cells were
incubated at 37°C. At 36 h post-infection, puromycin (2
µg/ml) was added to cells and selection was allowed for 3
days. Stable knockdown was validated by western blot. Cells were
used in the experiments or split for propagation. Selected cells
were maintained in complete medium with puromycin (1
µg/ml).
The sequence of plasmid inserted in cells clone 1
(ShA) was: 5′-CCG GCA CCG ATT T-GA AGA TTC AA T ACT CGA GTA TTG AAT
CTT CAA ATC GGT GTT TTTG.
The sequence of plasmid inserted in cells clone 2
(ShB) was: 5′-CCG GCT CTA GCT TTG TCA TAG TTA TCT CGA GAT AAC TAT
GAC AAA GCT AGA G-TT TTTG.
To generate stable ALDH1A1 overexpressed
(ALDH1A1+) cultures, 1.5×105 melanoma cells
were seeded on 6-multiplates and infected with lentiviral particles
containing nucleotide sequences encoding for ALDH1A1 (Origene
RC200723 LentiORF particles, ALDH1A1 (Myc-DDK tagged) - Human). The
empty vector for overexpression was from OriGene Technologies, Inc.
(cat. no. PS100001). A MOI of 10 was used. The cells were incubated
at 37°C. At 36 h post-infection, medium was replaced with complete
culture medium containing G418 (400 µg/ml).
ALDH1A1+ cells were generated by G418 selection for 10
days. Stable overexpression was validated by western blot. Cells
were used in the experiments or split for propagation. Selected
cells were maintained in a complete medium with G418 (400
µg/ml).
The clones were expanded and used until 20
passages.
In vivo tumor xenograft
The present study was conducted in accordance with
the ethical standards and according to the Declaration of Helsinki
and the Italian law (Legislative Decree no.26, 4 March 2014), which
acknowledges the European Directive 2010/63/UE, being approved by
the animal welfare board of University of Siena and the Italian
Ministry of Health (authorization n. 62/2014-B). To assess the
involvement of ALDH1A1 in tumor growth and angiogenesis,
immunodeficient mice (5 week-old female athymic mice, 20-25 g,
Envigo) were subcutaneously (s.c.) inoculated in the right flank
with 1×107 A375 cells/100 µl (50 µl of
cells and 50 µl of Matrigel).
The mice were kept in temperature- and
humidity-controlled rooms (22°C and 50%) with a 12-h light/dark
cycle and water and food available ad libitum. A total of 18
different mice were randomly assigned to three different groups of
six mice. In the first group, mice were injected with A375
ALDH1A1SC. A375 ALDH1A1KD and A375 ALDH1A1+ were
injected respectively in the second and third group. Mice were
observed daily. No side effects such as changes in body weight,
behavioral changes or other signs of discomfort were observed. The
duration of experiment was 23 days, a coherent time to study tumor
angiogenesis. At the end of the experiment the animals were
euthanized by carbonic dioxide inhalation. The volume displacement
for euthanasia of the animals was 30-70% vol/min. Death was
ascertained by respiratory arrest. Each tumor was embedded in
Tissue-Tek O.C.T., cooled in isopentane and frozen in liquid
nitrogen for histology.
Immunofluorescence staining on O.C.T.
sections
Cryostat sections (7 µm) from tissue samples
were used for immunofluorescence staining with anti-CD31 antibody.
Sections were rehydrated with PBS and fixed with 4%
paraformaldehyde for 20 min at room temperature. Subsequently,
sections were washed and permeabilized with 0.2% Triton-X100 in PBS
for 20 min. After the washes (3×5 min) with PBS, sections were
blocked with 5% goat serum in PBS at room temperature. Samples were
incubated for 18 h (at 4°C) with anti-CD31 (BD Biosciences, cat.
no: 550274) in 5% goat serum in PBS (dilution 1:100). After washes
(3×5 min) with PBS, secondary antibody (goat anti-rat Alexa Fluor
568) in 5% goat serum in PBS (dilution 1:200) were applied for 60
min in the dark at room temperature. Samples were washed (3×5 min)
with PBS and incubated with DAPI in PBS (1:5,000). Sections were
washed (3×5 min) with PBS and mounted in Eukitt. Quantification of
CD31 positive vessels was performed counting by fluorescence
microscope (Nikon Eclipse TE 300) five random fields for section,
each slide having five sections (magnification, ×20). Analysis was
performed by GraphPad Prism 7 (GraphPad Software, Inc.).
In vitro multicellular skin spheroid and
melanoma tumorspheres
Commercially available cell lines were mixed as
described to recreate in vitro multicellular skin and
melanoma tumorspheres (19,20). For multicellular 3D skin
spheroids, HaCaT, NHDF and HUVECs were mixed in equal proportions
(1×103 cells each). To create multicellular melanoma 3D
tumorspheres A375 (SC, ALDH1A1KD or ALDH1A1+) were
cultured with HaCaT, NHDF and HUVECs (1×103 cells each).
All tumorspheres were seeded in ultralow attachment plates
(Corning, Inc.) and grown in endothelial growth medium (EGM-2),
containing VEGF, R3-IGF-1, hEGF, hFGF, hydrocortisone, ascorbic
acid, heparin and GA-1000 (Lonza Group, Ltd.), 10% FBS, 2 mM
glutamine, 100 units/ml penicillin and 0.1 mg/ml streptomycin
(Merck KGaA). Tumorspheres were cultured at 37°C in 5%
CO2 for 6 days. At the end of experiment tumorspheres
were analyzed with a confocal microscope (Zeiss LSM700; Zeiss
GmbH).
Tumorspheres fluorescence analyses
3D tumorspheres were harvested and fixed in 4%
paraformaldehyde for 18 h at 4°C as previously reported (21) after three washes with PBS,
tumorspheres were permeabilized with 0.2% Triton-X100 in PBS for 20
min at room temperature and then nuclei were labeled with DAPI
(1:5,000 for 10 min at room temperature). Spheroids were washed
(3×5 min) with PBS and mounted in Fluoromount aqueous mounting
medium. Images were captured using Nikon Eclipse TE 300 (Nikon
Corporation; magnification, ×20).
GFP-HUVECS sorting
To isolate GFP-HUVECS from melanoma 3D tumorspheres,
spheroids were harvested and trypsinized with 5 mM EDTA and 2.5%
trypsin at room temperature. Cells were centrifuged (5 min at 4°C
and 300 × g) and resuspended in PBS. GFP-HUVECS were then isolated
by using BD FACSAria Fusion (BD Biosciences).
RNA isolation and reverse
transcription-quantitative (RT-q) PCR
Total RNA was prepared using RNeasy Plus kit (cat.
no. 74134 Qiagen GmbH) following the manufacturer's instructions.
The quality and quantity of the purified RNA were redetermined by
measuring the absorbance at 260/280 nm (A260/A280) using Infinite
F200 Pro, (Tecan Group, Ltd.). A total of 500 ng (for GFP-HUVECS)
or 1 µg (for tumor cells) of RNA were reverse transcribed
using QuantiTect Reverse Transcription kit (cat. no. 205313 Qiagen
GmbH; 42°C for 30 min and the reaction was then terminated by
incubating the tube at 95°C for 3 min).
RT-qPCR was performed using QuantiNova SYBR Green
PCR kit (cat. no. 208056 Qiagen GmbH) in a RotorGene qPCR machine
(Qiagen GmbH) under the following conditions: 95°C for 5 min,
followed by 45 cycles of 95°C for 20 sec, 60°C for 20 sec and then
at 72°C for 20 sec.
Fold change expression was determined by the
comparative Ct method (ΔCt) normalized to 60S ribosomal protein L19
expression. RT-qPCR data were represented as Ct value (cycle
threshold) or fold increase relative to GFP-HUVECS from melanoma
tumorspheres ALDH1A1SC or ALDH1A1KD, assigned to 1 (22).
The primer sequences were: DLL4 forward,
5′-AAT GGA GGC AGC TGT AAG GA-3′ and reverse, 5′-CAT AGT AGC CCG
GAG GAC AC-3′; NOTCH1 forward, 5′-GGC AAT CCG AGG ACT ATG
AG-3′ and reverse, 5′-CAG AAC GCA CTC GTT GAT GT-3′; NOTCH3
forward, 5′-AGG CCA TGG TCT TCC CTT AC-3′ and reverse, 5′-TCA ATC
TCC AGC ATT ACT ACC G-3′; ADAM17 forward, 5′-AAC AGC GAC TGC
ACG TTG AAG G-3′ and reverse, 5′-CTG TGC AGT AGG ACA CGC CTT T-3′;
Bcl-2 forward 5′-TTG TGG CCT TCT TTG AGT TCG GTG-3′ and
reverse 5′-GGT GCC GGT TCA GGT ACT CAG TCA-3′; Bax forward
5′-CCT GTG CAC CAA GGT GCC GGA ACT-3′ and reverse 5′-CCA CCC TGG
TCT TGG ATC CAG CCC-3′; IL-8 forward, 5′-GAG CAC TCC ATA AGG
CAC AAA-3′ and reverse 5′-ATG GTT CCT TCC GGT GGT-3′; RPL19
forward 5′-GAT GCC GGA AAA ACA CCT TG-3′ and reverse 5′-TGG CTG TAC
CCT TCC GCT T-3′. All primers were from Merck KGaA.
Proliferation of HUVECs in co-culture
with melanoma cells
TME communication between melanoma and ECs was
reconstructed in vitro using a Transwell system (Corning,
Inc.) (17). Transwells provide
2D-co-culture without contact between two cell types. HUVECs
(5×103 cells) were seeded on the bottom of 24
multiplates precoated with gelatin. Melanoma cells were plated on
the polyester membrane of the Transwell (2×104 cells).
After 24 h, tumor cells were pre-treated for 1 h with CM037 (10
µM; in DMEM with 1% FBS, 37°C) and then co-cultured with
HUVECs in the same 24 multiplates for 48 h in the presence of EBM
medium (without growth factors) with 1% FBS. Anti-IL-8 neutralizing
antibody was added at 80 ng/ml, where appropriate. Cells were then
fixed using Fixing for fast staining (methanol based) (Panoptic No.
1) for 15 min at room temperature and then stained using Eosin for
fast staining (Panoptic No. 2) and Blue for fast staining (Panoptic
No. 3; Azur B based; 15 min each; PanReac AppliChem). Cells were
randomly counted at original magnification of ×20 in five fields as
previously reported (17).
Analysis was performed by GraphPad Prism 7 (GraphPad Software,
Inc.).
Scratch assay in HUVECs co-cultured with
melanoma cells
HUVECs (1×105 cells) were seeded on the
bottom of 12-well multiplates pre-coated with gelatin. Once HUVECs
reached confluence, cells were scratched using a sterile 100-1,000
µl micropipette tip to create a wound ±500 µm across
the monolayer and Transwells were put in the same 12-well
multiplates for 18 h of co-culture in EBM medium (without growth
factors, but with 1% FBS). ARA-C (2.5 µg/ml) was then added
to all the wells to control cell proliferation. Where appropriate,
tumor cells were pre-treated for 1 h (at 37°C) with CM037 (10
µM) and then co-cultured with HUVECs (at 37°C) as described
above. Where indicated, co-culture was treated with the
neutralizing anti-IL-8 antibody at 80 ng/ml. Images of the wound in
each well were acquired from 0-18 h under a phase contrast
microscope (Nikon Eclipse TE 300, Nikon), at ×20 magnification. The
rate of scratch area was measured by quantifying the uncovered area
of the wound that HUVECs covered starting from the edge of the
scratch. All quantifications were done with Fiji software (64-bit
Java 1.8.0_172). Results are expressed as a percentage of the area
of the wound at 18 h respect to time 0 (23). Analysis was performed by GraphPad
Prism 7 (GraphPad Software, Inc.).
Permeability assay in HUVECs co-cultured
with melanoma cells
Permeability assay was performed in endothelial
monolayers as previously described (17). Briefly, melanoma (SC, ALDH1A1KD
and ALDH1A1+) were seeded at a density of
3×104 on the bottom of 12-well multiplates. HUVECs
(8×104 cells) were seeded on the top of the
polycarbonate membrane with 0.4 µm pores, pre-coated with
gelatin. After 24 h incubation necessary for cell adherence,
Transwells were put in the same 12-well multi-plates with medium
supplemented with 1% FBS until HUVECs confluence (at 37°C).
Anti-IL-8 anti-body was added, where indicated, at 80 ng/ml.
Fluorescein isothiocyanatedextran (FITC-Dextran, 3 kDa; added at 10
µM concentration in the upper compartment of the Transwell,
at 37°C) was used as a fluorescent marker of paracellular
permeability, which was evaluated after 15 min by measuring the
fluorescence of medium in the lower compartment in a multi-plate
reader (Infinite F200 Pro; Tecan Group, Ltd.) at 485 and 535 nm
excitation and emission, respectively. Data are reported as
fluorescence units.
Tube formation assay by HUVECs
co-cultured with melanoma cells
Tumor cells (3×104 cells) were cultured
on Transwell inserts. After 24 h the inserts were transferred on
top of HUVECs plated on Matrigel (1.5×105 cells in
12-well multiplates) at 37°C. Where appropriate, tumor cells were
pre-treated for 1 h with CM037 (10 µM) and then co-cultured
with HUVEC. Anti-IL-8 antibody was added at 80 ng/ml.
After 18 h of incubation (at 37°C), images of the
ECs were captured and network formation on Matrigel was quantified
by using Fiji Software using the number of branching points under a
light microscope (five random fields/well; Nikon Eclipse E400 and
camera Nikon DS5MC; Nikon Corporation) at magnification ×20
(24).
Human cytokine ELISA plate array
Human Cytokine ELISA Plate Array (cat. no. EA-4001,
Signosis, Inc.) was performed for quantitative comparison of 31
cytokines on supernatants of melanoma cells treated with CM037 (a
selective ALDH1A1 enzyme blocker). Cells (3×103) were
exposed to a medium with 1% FBS in the presence/absence of CM037 (1
µM) for 48 h (with CM037 treatment every 24 h). The cell
culture supernatants from each sample were incubated for 2 h at
room temperature in the wells of the cytokine ELISA plate and the
captured cytokine proteins were subsequently detected with a
cocktail of biotinylated detection antibodies against 31 human
cytokines The test sample was allowed to react with a pair of
antibodies, resulting in the cytokines being sandwiched between the
solid phase and enzyme-linked antibodies. After incubation at room
temperature (2 h), the wells were washed to remove unbound-labeled
antibodies. The plate was further detected with horseradish
peroxidase (HRP) luminescent substrate (cat. no. EA-4001; Signosis,
Inc.). The level of expression for each specific cytokine is
directly proportional to the luminescence intensity. Data were
reported as percentage of fold change vs. untreated cells (Ctr).
The experiment was performed twice in duplicate.
Human Notch signaling real time-based
array analysis
The human Notch signaling RT Profiler PCR array
(PAHS-059Y, Qiagen GmbH) was used to profile a panel of 84 genes
representative of the Notch pathway in HUVECs co-cultured with A375
cells (A375 ALDH1A1SC, ALDH1A1KD and ALDH1A1+) for 6
days at 37°C. HUVECs were seeded on the bottom of 6-well
multiplates (8×104 cells) and tumor cells on the top of
the Transwell (5×104 cells). The medium was changed
every two days.
RNA was isolated from HUVECs using a RNeasy Plus kit
at room temperature (cat. no. 74134 Qiagen GmbH) and then reverse
transcribed using QuantiTect Reverse Transcription kit (cat. no.
205313 Qiagen GmbH; 42°C for 30 min and the reaction was then
terminated by incubating the tube at 95°C for 3 min). The cDNA was
used on the real-time RT Profiler PCR array (PAHS-059Y, Qiagen
GmbH). The experiment was performed twice.
Western blotting
Western blotting was performed on HUVECs co-cultured
with A375 cells for 6 days at 37°C. HUVECs (1.5×105
cells) were seeded on the bottom of 6-well multi-plates pre-coated
with gelatin. A375 cells were plated on the polyester membrane of
the Transwell (8×104 cells). After 24 h, tumor cells
were co-incubated with ECs in the same 6-well multiplates for 6
days in the presence of EBM medium (without growth factors) with 1%
FBS. Where indicated, co-cultures were treated with the
neutralizing anti-IL-8 antibody at 80 ng/ml (the treatment was
repeated every 48 h). Melanoma cells (3×105) were seeded
in 60 mm Petri dishes. After adherence, cells were starved for 4 h
and then treated with retinoic acid (1 µM, 24 h) or 2% FBS
for 24 h at 37°C. Proteins were isolated and western blotting were
performed as previously described. Proteins were isolated and
western blotting were performed as previously described (25). After collecting the cells,
proteins were extracted using CelLytic MT supplemented with 2 mM
Na3VO4 and 1X Protease inhibitor cocktail for
mammalian cells (Merck KGaA). The protein concentration was
measured using a Bradford assay. Proteins (50 µg) were
separated by polyacrylamide gel electrophoresis (Bolt 4 to 12%,
Bis-Tris, 1.0 mm, Mini Protein Gel, 10-well, from Thermo Fisher
Scientific, Inc.) and then transferred onto a nitrocellulose
membrane (iBlot 2 Transfer Stacks, nitrocellulose, regular size
from Thermo Fisher Scientific, Inc.). After blocking with 5 %
nonfat dried milk (1 h at room temperature) (cat. no. 1706404;
Bio-Rad Laboratories, Inc.), the membranes were incubated at 4°C
overnight with primary antibody including: anti-ALDH1A1 (rabbit,
1:1,000, cat. no. 54135), anti-delta-like canonical Notch ligand 4
(Dll4; rabbit, 1:1,000, cat. no. 2589), anti-Notch1 (rabbit,
1:1,000, cat. no. 3608), anti-recombining binding protein,
suppressor of hairless (RBPSUH; rabbit, 1:1,000, cat. no. 5313) and
anti-A disintegrin and metalloproteinase-17 (ADAM17; rabbit,
1:1,000, cat. no. 6978) antibodies were from Cell Signaling
Technology, Inc. Anti-β-actin antibody (mouse, 1:10,000, cat. no.
MABT825) was from Merck KGaA. Anti-NF-κB p65 (rabbit, 1:1,000, cat.
no. sc-8008) was from Santa Cruz Biotechnology, Inc. The membranes
were than incubated with secondary antibodies IgG (H+L), HRP
conjugate (anti-rabbit, 1:2,500, cat. no. W401B; anti-mouse
1:2,500, cat. no. W402B, both from Promega Corporation) at room
temperature for 1 h and washed with PBS and Tween20 0.5 % three
times. Protein bands were analyzed by ChemiDoc XRS (Bio-Rad
Laboratories, Inc.) following incubation at room temperature for 2
min with enhanced chemiluminescent substrate (Euroclone SpA).
ALDH1A1 enzymatic activity
ALDH1A1 enzymatic activity was determined by
measuring the conversion of acetaldehyde to acetic acid, as
previously reported (17).
Briefly, cells were scraped into 600 µl lysis buffer (100 mM
Tris-HCl pH 8.0, 10 mM DTT, 20% glycerol, 1% Triton X-100) and
centrifuged at 16,000 × g for 20 min at 4°C. The supernatant was
used to detect ALDH activity at 25°C by monitoring NADH formation
from NAD+ at 340 nm in a spectrophotometer (Infinite F200 Pro;
Tecan Group, Ltd.). The assay mixture (0.8 ml) contained 100 mM
sodium pyrophosphate pH 9.0, 10 mM NAD+ and 600 µg of sample
protein. The reaction was initiated by adding acetaldehyde (10 mM)
to the cuvette. The enzyme-specific activity was expressed as nmol
NADH/minute/mg protein.
MTT assay
Cell survival was quantified by MTT (Thiazolyl Blue
Tetrazolium Bromide, Sigma Aldrich) (26). Briefly, cells (3×103)
were seeded in 96-multiwell plates in medium with 10% serum for 24
h and then grown for 48 h in complete medium with 10% FBS at 37°C
in the presence or not of CM037 (1-10 µM) every 24 h.
Dimethyl sulfoxide (DMSO) was used to dissolve the formazan salt.
Data are reported as absorbance measured at 540 nm/well.
Immunofluorescence analysis
NF-κB p65 (anti-rabbit; 1:50; cat. no. sc-8008;
Santa Cruz Biotechnology, Inc.) localization was monitored by
fluorescence microscope. A total of 3×104 A375
(ALDH1A1SC, ALDH1A1KD and ALDH1A1+) were seeded on 1-cm
circular glass coverslips. After 24 h incubation at 37°C, cells
were starved for 4 h and then treated with retinoic acid (1
µM, for 1 h) or DMEM supplemented with 2% FBS at 37°C.
Immunofluorescence analysis was performed on ECs as previously
reported (17). Briefly, the
cells were fixed at room temperature with cold acetone for 15 min
and washed three times with PBS. After blocking with 5% goat serum
(at room temperature) for 1 h, the cells were incubated with
primary antibody diluted with goat serum and incubated at 4°C for
18 h. After three washes with PBS, secondary antibody was added
(goat anti-rat Alexa Fluor 488) for 1 h at room temperature in the
dark. Cells were washed with PBS and incubated with DAPI (1:5,000
at room temperature for 20 min). After washing, they were mounted
on specimen slides.
HUVECs proliferation
A total of 1,000 cells/well (of a 96-well
multiplate) were left to adhere in an incubator at 37°C in 10%
serum for 24 h and then IL-8 (used at increasing concentrations in
the range 0.1-100 ng/ml) was added in a medium with 1% serum which
represented the basal control condition. All experimental points
using cells from the single culture plate were run in triplicate.
After 48 h, cells were fixed using 100% Fixing for fast staining
(Panoptic No. 1) for 15 min at room temperature and then stained
using Eosin for fast staining (Panoptic No. 2) and Blue for fast
staining (Panoptic No. 3; 15 min each) and randomly counted under a
light microscope ×20 original magnification in five fields. Data
are reported as number of cells counted/well.
Data analysis and statistical
procedures
Results are either representative or the average of
at ≤3 independent experiments performed in triplicate. Statistical
analysis was performed using one-way ANOVA test followed by the
Bonferroni test and the unpaired Student t-test when appropriate
(GraphPad Prism 7; GraphPad Software, Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Melanoma ALDH1A1 promotes angiogenic
recruitment and Notch signaling modulation in vivo and in a model
of 3D tumor spheroids
Intrinsic ALDH1A1 serves an important role in the
stemness and progression of several solid tumors, including
melanoma (27). Since tumor
progression is closely related to the crosstalk that tumor cells
establish with the TME components, the present study investigated
the phenotype of ECs in terms of angiogenic functions and
remodulation of gene expression when exposed to melanoma cells
expressing ALDH1A1. The present study employed the following cell
lines: A375 mutated BRAF(V600E) and WM-266-4 metastatic BRAF wild
type melanoma cells. To strengthen the contribution of melanoma
ALDH1A1 in regulating the pro-angiogenic phenotype of ECs, we used
loss- and gain-of-function strategies. The present study created
cellular models knocked down for ALDH1A1 (ALDH1A1KD clones sh A and
B) in A375 and WM-266-4 cells. For gain-of function cells, A375
ALDH1A1+ and WM-266-4 ALDH1A1+ cells were
generated. Melanoma cells infected with a scrambled sequence were
reported as ALDH1A1SC cells and used as control. As expected,
compared with ALDH1A1SC melanoma cells, ALDH1A1KD melanoma cells
showed a reduction of ALDH1A1 protein expression (Fig. 1A and B) and impairment of enzyme
function, evaluated by ALDH1A1 enzymatic activity assay (Fig. 1C and D). By contrast, A375
ALDH1A1+ and WM-266-4 ALDH1A1+ melanoma cells
demonstrated increased ALDH1A1 activity (Fig. 1C and D) and expression (Fig. 1E and F).
 | Figure 1Melanoma ALDH1A1 promoted HUVECs
recruitment and modulated endothelial Notch signaling in a model of
3D multicellular melanoma tumorspheres. Loss-of function and
gain-of function validation was used to study ALDH1A1 expression in
melanoma cells. Western blot analysis of (A) A375 and (B) WM-266-4
(ALDH1A1SC and ALDH1A1KD, clones shA and shB) cultured in 10% FBS
for 48 h. Enzymatic activity in (C) A375 and (D) WM-266-4 evaluated
by NADH production. ***P<0.001 vs. ALDH1A1SC;
###P<0.001 vs. ALDH1A1KD; °P<0.05 and °°P<0.01
vs. ALDH1A1+. Western blot analysis of (E) A375 and (F)
WM-266-4 cultured in 10% FBS for 48 h. β-actin was used as loading
control. Blot representative of three experiments. (G)
Quantification of microvessel density by CD31 staining was
performed counting 5 random fields for section, each slide having
five sections (magnification, ×2). #P<0.05 vs.
ALDH1A1KD group. (H) Representative images of immunostaining for
CD31 (red) and DAPI (blue) in tumor sections from ALDH1A1SC (top),
ALDH1A1KD (center) or ALDH1A1+ (bottom) mice. Images show different
vessel densities in tumors. Magnification, ×20. (I) Bright-field
image (magnification, ×4) of skin 3D spheroids and multicellular
melanoma 3D tumorspheres consisting in A375 cells, HaCaT, NHDF and
HUVECs co-cultured in ultralow attachment plates for 6 days. Scale
bar=100 µm. Quantification of 3D multicellular spheres. The
area occupied by spheres was calculated using Fiji Software and
three images for each well were quantified. Spheres >100 pixel
square were considered. ##P<0.01 vs. ALDH1A1KD. (J)
Bright-field images obtained at magnification ×20 after fixation in
paraformaldehyde and mounting in Fluoromount aqueous mounting
medium. Scale bar=50 µm. (K) GFP-HUVECS localization in skin
and melanoma 3D models. Fluorescence imaging demonstrated
distribution of ECs (green) inside 3D spheroids. Merged images show
blood vessel-like tubules of green HUVECs (×20 magnification).
Scale bar=50 µm. (L) Z-stack images obtained through a
confocal microscope show bifurcation of HUVECs tubules in A375
ALDH1A1+ 3D tumorspheres (magnification, ×63; scale
bar=50 µm). (M) Notch pathway genes in HUVECs co-cultured
with A375 ALDH1A1+ vs. HUVECs co-cultured with A375
ALDH1A1SC sorted from multicellular melanoma 3D tumorspheres. Data
are reported as fold change relative to A375 ALDH1A1SC, assigned to
1. **P<0.01 and ***P<0.001 vs
ALDH1A1SC. (N) Notch pathway genes under/overexpressed in HUVECs
co-cultured with the A375 ALDH1A1+ cells vs. HUVECs
co-cultured with the A375 ALDH1A1KD cells sorted from multicellular
melanoma 3D tumorspheres. **P<0.01 and
***P<0.001 vs ALDH1A1KD. Data are reported as fold
change relative to A375 ALDH1A1KD, assigned to 1. HUVECs, human
umbilical vein endothelial cells; SC, scrambled control; GFP, green
fluorescent protein; ECs, endothelial cells. |
It was determined whether the reduced expression and
activity of ALDH1A1 in melanoma cancer cells influenced in
vivo tumor angiogenesis. A375 ALDH1A1KD, A375
ALDH1A1+ and A375ALDH1A1SC were implanted s.c. in nude
mice and the density of angiogenic microcapillaries in A375 tumors
evaluated. By immunostaining for CD31, a significant increase of
vessels in ALDH1A1+ tumors was found compared with
ALDH1A1KD ones (Fig. 1G and
H).
To investigate the contribution of the concerted
interactions of melanoma multiple cell components on ECs phenotype,
multicellular melanoma 3D tumorspheres were developed with close
contacts among different cell types (19,20). The present study focused in
particular on the communication of tumor cells and ECs and how
melanoma cells expressing different levels of ALDH1A1 enzyme
'corrupt' normal ECs. A375 (ALDH1A1SC, ALDH1A1KD and
ALDH1A1+) were seeded with keratinocytes HaCat, dermal
fibroblasts NHDF and GFP-HUVECS in equal proportions to generate
multicellular melanoma 3D tumorspheres. As a control, a model of
skin spheroids (HaCat, NHDF and GFP-HUVECS) was used. Spheroids
were grown for 6 days to favor cell-cell interaction and matrix
deposition to obtain multicellular structurally established
spheroids. Fig. 1I and J show
that the skin spheroids presented a circular shape with regular and
homogeneous edges. Instead, multicellular melanoma tumorspheres had
an irregular shape, were more aggregated and presented a less
homogeneous structure (Fig. 1I and
J). The dimension and number of ALDH1A1+
tumorspheres were higher in respect of ALDHKD and skins spheroids
(Fig. 1I).
Focusing on tumor angiogenesis, the recruitment of
GFP-HUVECS in skin 3D spheroids and multicellular melanoma
tumorspheres was determined. Fluorescence analysis showed the
presence of a cluster of GFP-HUVECS in the central region of the
tumorspheres (Fig. 1K). The
multicellular melanoma tumorspheres showed more significant
infiltration of GFP-HUVECS compared with skin spheroids. Among the
tumorspheres, A375 ALDH1A1KD tumorspheres displayed the lowest
ability to recruit ECs. On the other hand, the Z-stacks analysis on
confocal microscopy images revealed a 3D organization of
endothelium in multicellular A375 ALDH1A1+ tumorspheres,
showing vessel-like tubule structures of green ECs with bifurcation
(Fig. 1L). Whether tumor cells
harboring different levels of ALDH1A1 might influence canonical
angiogenesis pathways in ECs was then investigated, focusing on
Notch signaling, which is known to contribute to vascular
development and remodeling (28-30). First, to investigate whether
co-culture of HUVECs in 3D organization with tumor cells affected
HUVECs viability, qPCR analysis was performed to study
apoptosis-related genes. The Bax/Bcl-2 ratio was analyzed. As shown
in Fig. S1, compared with
HUVECs grown in monolayer, no differences in the Bax/Bcl-2 ratio
were observed. Then, the Notch signaling pathway in GFP-HUVECS
sorted by BD FACSAria Fusion from A375 ALDH1A1KD, A375
ALDH1A1+ and ALDH1A1SC tumorspheres was investigated.
RT-qPCR analysis of these cells revealed an increase of DLL4 gene
transcription in endothelium derived from A375 ALDH1A1+
3D tumorspheres compared with HUVECs recovered from A375 ALDH1A1SC
(Fig. 1M) and A375 ALDH1A1KD 3D
spheres (Fig. 1N). An increased
expression was observed for DLL4, NOTCH1 and ADAM17 transcripts,
exclusively in HUVECs derived from A375 ALDH1A1+
compared with A375 ALDH1A1KD 3D tumorspheres (Fig. 1N).
These results indicated that tumor ALDH1A1 levels
correlated with angiogenic phenotype and increased microvessel
density in vivo. In TME in vitro settings, a genetic
modulation of Notch signaling in ECs was observed, suggesting an
enrichment of angiogenic factors in the TME driven by melanoma
ALDH1A1.
ALDH1A1 expression and activity in
melanoma cells regulate IL-8 release and control ECs proliferation,
migration, tube formation and permeability
In light of the changes of ECs elicited by ALDH1A1
in melanoma cells, the present study explored the release of
cytokines involved in the angiogenesis process from melanoma cells
pre-treated with CM037, a selective ALDH1A1 blocker. CM037 (10
µM) produced a substantial decline of enzymatic activity in
both A375 and WM-266-4 cells (Fig.
S2A and S2B), while it did not affect cell survival (Fig. S2C and D).
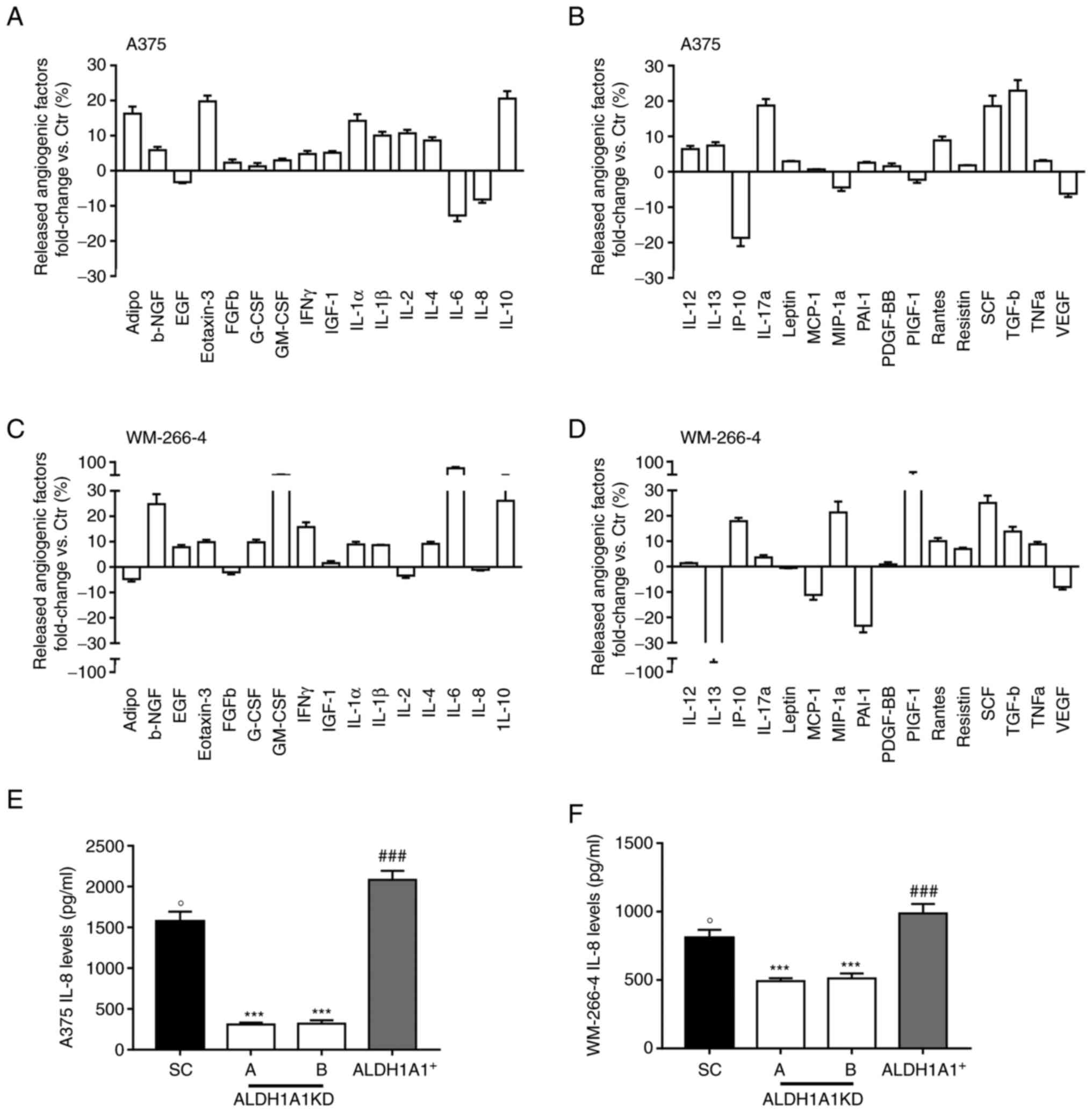
When ALDH1A1 was inhibited, in both melanoma cell
lines, an altered secretion of angiogenesis inducers with a drastic
reduction of VEGF (-6.18 fold and -8.04 fold in A375 and WM-266-4,
respectively) (Fig. 2B and D)
and IL-8 release (-8.18 fold and -5.06 fold in A375 and WM-266-4,
respectively) was found (Fig. 2A and
C). IL-8 has been demonstrated to positively influence the
melanoma microenvironment and ECs in an autocrine and paracrine
manner (31). By contrast, in
CM037-treated A375 a relevant increase of anti-angiogenic factors
such as IL-12 and plasminogen activator inhibitor, type I (PAI-1)
(+6.41 and +2.56 fold, respectively) was observed (Fig. 2B). Similarly, in CM037 treated
WM-266-4 a substantial increase in the anti-angiogenic IL-6 (+67.03
fold) and IL-12 (+2,34 fold) compared with untreated cells was
observed (Fig. 2C and D),
highlighting the contribution of the ALDH1A1 enzyme activity to
melanoma angiogenic phenotype. Based on these results, the present
study focused on IL-8. Indeed, overexpression of IL-8 is related to
melanoma angiogenesis and metastases (32,33). In melanoma cells expressing
different levels of ALDH1A1, it was found that high ALDH1A1
expression was associated with a significant increase of soluble
IL-8 levels compared with melanoma ALDH1A1KD (Fig. 2E and F). In agreement with array
panel results, A375 showed greater IL-8 release compared with
WM-266-4.
High protein levels were associated with increased
IL-8 gene expression in A375 ALDH1A1+ compared with A375
ALDH1A1KD, indicating that ALDH1A1 modulates IL-8 levels at the
transcriptional level (Fig.
S3A). As the ALDH1 family is required for retinoic acid (RA)
biosynthesis, the present study investigated whether RA signaling
might mediate the observed ALDH1A1-dependent IL-8 regulation in
A375. Exposure of A375 ALDH1A1SC and A375 ALDH1A1+ to RA
receptor inhibitor (AGN 193109) reduced IL-8 gene expression
(Fig. S3B), while exposure of
A375 ALDH1A1KD to exogenous RA increased it (Fig. S3C). Furthermore, in A375
ALDH1A1KD, RA promoted nuclear localization of NF-κB, one of the
transcription factors of IL-8 genes (Fig. S3D) and increased NF-κB-p65
subunit expression (Fig. S3E),
suggesting the existence of an RA-NF-κB axis in ALDH1A1-mediated
IL-8 expression.
Next, whether and how ALDH1A1 expression in melanoma
cells influenced endothelium was investigated, specifically
focusing on the communication between melanoma and ECs. The present
study generated a 2D co-culture model of ECs and melanoma cells
cultured in the Transwell system. HUVECs proliferation, scratch
assay, tube formation and permeability and cellular features of
angiogenesis were assessed.
Fig. 3A and B
show a decreased proliferation of HUVECs when co-cultured with A375
ALDH1A1KD and WM-266-4 ALDH1A1KD, respectively, compared with that
from ALDH1A1SC cells. Similar results were obtained when ALDH1A1 in
melanoma cells was blocked by CM037 (Fig. S4A and B). By contrast, the
co-incubation of HUVECs with A375 ALDH1A1+ and WM-266-4
ALDH1A1+ cells enhanced proliferation of ECs (Fig. 3A and B) and IL-8 neutralizing
antibody blunted this effect. EC migration was then analyzed by
scratch assay and a significant reduction of HUVECs motility when
co-cultured with A375 ALDH1A1KD (Fig. 3C) and WM-266-4 ALDH1A1KD cells
(Fig. 3D) was found compared
with ALDH1A1SC melanoma cells. In the same experiments, melanoma
cells ALDH1A1+ provided a pro-migratory stimulus for
endothelium (Fig. 3C and D),
mitigated by IL-8 neutralizing antibody addition. In addition, the
ability of melanoma cells to induce HUVECs motility was abolished
when CM037 inhibited ALDH1A1 (Fig.
S4C for A375 and S4D for
WM-266-4 cells).
Tube formation assay results further corroborated
these effects. When co-cultured with A375 ALDH1A1+
(Fig. 3E) and WM-266-4
ALDH1A1+ (Fig. 3F),
HUVECs showed a strong ability to form net-like structures, which
was significantly reduced when co-cultured in the presence of IL-8
neutralizing antibody. A marked reduction in tube formation was
also observed when HUVECs were incubated with A375 ALDH1A1KD and
WM-266-4 ALDH1A1KD cells (Fig. 3E
and F). Similarly, pharmacological inhibition of ALDH1A1
resulted in an impairment of angiogenic sprouting of HUVECs
co-cultured with both melanoma cell lines (A375, Fig. S4E and WM-266-4 S4F).
Similar results were obtained by measuring
endothelial permeability. HUVECs co-cultured with melanoma
ALDH1A1+ cells were more permeable compared with HUVECs
co-cultured with ALDH1A1KD cells counterpart (Fig. 3G for A375 and Fig. 3H for WM-266-4). A neutralizing
antibody to IL-8 significantly inhibited the permeability of HUVECs
(Fig. 3G and H).
Collectively, these findings demonstrated the
existence of dynamic crosstalk between melanoma and ECs which
acquired an angiogenic phenotype, promoted by tumor ALDH1A1
expression and activity through, at least in part, the release of
NF-κB associated IL-8. Of note, in HUVECs, at concentration similar
to that measured in the medium of melanoma cells, exogenous IL-8
was able to promote cell proliferation (Fig. S4G).
Tumor ALDH1A1 effects Notch signaling in
ECs through IL-8
To investigate the underlying mechanisms by which
tumor ALDH1A1 affects the endothelial angiogenic program, a Notch
signaling gene array was performed on ECs co-cultured with melanoma
cells using the Transwell system. A long-term 2D co-culture (6
days) was performed with HUVECs and A375 cells to mimic the
co-existence of melanoma and ECs in 3D tumorspheres. Profiling the
expression of 84 genes involved in Notch signaling, an impressive
gene rearrangement between endothelium grown alone and co-cultured
with melanoma cells was found (Fig.
S5). The present study focused on Notch ligands, receptors and
downstream effectors involved in angiogenesis. Delta-like ligand 4
(DLL-4) is the most important Notch ligand for early vascular
development and angiogenesis (34). DLL-4 gene expression was strongly
induced in HUVECs incubated with A375 ALDH1A1+ compared
with HUVECs co-cultured with A375 ALDH1A1SC and A375 ALDH1A1KD
(>133 and 128 fold, respectively) (Fig. 4A and B). An important increase of
DLL-3 was observed in ECs derived from 2D co-cultures with melanoma
ALDH1A1+ compared with other settings. By contrast,
another Notch ligand gene, Jagged1 (Jag1), which can compete with
DLL4 to negatively regulate angiogenesis (34), was markedly downregulated in
HUVECs co-cultured with A375ALDH1A1+ when compared with
A375 ALDH1A1SC (-24.29 fold) and A375 ALDH1A1KD (-78.76 fold;
Fig. 4A and B).
As with the ligands, the Notch receptors also serve
important roles in angiogenesis (35). Expression of all three Notch1,
Notch2 and Notch3 genes was upregulated in HUVECs co-cultured with
A375 ALDH1A1+ when compared with HUVECs co-cultured with
A375 ALDH1A1SC and A375 ALDH1A1KD. Notch1 is the most important
receptor of the Notch signaling cascade involved in early
angiogenesis. A +19.98 and +16.71 fold increase in HUVECs
co-cultured with A375 ALDH1A1+ was observed compared
with HUVECs co-cultured with A375 ALDH1A1SC and A375 ALDH1A1KD,
respectively. Similar regulation was observed for the Notch2 gene
(+14.92 fold in HUVECs co-cultured with A375 ALDH1A1+
vs. A375 ALDH1A1KD, +15.45 fold vs. A375 ALDH1A1SC) and Notch3 gene
(+11.89 fold in HUVECs co-cultured with A375 ALDH1A1+
vs. A375 ALDH1A1KD and +8.87 fold vs. ALDH1A1SC) (Fig. 4A and B).
The binding of Notch receptors with ligands promotes
the proteolytic cleavage of the Notch receptors, mediated by
ADAM-family metalloproteases (36). The array showed an upregulation
in gene expression of ADAM17, one of the main proteases involved in
Notch signaling, in endothelium co-cultured with A375
ALDH1A1+ when compared with A375 ALDH1A1SC (+4.26 fold)
and A375 ALDH1A1KD (+4.10 fold; Fig.
4A and B).
The canonical Notch target genes, such as the HEY
family, were also significantly affected by tumor ALDH1A1 levels.
HEY1 showed an increase of expression by 253 fold in HUVECs
co-cultured with A375 ALDH1A+ compared with HUVECs
co-cultured with A375 ALDH1A1SC and reached >300 fold when
compared with HUVECs incubated with A375 ALDH1A1KD (Fig. 4A and B). HEY2 gene showed a lower
variation within different experimental settings. This gene was
overexpressed ~2.3 fold in HUVECs co-cultured with A375
ALDH1A1+ vs. HUVECs incubated with A375 ALDH1A1KD
(Fig. 4A and B). An increase of
~20.01 fold was observed in HUVECs co-cultured with A375 ALDH1A1SC
vs. HUVECs grown with A375 ALDH1A1+ (Fig. 4A and B).
By contrast, the present study found a decrease of
NUMB gene, an inhibitory Notch pathway regulator, in HUVECs
co-cultured with A375 ALDH1A1+ compared with both A375
ALDH1A1SC (- 10.59 fold) and A375 ALDH1A1KD (-10.87 fold; Fig. 4A and B).
Evidence supports the idea that pro-inflammatory
stimuli can activate Notch signaling in different cellular contexts
(37). Thus, the present study
explored the involvement of IL-8 in the modulation of protein
expression of Notch pathway driven by melanoma ALDH1A1 in the 2D
co-culture model of ECs and A375 cells in the presence of
IL-8-neutralising antibody. Western blot analysis in ECs
corroborated the gene array results (Fig. 4C). An increase of DLL4 protein
expression in ECs co-cultured with A375 ALDH1A1+ was
found compared with endothelium incubated with A375ALDH1A1KD and
treatment of co-cultures with IL-8-neutralising antibody
significantly reduced DLL4 expression. Furthermore, Notch1 and
ADAM17 expression in ECs was also influenced by melanoma ALDH1A1
levels and IL-8 treatment, indicating that IL-8 released by
melanoma cells contributes to Notch signaling modification
(Fig. 4C). Finally, the present
study analyzed the RBPSUH expression. When Notch signaling is
activated, BPSUH promotes genes transcription leading to activation
of Notch target genes (38). The
present study found a high RBPSUH expression in HUVECs co-cultured
with A375 ALDH1A1+ compared with HUVECs co-cultured with
A375 ALDH1A1KD and IL-8 neutralizing antibody blunted this
effect.
Altogether, these data linked tumor ALDH1A1 levels
and secreted IL-8 to Notch signaling activation in the
endothelium.
Discussion
The present study aimed to assess the contribution
of melanoma ALDH1A1 on tumor angiogenesis and to characterize the
molecular signature of endothelium co-cultured with melanoma cells
in 2D and 3D in vitro settings. The results showed dynamic
crosstalk between tumor melanoma cells and endothelium, partly
mediated by IL-8 release from melanoma cells and favored by tumor
ALDH1A1 overexpression and activity. In particular, IL-8 induced
the expression in normal ECs of key mediators involved in the
activation of Notch signaling associated with the angiogenic
phenotype.
Increased metabolism of toxic aldehydes through
ALDH upregulation promotes cancer progression and therapy
resistance. ALDH1A1 is a cytosolic enzyme expressed in several
solid tumors (39), where it
confers stem-like phenotype and aggressive features. We and other
research groups have demonstrated that the acquisition of stem-like
phenotype driven by ALDH1A1 in breast cancer cells is implicated in
tumor vascularization (40,41) and angiogenesis through tumor
HIF-1α/VEGF signaling pathway activation and VEGF paracrine action
on ECs (17). A role of ALDH1A1
in melanoma pathogenesis has also been suggested in recent studies
(42,43), but no direct evidence for a
functional role in melanoma angiogenesis has been reported. In the
present study, in vivo experiments demonstrated a role of
ALDH1A1 overexpression with increased microvessel density in tumor
xenografts. Moreover, by using a 2D ECs-melanoma model and a 3D
model of multicellular tumorspheres, the present study found a
differential endothelial angiogenic phenotype mediated by tumor
ALDH1A1 expression levels and IL-8 release. Multicellular melanoma
tumorspheres were generated to recapitulate the global tumor tissue
organization and create a more complex TME than 2D culture
(20,44). Normal ECs and fibroblasts were
included in the 3D constructs to assess the role of variable
ALDH1A1 expression in tumor cells in conditioning the other cells,
focusing on the endothelium. Increased infiltration and
organization of HUVECs were found in the core of A375
ALDH1A1+ tumorspheres, presumably linked with activation
of HIF-1α signaling mediated by ALDH1A1 (17). Indeed, HIF-1α is a master
transcriptional factor for angiogenesis and metabolic remodulation
of tumor cells (45).
The crosstalk between tumor cells and their
microenvironment is crucial for cancer cell self-renewal, tumor
growth and metastasis (46).
Nevertheless, the metabolic modulation of the endothelium and the
formation of a deregulated and aberrant tumor vasculature serve a
critical role in maintaining the stem-like status in tumors
(47). Notably, Notch pathway
activation by signals within the TME has been proposed as an
additional mechanism by which endothelium controls the activity of
stem-like cells, which in turn influence the pro-angiogenic status
of ECs in a vicious cycle (48).
In the 3D co-culture model of the present study,
HUVECs from multicellular A375 ALDH1A1+ tumorspheres
expressed higher levels of Notch1 and DLL4 genes when
compared with A375 ALDH1A1KD. DLL4-Notch signaling has been
implicated in the specification of the endothelial tip cells
(48) and tumor vasculature has
been shown to overexpress DLL4, as endothelial-specific loss of
DLL-4 resulted in tumor vessel regression along with a reduction in
both epithelial-mesenchymal transition and stem-like features in
tumor cells (49).
In agreement with the evidence gathered in
tumorspheres, high ALDH1A1 expression in A375 and WM-266-4 cells
promoted HUVECs proliferation, migration, tube formation and
hyperpermeability in a 2D co-culture model. Conversely, loss of
function experiments produced a marked decrease of endothelial
pro-angiogenic functions when co-cultured with melanoma cells
silenced for ALDH1A1. Moreover, exposure of melanoma cells to the
enzyme inhibitor CM037, significantly impaired the angiogenic
features of the endothelium, suggesting that both expression and
activity of melanoma ALDH1A1 are critical for the acquisition of
ECs of an angiogenic phenotype.
The present study identified IL-8 as one of the
downstream target of ALDH1A1, responsible for endothelium phenotype
remodeling. IL-8 is constitutively expressed in melanoma (50); however, it is unknown which
factors mediate its upregulation in tumors. A hypothetical
mechanism might involve tumor hypoxia. In vivo studies in
human melanoma and other tumors show an increase in IL-8 production
mediated by hypoxia and acidosis of the microenvironment (51,52). Another mechanism may involve the
signaling of RA (53,54), one of the main products of
ALDH1A1 activity (39). Although
the RA receptor (RAR) may regulate the transcription of cytokines
genes, the promoter regions of a number of cytokines do not contain
any RA responsive elements, supposing an indirect role of RAR in
genes regulation (55) through
the activation of transcription factors. The binding of NF-κB to
the IL-8 promoter is required for triggering IL-8 gene
transcription, with p65 as the subunit responsible for binding to
the promoter (56-59). Furthermore, in a model of
melanoma cell line, RA in combination with TNFα, is able to induce
IL-8 expression with the contribution of NF-κB (60,61). The present study showed that
exogenous RA promoted IL-8 expression in A375 ALDH1A1SC, as well
NF-κB nuclear translocation and NF-κB-p65 subunit expression, while
RAR inhibition in A375 ALDH1A1SC and ALDH1A1+ decreased
IL-8 expression, suggesting that ALDH1A1 mediated IL-8 expression
and production by RA-NF-κB signaling pathway. However, whether
alternative and/or complementary pathways are involved, requires
further investigations. A critical function for this chemokine in
establishing stem-like properties of a number of solid tumors has
been reported (62) and
involvement of IL-8 in tumor progression has also been demonstrated
(63).
IL-8 influences tumor growth, angiogenesis,
invasion and metastasis through autocrine and paracrine signaling
(32,33,64) by binding two cell-surface G
protein-coupled receptors (CXCR1 and CXCR2) (65). Cytokines, including IL-8, can
induce Notch signaling (66),
but the mechanism remains unclear. Notch signaling serves a
critical role in the overall regulation of tumor angiogenesis
(67). The present study found a
differential Notch pathway gene and protein expression profile in
ECs co-cultured with melanoma cells, depending on melanoma ALDH1A1
expression and activity and IL-8 release and paracrine activity on
ECs. By comparing the expression of Notch mediators in ECs derived
from different multicellular 3D tumorspheres, it was found that
high ALDH1A1 expression in melanoma cells was associated with
higher expression of ligands DLL3 and DLL4. DLL4 is a
critical Notch ligand for stimulating angiogenesis (68). Contrarily, the gene expression of
another Notch ligand, Jag1, which can compete with DLL4 to
negatively regulate angiogenesis (69), was drastically reduced. In the
models of the present study, the Notch receptors and downstream
effectors were influenced by melanoma ALDH1A1. The expression of
Notch1, Notch2 and Notch3 receptors and ADAM17 (which cleaves the
receptors), as well as the Notch target genes HEY1 and HEY2, were
induced in ECs grown with melanoma ALDH1A1+.
Consistently, a reduction of NUMB gene expression, an inhibitory
Notch pathway regulator, was observed. When protein analysis of the
Notch pathway was performed in ECs co-cultured in the presence of
IL-8 neutralizing antibody, a reduction of DLL4, Notch1 and ADAM17
protein expression was observed. These findings suggested that
ALDH1A1 possibly through IL-8 release, remodels TME, controlling
ECs pro-angiogenic phenotype through Notch signaling regulation.
The mechanism by which IL-8 activates the Notch pathway remains to
be elucidated. Data suggest that IL-8 may interact with VEGF
receptor 2 (VEGFR2, the receptor primarily involved in Notch
signaling activation), promoting angiogenesis through receptor
transactivation (70,71). However, whether IL-8 promotes
activation of the Notch pathway in ECs through transactivation of
VEGFR2 or direct activation of CXCR1 and CXCR2 remains to be
elucidated. Nevertheless, that the modulation of the gene and
protein expression profile of Notch pathway mediators in the
endothelium in 3D tumorspheres may be partly attributed to direct
cell-cell and cell-matrix activity mediated by melanoma cells
expressing different levels of ALDH1A1 cannot be excluded.
Together, the present study provided further
insight into the mechanisms underlying angiogenesis and tumor
progression driven by ALDH1A1 in melanoma cancer cells and
described the concerted flow of signals from tumor to ECs.
Considering the functional changes in endothelium caused by ALDH1A1
overexpression in melanoma cell lines, this enzyme may cause an
extensive remodeling on TME to sustain tumor progression and
maintain stem-like phenotype.
In conclusion, ALDH1A1 is a cytosolic enzyme
upregulated in tumor cells and involved in detoxifying cells from
reactive aldehydes, such as retinaldehyde. This enzyme also engages
in acquired resistance to chemotherapeutic drugs, such as
oxazolidine, taxanes and platinum derivatives. It is a marker of
stemness in several solid tumors and correlates with poor clinical
outcome in a number of cancers (72,73). The present study showed that
there is also a relationship between ALDH1A1 expression and
activity and tumor angiogenesis, through the upregulation of
several pro-angiogenic mediators, including the chemokine IL-8.
RA-derived ALDH1A1 appeared to be involved in IL-8 expression
through NF-κB activation and expression. In TME, tumor-derived IL-8
activated Notch signaling on ECs and promotes the acquisition of a
pro-angiogenic phenotype. Based on the role of ALDH1A1 in the
control of TME by melanoma, the enzyme is a promising marker of
cross-talk between tumor (stem) cells and ECs, which in turn could
be an interesting target for development of new treatments.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SD, MZ and VC conceived the present study. VC, ET,
AF and ER performed methodology, validation, investigation, data
analysis. VC wrote the original draft of the manuscript. SD, LM and
MZ wrote reviewed and edited the manuscript. SD supervised and
administered the project and acquired funding. All authors reviewed
and approved the final manuscript.
Ethics approval and consent to
participate
The animal study was conducted according to the
guidelines of the Declaration of Helsinki and the Italian law
(Legislative Decree no. 26, 4 March 2014), which acknowledges the
European Directive 2010/63/UE and was approved by the animal
welfare board of University of Siena and the Italian Ministry of
Health (authorization no. 62/2014-B).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professor Ambra
Grolla, Department of Drug Science, University of Piemonte
Orientale A. Avogadro, Novara (Italy) who generously provided
cells. The authors would also like to thank Professor Donata
Medaglini and Dr Annalisa Ciabattini, Department of Medical
Biotechnology, University of Siena, Siena (Italy) for their
technical support.
Funding
This work was funded by Regione Toscana-Bando Ricerca Salute
2018_CORELAB and Fondazione Umberto Veronesi, Milan, Italy
(Post-doctoral Fellowship 2022).
References
|
1
|
Tripp MK, Watson M, Balk SJ, Swetter SM
and Gershenwald JE: State of the science on prevention and
screening to reduce melanoma incidence and mortality: The time is
now. CA Cancer J Clin. 66:460–480. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rambow F, Marine JC and Goding CR:
Melanoma plasticity and phenotypic diversity: Therapeutic barriers
and opportunities. Genes Dev. 33:1295–1318. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Klemen ND, Wang M, Feingold PL, Cooper K,
Pavri SN, Han D, Detterbeck FC, Boffa DJ, Khan SA, Olino K, et al:
Patterns of failure after immunotherapy with checkpoint inhibitors
predict durable progression-free survival after local therapy for
metastatic melanoma. J Immunother Cancer. 7:1962019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li F and Simon MC: Cancer cells don't live
alone: Metabolic communication within tumor microenvironments. Dev
Cell. 54:183–195. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hass R, von der Ohe J and Ungefroren H:
Impact of the tumor microenvironment on tumor heterogeneity and
consequences for cancer cell plasticity and stemness. Cancers
(Basel). 12:37162020. View Article : Google Scholar
|
|
6
|
Qin S, Jiang J, Lu Y, Nice EC, Huang C,
Zhang J and He W: Emerging role of tumor cell plasticity in
modifying therapeutic response. Signal Transduct Target Ther.
5:1–36. 2020. View Article : Google Scholar
|
|
7
|
Maishi N and Hida K: Tumor endothelial
cells accelerate tumor metastasis. Cancer Sci. 108:1921–1926. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Howard JD, Moriarty WF, Park J, Riedy K,
Panova IP, Chung CH, Suh KY, Levchenko A and Alani RM: Notch
signaling mediates melanoma-endothelial cell communication and
melanoma cell migration. Pigment Cell Melanoma Res. 26:697–707.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Long X, Ye Y, Zhang L, Liu P, Yu W, Wei F,
Ren X and Yu J: IL-8, a novel messenger to cross-link inflammation
and tumor emt via autocrine and paracrine pathways (Review). Int J
Oncol. 48:5–12. 2016. View Article : Google Scholar
|
|
10
|
Weinberg F, Ramnath N and Nagrath D:
Reactive oxygen species in the tumor microenvironment: An overview.
Cancers (Basel). 11:11912019. View Article : Google Scholar
|
|
11
|
Tasdogan A, Faubert B, Ramesh V,
Ubellacker JM, Shen B, Solmonson A, Murphy MM, Gu Z, Gu W, Martin
M, et al: Metabolic heterogeneity confers differences in melanoma
metastatic potential. Nature. 577:115–120. 2020. View Article : Google Scholar
|
|
12
|
Barrera G: Oxidative stress and lipid
peroxidation products in cancer progression and therapy. ISRN
Oncol. 2012:1372892012.PubMed/NCBI
|
|
13
|
Luo Y, Dallaglio K, Chen Y, Robinson WA,
Robinson SE, McCarter MD, Wang J, Gonzalez R, Thompson DC, Norris
DA, et al: ALDH1A isozymes are markers of human melanoma stem cells
and potential therapeutic targets. Stem Cells. 30:2100–2113. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma I and Allan AL: The role of human
aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell
Rev Rep. 7:292–306. 2011. View Article : Google Scholar
|
|
15
|
Moreb JS, Baker HV, Chang LJ, Amaya M,
Lopez MC, Ostmark B and Chou W: ALDH isozymes downregulation
affects cell growth, cell motility and gene expression in lung
cancer cells. Mol Cancer. 7:872008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Charafe-Jauffret E, Ginestier C, Iovino F,
Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci
F, Jacquemier J, et al: Aldehyde dehydrogenase 1-positive cancer
stem cells mediate metastasis and poor clinical outcome in
inflammatory breast cancer. Clin Cancer Res. 16:45–55. 2010.
View Article : Google Scholar
|
|
17
|
Ciccone V, Terzuoli E, Donnini S,
Giachetti A, Morbidelli L and Ziche M: Correction to: Stemness
marker ALDH1A1 promotes tumor angiogenesis via retinoic
acid/HIF-1α/VEGF signalling in MCF-7 breast cancer cells. J Exp
Clin Cancer Res. 37:3112018. View Article : Google Scholar
|
|
18
|
Terzuoli E, Bellan C, Aversa S, Ciccone V,
Morbidelli L, Giachetti A, Donnini S and Ziche M: ALDH3A1
overexpression in melanoma and lung tumors drives cancer stem cell
expansion, impairing immune surveillance through enhanced PD-L1
output. Cancers (Basel). 11:19632019. View Article : Google Scholar
|
|
19
|
Ramamoorthy P, Thomas SM, Kaushik G,
Subramaniam D, Chastain KM, Dhar A, Tawfik O, Kasi A, Sun W,
Ramalingam S, et al: Metastatic tumor-in-a-dish, a novel
multicellular organoid to study lung colonization and predict
therapeutic response. Cancer Res. 79:1681–1695. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han SJ, Kwon S and Kim KS: Challenges of
applying multicellular tumor spheroids in preclinical phase. Cancer
Cell Int. 21:1522021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerebral Organoid Cryopreservation and
Immunofluorescence. https://www.stemcell.com/cerebral-organoid-cryosectioning-immunofluorescence.html.
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Ciccone V, Zazzetta M and Morbidelli L:
Comparison of the effect of two hyaluronic acid preparations on
fibroblast and endothelial cell functions related to angiogenesis.
Cells. 8:14792019. View Article : Google Scholar
|
|
24
|
Ciccone V, Monti M, Monzani E, Casella L
and Morbidelli L: The metal-nonoate Ni(SalPipNONO) inhibits in
vitro tumor growth, invasiveness and angiogenesis. Oncotarget.
9:13353–13365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Flori L, Macaluso M, Taglieri I, Sanmartin
C, Sgherri C, Leo MD, Ciccone V, Donnini S, Venturi F, Pistelli L,
et al: Development of fortified citrus olive oils: From their
production to their nutraceutical properties on the cardiovascular
system. Nutrients. 12:15572020. View Article : Google Scholar :
|
|
26
|
Ciccone V, Monti M, Antonini G, Mattoli L,
Burico M, Marini F, Maidecchi A and Morbidelli L: Efficacy of
AdipoDren® in reducing interleukin-1-induced lymphatic
endothelial hyperpermeability. J Vasc Res. 53:255–268. 2016.
View Article : Google Scholar
|
|
27
|
Samson JM, Menon DR, Smith DE, Baird E,
Kitano T, Gao D, Tan AC and Fujita M: Clinical implications of
ALDH1A1 and ALDH1A3 mRNA expression in melanoma subtypes. Chem Biol
Interact. 314:1088222019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gridley T: Notch signaling in vascular
development and physiology. Development. 134:2709–2718. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rostama B, Peterson SM, Vary CPH and Liaw
L: Notch signal integration in the vasculature during remodeling.
Vascul Pharmacol. 63:97–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Welti J, Loges S, Dimmeler S and Carmeliet
P: Recent molecular discoveries in angiogenesis and antiangiogenic
therapies in cancer. J Clin Invest. 123:3190–3200. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Srivastava SK, Bhardwaj A, Arora S, Tyagi
N, Singh AP, Carter JE, Scammell JG, Fodstad Ø and Singh S:
Interleukin-8 is a key mediator of FKBP51-induced melanoma growth,
angiogenesis and metastasis. Br J Cancer. 112:1772–1781. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li A, Dubey S, Varney ML, Dave BJ and
Singh RK: IL-8 directly enhanced endothelial cell survival,
proliferation, and matrix metalloproteinases production and
regulated angiogenesis. J Immunol. 170:3369–3376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu S, Singh S, Varney ML, Kindle S and
Singh RK: Modulation of CXCL-8 expression in human melanoma cells
regulates tumor growth, angiogenesis, invasion, and metastasis.
Cancer Med. 1:306–317. 2012. View Article : Google Scholar
|
|
34
|
Fernández-Chacón M, García-González I,
Mühleder S and Benedito R: Role of notch in endothelial biology.
Angiogenesis. 24:237–250. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Taslimi S and Das S: Angiogenesis and
angiogenesis inhibitors in brain tumors. Handbook of Brain Tumor
Chemotherapy, Molecular Therapeutics, and Immunotherapy. Newton HB:
2nd edition. Academic Press; Cambridge, MA: pp. 361–371. 2018
|
|
36
|
Groot AJ and Vooijs MA: The role of adams
in notch signaling. Adv Exp Med Biol. 727:15–36. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fazio C and Ricciardiello L: Inflammation
and notch signaling: A crosstalk with opposite effects on
tumorigenesis. Cell Death Dis. 7:e25152016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Christopoulos PF, Gjølberg TT, Krüger S,
Haraldsen G, Andersen JT and Sundlisæter E: Targeting the notch
signaling pathway in chronic inflammatory diseases. Front Immunol.
12:6682072021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tomita H, Tanaka K, Tanaka T and Hara A:
Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget.
7:11018–11032. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ribatti D: Cancer stem cells and tumor
angiogenesis. Cancer Lett. 321:13–17. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fan YL, Zheng M, Tang YL and Liang XH: A
new perspective of vasculogenic mimicry: EMT and cancer stem cells
(Review). Oncol Lett. 6:1174–1180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dinavahi SS, Gowda R, Gowda K, Bazewicz
CG, Chirasani VR, Battu MB, Berg A, Dokholyan NV, Amin S and
Robertson GP: Development of a novel multi-isoform ALDH inhibitor
effective as an antimelanoma agent. Mol Cancer Ther. 19:447–459.
2020. View Article : Google Scholar
|
|
43
|
Yue L, Huang ZM, Fong S, Leong S, Jakowatz
JG, Charruyer-Reinwald A, Wei M and Ghadially R: Targeting ALDH1 to
decrease tumorigenicity, growth and metastasis of human melanoma.
Melanoma Res. 25:138–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Brassard-Jollive N, Monnot C, Muller L and
Germain S: In vitro 3D systems to model tumor angiogenesis and
interactions with stromal cells. Front Cell Dev Biol. 8:5949032020.
View Article : Google Scholar :
|
|
45
|
LaGory EL and Giaccia AJ: The
ever-expanding role of HIF in tumour and stromal biology. Nat Cell
Biol. 18:356–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Whiteside TL: The tumor microenvironment
and its role in promoting tumor growth. Oncogene. 27:5904–5912.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ingangi V, Minopoli M, Ragone C, Motti ML
and Carriero MV: Role of microenvironment on the fate of
disseminating cancer stem cells. Front Oncol. 9:822019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jakobsson L, Franco CA, Bentley K, Collins
RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G,
Medvinsky A, et al: Endothelial cells dynamically compete for the
tip cell position during angiogenic sprouting. Nat Cell Biol.
12:943–953. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Patel NS, Li JL, Generali D, Poulsom R,
Cranston DW and Harris AL: Up-regulation of delta-like 4 ligand in
human tumor vasculature and the role of basal expression in
endothelial cell function. Cancer Res. 65:8690–8697. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Peng HH, Liang S, Henderson AJ and Dong C:
Regulation of interleukin-8 expression in melanoma-stimulated
neutrophil inflammatory response. Exp Cell Res. 313:551–559. 2007.
View Article : Google Scholar
|
|
51
|
Rofstad EK and Halsør EF:
Hypoxia-associated spontaneous pulmonary metastasis in human
melanoma xenografts: Involvement of microvascular hot spots induced
in hypoxic foci by interleukin 8. Br J Cancer. 86:301–308. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Korbecki J, Kojder K, Kapczuk P, Kupnicka
P, Gawrońska-Szklarz B, Gutowska I, Chlubek D and
Baranowska-Bosiacka I: The effect of hypoxia on the expression of
CXC chemokines and CXC chemokine receptors-a review of literature.
Int J Mol Sci. 22:8432021. View Article : Google Scholar
|
|
53
|
Chang MM, Harper R, Hyde DM and Wu R: A
novel mechanism of retinoic acid-enhanced interleukin-8 gene
expression in airway epithelium. Am J Respir Cell Mol Biol.
22:502–510. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mukherjee S, Date A, Patravale V, Korting
HC, Roeder A and Weindl G: Retinoids in the treatment of skin
aging: An overview of clinical efficacy and safety. Clin Interv
Aging. 1:327–348. 2006. View Article : Google Scholar
|
|
55
|
Duong V and Rochette-Egly C: The molecular
physiology of nuclear retinoic acid receptors. from health to
disease. Biochim Biophys Acta. 1812:1023–1031. 2011. View Article : Google Scholar
|
|
56
|
Dai X, Yamasaki K, Shirakata Y, Sayama K
and Hashimoto K: All-trans-retinoic acid induces interleukin-8 via
the nuclear factor-KappaB and P38 mitogen-activated protein kinase
pathways in normal human keratinocytes. J Invest Dermatol.
123:1078–1085. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hoffmann E, Dittrich-Breiholz O, Holtmann
H and Kracht M: Multiple control of interleukin-8 gene expression.
J Leukoc Biol. 72:847–855. 2002.PubMed/NCBI
|
|
58
|
Kunsch C, Lang RK, Rosen CA and Shannon
MF: Synergistic transcriptional activation of the IL-8 gene by
NF-Kappa B P65 (RelA) and NF-IL-6. J Immunol. 153:153–164.
1994.PubMed/NCBI
|
|
59
|
Kunsch C and Rosen CA: NF-Kappa B
subunit-specific regulation of the interleukin-8 promoter. Mol Cell
Biol. 13:6137–6146. 1993.PubMed/NCBI
|
|
60
|
Harant H, de Martin R, Andrew PJ, Foglar
E, Dittrich C and Lindley IJ: Synergistic activation of
interleukin-8 gene transcription by all-trans-retinoic acid and
tumor necrosis factor-alpha involves the transcription factor
NF-KappaB. J Biol Chem. 271:26954–26961. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Jambrovics K, Uray IP, Keresztessy Z,
Keillor JW, Fésüs L and Balajthy Z: Transglutaminase 2 programs
differentiating acute promyelocytic leukemia cells in all-trans
retinoic acid treatment to inflammatory stage through NF-KB
activation. Haematologica. 104:505–515. 2019. View Article : Google Scholar :
|
|
62
|
Alfaro C, Sanmamed MF, Rodríguez-Ruiz ME,
Teijeira Á, Oñate C, González Á, Ponz M, Schalper KA, Pérez-Gracia
JL and Melero I: Interleukin-8 in cancer pathogenesis, treatment
and follow-up. Cancer Treat Rev. 60:24–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yuan A, Chen JJW, Yao PL and Yang PC: The
role of interleukin-8 in cancer cells and microenvironment
interaction. Front Biosci. 10:853–865. 2005. View Article : Google Scholar
|
|
64
|
Singh S, Wu S, Varney M, Singh AP and
Singh RK: CXCR1 and CXCR2 silencing modulates CXCL8-dependent
endothelial cell proliferation, migration and capillary-like
structure formation. Microvasc Res. 82:318–325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Waugh DJJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang H, Tian Y, Wang J, Phillips KLE,
Binch ALA, Dunn S, Cross A, Chiverton N, Zheng Z, Shapiro IM, et
al: Inflammatory cytokines induce NOTCH signaling in nucleus
pulposus cells. J Biol Chem. 288:16761–16774. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Siekmann AF and Lawson ND: Notch
signalling and the regulation of angiogenesis. Cell Adh Migr.
1:104–106. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Benedito R, Roca C, Sörensen I, Adams S,
Gossler A, Fruttiger M and Adams RH: The notch ligands Dll4 and
jagged1 have opposing effects on angiogenesis. Cell. 137:1124–1135.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Akil A, Gutiérrez-García AK, Guenter R,
Rose JB, Beck AW, Chen H and Ren B: Notch signaling in vascular
endothelial cells, angiogenesis, and tumor progression: An update
and prospective. Front Cell Dev Biol. 9:6423522021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Petreaca ML, Yao M, Liu Y, Defea K and
Martins-Green M: Transactivation of vascular endothelial growth
factor receptor-2 by interleukin-8 (IL-8/CXCL8) is required for
IL-8/CXCL8-induced endothelial permeability. Mol Biol Cell.
18:5014–5023. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Martin D, Galisteo R and Gutkind JS:
CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF)
expression and the autocrine activation of VEGFR2 in endothelial
cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1)
complex. J Biol Chem. 284:6038–6042. 2009. View Article : Google Scholar :
|
|
72
|
Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang
J, Zhang G, Wang X, Dong Z, Chen F and Cui H: Targeting cancer stem
cell pathways for cancer therapy. Signal Transduct Target Ther.
5:82020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ciccone V, Morbidelli L, Ziche M and
Donnini S: How to conjugate the stemness marker ALDH1A1 with tumor
angiogenesis, progression, and drug resistance. Cancer Drug
Resistance. 3:26–37. 2020.PubMed/NCBI
|