Introduction
Bladder cancer is the most common malignant tumor of
the urinary system and is rated as the tenth most common form of
cancer and the ninth leading cause of cancer-associated mortality
worldwide in 2018, occurring in the mucous membrane of the bladder
tissues (1,2). In China, its incidence ranks first
among urogenital system tumors (3). Bladder cancer mainly includes
bladder urothelial carcinoma, bladder squamous cell carcinoma and
bladder adenocarcinoma (4). Early
bladder cancer lacks significant symptoms, and a small number of
patients develop hematuria, which is often in the advanced stage at
the time of diagnosis (5). The
main treatment methods for bladder cancer include surgical
resection, chemoradiotherapy and targeted therapy, among which
targeted therapy is the most effective and promising treatment for
patients with advanced bladder cancer (6). To combat this disease and improve
the prognosis of patients with bladder cancer, more therapeutic
targets are still urgently needed.
Lamin B2 (LMNB2) is a type of nuclear lamina
filament protein, which is involved in multiple cellular processes,
such as transcription regulation and mitosis (7,8).
During normal mitosis, LMNB2 regulates chromosome stability by
ensuring proper mitotic chromosome segregation (9). Additionally, LMNB2 mediates
nucleolar morphology, affects cardiomyocyte polyploidization, and
promotes myocardial regeneration (10). It has also been reported that
LMNB2 promotes retinal development, and the mutations of LMNB2 lead
to neurodevelopmental and nuclear morphology defects (11,12). LMNB2 variants were revealed to
cause primary microcephaly and define a novel laminopathy (12).
Notably, LMNB2 is recognized as an oncogene
affecting the progression of multiple types of cancers, such as
ovarian, lung, and liver cancer (13-15). LMNB2 was revealed to be aberrantly
highly expressed in tumor tissues, and correlated with the
prognosis of several cancers, such as breast, liver, and lung
cancer (15-17). In a previous study, LMNB2 bound to
MCM7 and promoted the activity of MCM7 helicase, thus promoting the
proliferation of lung cancer cells (16). Additionally, in another study,
LMNB2 promoted the dimethylation of histone 3 lysine 9 (H3K9), and
thus promoted the malignant phenotype of non-small cell lung cancer
(NSCLC) (14). Although the
multiple effects of LMNB2 on cancer progression have been
elucidated, its possible role in bladder cancer remains
unclear.
In the present study, the expression levels of LMNB2
in human bladder cancer tissues were assessed and its correlation
with the prognosis and clinical features of patients was
investigated. The involvement of LMNB2 in the regulation of bladder
cancer cells in vitro and in mice was further demonstrated,
and the molecular mechanism was clarified. It is therefore
suggested that LMNB2 could serve as a promising therapeutic target
for bladder cancer treatment.
Materials and methods
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/) was used
to analyze the data in The Cancer Genome Atlas (TCGA; https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
to determine the mRNA levels and the effects of LMNB2 and cell
division cycle-associated protein 3 (CDCA3) on the survival rates
of patients with bladder cancer. The expression of LMNB2 in several
types of cancers was analyzed on the web of The Human Protein Atlas
(https://www.proteinatlas.org/).
Human tissue samples
A total of 107 bladder cancer tissues and adjacent
tissues were collected from patients who received surgical therapy
at the College of Clinical Medicine of Henan University of Science
and Technology (Luoyang, China). The present study was approved by
the Ethics Committee of the College of Clinical Medicine of Henan
University of Science and Technology (approval no. AMI-2020-073).
The clinicopathological features of patients with bladder cancer,
such as age, sex, tumor grade, and recurrence were analyzed and are
presented in Tables I and
II. The age distribution of the
patients was 47-82 years (median 66 years).
 | Table IAssociations between the expression
of LMNB2 and clinicopathological characteristics in 107 patients
with bladder cancer. |
Table I
Associations between the expression
of LMNB2 and clinicopathological characteristics in 107 patients
with bladder cancer.
| Features | Total no. of
patients (n=107) | LMNB2 expression
| χ2 | P-value |
|---|
Low
| High
|
|---|
| n=32 | n=75 |
|---|
| Age (years) | | | | 0.824 | 0.364 |
| <65 | 54 | 14 | 40 | | |
| ≥65 | 53 | 18 | 35 | | |
| Sex | | | | 0.077 | 0.782 |
| Male | 58 | 18 | 40 | | |
| Female | 49 | 14 | 35 | | |
| Tumor stage | | | | 10.449 | 0.001a |
| T2 | 36 | 18 | 18 | | |
| T3/T4 | 71 | 14 | 57 | | |
| Tumor grade | | | | 1.222 | 0.269 |
| Low | 29 | 11 | 18 | | |
| High | 78 | 21 | 57 | | |
| Lymph node
metastasis | | | | 1.195 | 0.274 |
| Yes | 20 | 8 | 12 | | |
| No | 87 | 24 | 63 | | |
| Recurrence | | | | 7.422 | 0.006a |
| Yes | 55 | 10 | 45 | | |
| No | 52 | 22 | 30 | | |
 | Table IIAssociations between the expression
of CDCA3 and clinicopathological characteristics in 107 patients
with bladder cancer. |
Table II
Associations between the expression
of CDCA3 and clinicopathological characteristics in 107 patients
with bladder cancer.
| Features | Total no. of
patients (n=107) | CDCA3 expression
| χ2 | P-value |
|---|
Low
| High
|
|---|
| n=35 | n=72 |
|---|
| Age (years) | | | | 2.280 | 0.131 |
| <65 | 54 | 14 | 40 | | |
| ≥65 | 53 | 21 | 32 | | |
| Sex | | | | 0.704 | 0.402 |
| Male | 58 | 21 | 37 | | |
| Female | 49 | 14 | 35 | | |
| Tumor stage | | | | 7.368 | 0.007a |
| T2 | 36 | 18 | 18 | | |
| T3/T4 | 71 | 17 | 54 | | |
| Tumor grade | | | | 1.328 | 0.249 |
| Low | 29 | 7 | 22 | | |
| High | 78 | 28 | 50 | | |
| Lymph node
metastasis | | | | 0.594 | 0.441 |
| Yes | 20 | 8 | 12 | | |
| No | 87 | 27 | 60 | | |
| Recurrence | | | | 10.853 | 0.001a |
| Yes | 55 | 10 | 45 | | |
| No | 52 | 25 | 27 | | |
The expression levels of LMNB2 and CDCA3 in tumor
and adjacent tissues were detected using immunohistochemistry
(IHC). All tissues were fixed in 4% formalin for 48 h at room
temperature and embedded in paraffin. Subsequently, 4-µm
sections were blocked using 5% BSA for 30 min at room temperature.
The blocked sections were then incubated with primary antibodies:
Anti-LMNB2 antibody (1:400; product code ab151735) and anti-CDCA3
antibody (1:200; product code ab166902; both from Abcam) for 2 h at
room temperature, and subsequently incubated with a biotinylated
secondary antibody kit (ready to use; cat. no. PV6000; ZSGB-BIO;
OriGene Technologies, Inc.) for another 1 h at 37°C according to
the manufacturer's instructions. Finally, DAB solution was applied
for color development.
The expression levels of LMNB2 and CDCA3 were
manually divided based on the staining intensity (0, negative
staining; 1, weak staining; 2, moderate staining; and 3 strong
staining). Meanwhile, the proportion of stained cells was as
follows (0, 0% positive-stained cells; 1, 1-30% positive-stained
cells; 2, 31-60% positive-stained cells; and 3, 61-100%
positive-stained cells). The total score was the staining intensity
x the score of the percentage of positive-stained cells, and <1
or =1 was considered as negative staining, whereas 2-4 was
considerate as weak staining and >4 was considered as strong
staining of CDCA3.
The sections from each patient were observed in at
least five light visual fields, and two experienced pathologists
examined the sections using a light microscope at magnifications of
×100 (scale bar, 100 µm) and ×200 (scale bar, 50
µm).
Cell culture and transfection
The human bladder cancer cells, including T24 (ATCC
no. HTB-4) and 5637 cells (ATCC no. HTB-9), and normal epithelial
cell SV-HUC-1 (ATCC no. CRL-9520) were all obtained from ATCC and
maintained in Dulbecco's modified Eagle's medium (DMEM; cat. no.
11971025; Thermo Fisher Scientific, Inc.) supplemented with 10% of
fetal bovine serum (FBS; cat. no. 04-001-1A; Biological
Industries), 1% penicillin-streptomycin (cat. no. P1400; Beijing
Solarbio Science & Technology Co., Ltd.) and incubated at 37°C
in a 5% CO2 incubator.
The indicated plasmids in the present study were
transfected into bladder cancer cells using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). LMNB2 knockdown was
confirmed in both T24 and 5637 cells, two of the most commonly used
in vitro models for bladder cancer.
All sequences were synthesized by Sangon Biotech
Co., Ltd. (https://www.sangon.com/). The shRNA
plasmids of LMNB2 and CDCA3 were constructed in our laboratory.
Short hairpin (sh)RNA LMNB2, 5′-GCA GAG TTG GAC GAG GTC AAC AAG
A-3′; shRNA CDCA3, 5′-CCG CTC TCC TAC TCT TGG TAT TGC A-3′; and
shRNA scramble, 5′-ACT CAA GAG TCT AGC AAG CCT GCA G-3′. The
overexpression plasmids of pcDNA3.1-vector (cat. no. 34706) were
obtained from Addgene, Inc. and pcDNA3.1-LMNB2 and pcDNA3.1-CDCA3
plasmids were constructed in our laboratory. Other plasmids,
pGL3-Basic Luciferase Reporter Vector (cat. no. E1751; Promega
Corporation) and pGL-CDCA3 plasmids were constructed in our
laboratory. For plasmid transfection, 2.5 µg plasmid
(OD260/280=1.8-2.0) were transfected into target cells
(50×104) using Lipofectamine 3000 (cat. no. L3000001;
Thermo Fisher Scientific, Inc.) for 15 min at room temperature.
After 2 days, subsequent experimentations were performed.
For lentivirus transduction, the plasmid PLKO.1 (3rd
generation; cat. no. 10878; Addgene, Inc.) was used to produce the
lentivirus according to the manufacturer's instructions. Simply, 6
µg pMDL (cat. no. 12251), 3 µg pVSV-G (cat. no.
138479), 2 µg pRSV-Rev (cat. no. 12253; all from Addgene,
Inc.) and 5 µg PLKO.1 were transfected into 293FT cells (80%
confluence; cat. no. R70007; Thermo Fisher Scientific, Inc.) using
Lipofectamine® 3000 (cat. no. L3000001; Thermo Fisher
Scientific, Inc.) for 15 min at room temperature. After 2 days, the
lentivirus was harvested from the supernatant by
ultracentrifugation at 72,000 × g at 4°C for 120 min. Subsequently,
2×105 T24 and 5637 cells were transfected with the
lentivirus for 36 h at 37°C and 5% CO2, with an MOI of
1:5. After 3 days, LMNB2-knockdown cells were selected using 2
µg/ml puromycin (cat. no. P8230; Beijing Solarbio Science
& Technology Co., Ltd.) for 5 days.
Reverse transcription-quantitative
(RT-q)PCR assays
Total RNA was extracted from bladder cancer cells
using TRIzol reagent (cat. no. 15596-018; Invitrogen; Thermo Fisher
Scientific, Inc.). Then total RNA was reverse-transcribed using
M-MLV reverse transcriptase kit (cat. no. M1701; Promega
Corporation) according to the manufacturer's instructions. RT-qPCR
was then performed using SYBR Green mixture (cat. no. RR420A;
Takara Bio, Inc.). The following thermocycling conditions were used
for qPCR: Initial denaturation at 95°C for 3 min; followed by 30
cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 30
sec and extension at 72°C for 30 sec. The 2−ΔΔCq method
was used to quantify the results (18). LMNB2 and CDCA3 expression levels
were normalized to the expression of GAPDH. The following primers
were used:
LMNB2 forward, 5′-TTT CCA CCA ACA GGG GGA C-3′ and
reverse, 5′-ACG TTC TGG CAG TTC GCT T-3′; CDCA3 forward, 5′-CAC CTA
GTG CTG GCA TCC TG-3′ and reverse, 5′-GGC AGA ACA GGC TCT CCA
CT-3′; GAPDH forward, 5′-ATG GGC AGC CGT TAG GAA AG-3′ and reverse,
5′-GCC CAA TAC GAC CAA ATC AGA GA-3′.
Western blot analysis
Bladder cancer cells were lysed using RIPA buffer
(product no. 9806S; Cell Signaling Technology, Inc.). All the cell
and tissue samples were isolated to extract the proteins and
determined using the BCA method. A total of 30 µg protein
per lane was separated by 10% SDS-PAGE, sequentially transferred
onto the PVDF membranes, followed by blocking with 5% fat-free milk
in TBST buffer at room temperature for 30 min. PVDF membranes were
then treated with primary antibodies targeting a series of proteins
at room temperature for 1.5 h. Subsequently the membranes were
incubated with secondary antibodies which were HRP-conjugated at
room temperature for 1 h. The secondary antibodies were as follows:
Goat anti-rabbit secondary antibody (cat. no. ZB-2301; 1:10,000;
ZSGB-BIO; OriGene Technologies, Inc.) and goat anti-mouse secondary
antibody (cat. no. G-21040; dilution, 1:10,000; Thermo Fisher
Scientific, Inc.). Signals were detected using an ECL kit (Novex
ECL Chemiluminescent Substrate Reagent kit; Thermo Fisher
Scientific, Inc.) and analyzed using ImageJ software (v1.32;
National Institutes of Health) according to the manufacturer's
protocol. The primary antibodies used for the various experiments
were as follows: Anti-LMNB2 antibody (1:400 dilution for IHC;
1:1,500 dilution for western blotting; and 1:200 dilution for CHIP
assays; product code ab151735), anti-CDCA3 antibody (1:200 dilution
for IHC; 1:2,000 dilution for western blotting; product code
ab166902), anti-Ki67 antibody (1:1,000 dilution; product code
ab16667), anti-proliferating cell nuclear antigen (PCNA) antibody
(1:500 dilution; product code ab29), anti-caspase-3 antibody
(1:1,000 dilution; product code ab32351), anti-caspase-8 antibody
(1:1,000 dilution; product code ab32397), and anti-β-actin antibody
(1:3,000 dilution; product code ab8226; all from Abcam).
Colony formation assay
A total of 1,000 bladder cancer cells were re-seeded
into 6-well plates in complete medium and maintained for nearly 2
weeks in a cell incubator, until the colonies were formed.
Subsequently, the colonies were fixed with 4% paraformaldehyde
(PFA) for 20 min and stained with 0.1% crystal violet at room
temperature for 20 min. Following staining, colonies (>1
mm2 was considered a cell colony) were captured and
analyzed by ImageJ (ImageJ 1.48v; National Institutes of
Health).
MTT assay
Bladder cancer cells were seeded into 96-well plates
with 5,000 cells/well and maintained for 12 h. On days 1, 2, 3, 4
and 5, cells were subsequently incubated with MTT for 2 h and then
washed with PBS. Cells were then isolated using 150 µl DMSO
and the OD570 was analyzed.
Cell Counting Kit-8 (CKK-8) assay
Bladder cancer cells were plated into 96-well plates
with a density of 5,000 cells and subsequently maintained for 12 h.
On days 1, 2, 3, 4 and 5, cells were then treated with 10 µl
CCK-8 (cat. no. 96992; Merck KGaA) for 2 h and the OD value was
measured at 490 nm.
Cell cycle assay
Bladder cancer cells (1×106) were fixed
using 70% ethylalcohol for 24 h at −20°C and then incubated with a
concentration of 100 µg/ml propidium iodide (PI) at 37°C for
15 min. Subsequently the samples (300 µl) were analyzed
using FACSCalibur flow cytometer (BD Biosciences) and FlowJo
software (FlowJo v10.6.2; FlowJo LLC). The percentage of cells at
different phases was compared.
Cell apoptosis assay
Bladder cancer cells (1×106) were
re-suspended and incubated with Annexin V-FITC and propidium iodide
kit (cat. no. V13242; Invitrogen; Thermo Fisher Scientific, Inc.)
at room temperature for 10 min according to the manufacturer's
instructions. Subsequently, the samples were analyzed using
FACSCalibur flow cytometer (BD Biosciences) and FlowJo software
(FlowJo v10.6.2; FlowJo LLC), and the apoptosis of cells in
different groups was analyzed and compared.
Tumor growth in vivo assay
The procedures used for the animal assays were all
approved by the Institutional Animal Care and Use Committee (IACUC)
of the College of Clinical Medicine of Henan University of Science
and Technology (approval no. SYXK 2020-0127). The female BALB/c
nude mice (8 weeks old; weight, 19-21 g) were purchased from
Beijing Vital River Laboratory Animal Technology Co., Ltd. All mice
in the present study were fed ad libitum with food and
water, and were maintained in specific pathogen-free conditions at
20°C, a 12-h light/dark cycle and 60% humidity. A total of 10 nude
mice were used in the present study, including control (n=5) and
LMNB2-knockdown mice (n=5). To measure tumor growth in vivo,
T24 cells were stably transfected with LMNB2 shRNA plasmids and
injected into the right flank of female nude mice. After 7 days,
tumors began to be established, and the volume of the tumors was
assessed every 7 days and calculated. After 42 days, the tumor
growth curves were calculated and compared between different
groups. The tumor volume was calculated as follows: Tumor volume
(mm3)=tumor length (mm) x tumor width
(mm)2/2. Humane endpoints were used during the animal
assays and the mice were sacrificed if they met one of humane
endpoints: Weight loss of >25%, maximum tumor diameter >20
mm. Mice were sacrificed by intraperitoneal injection of sodium
pentobarbital (100 mg/kg). Mortality was confirmed by cervical
dislocation. The maximum tumor volume observed in the xenograft
study was 700 mm3.
ChIP and luciferase assays
ChIP assays were performed using a CHIP Assay kit
(product code ab500; Abcam). A total number of 1×108 T24
cells were crosslinked, resuspended and lysed, then sonicated so as
to shear the DNA into a range of 600-900 bp. The chromatin fraction
was then immunoprecipitated using LMNB2 (1:200 dilution; product
code ab151735) or IgG targeted antibodies (1:200 dilution; product
code ab172730; both from Abcam), respectively, and the mix was
enriched by the use of protein A Agarose (product code ab193254;
Abcam). Beads were isolated and washed five times. DNA was finally
purified and quantified using Nanodrop spectrophotometer (NanoDrop
ND-1000; NanoDrop Technologies; Thermo Fisher Scientific, Inc.).
Next-generation sequencing (300 bp paired-end sequencing) and data
analysis were then conducted at Sangon Biotech Co. Ltd. Briefly, 20
pm DNA was used for sequencing using NovaSeq 6000 SP reagent kit
(100 cycles; cat. no. 2002746; Illumina Inc.). Quality-control and
data analysis were performed using FastQC software (version 0.11.9;
http://www.bioinformatics.babraham.ac.uk/projects/fastqc/),
Bowtie2 (version 2.2.4; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml),
and R (version 4.0.5; https://www.r-project.org/).
For luciferase assays, bladder cancer cells were
maintained and transfected with 2.5 µg pGL-CDCA3 (cat. no.
E1751; Promega Corporation), pGL-Basic, pEnter-LMNB2 plasmids for
48 h using Lipofectamine 3000. The cells were then washed twice and
lysed, and the luciferase activities were then detected using
Dual-Luciferase Reporter Assay System (cat. no. E1910; Promega
Corporation) according to the manufacturer's protocol. Renilla
reniformis luciferase activity was measured to normalize
transfection efficiency.
Statistical analysis
GraphPad 6.0 (GraphPad Software, Inc.) was used for
all statistical analyses. Data were expressed as the mean ± SEM in
the present study. The survival analysis between clinical features
and LMNB2 expression was performed using Kaplan-Meier method with
log-rank testing test. Student's t-test was used for statistical
comparisons between control and shRNA groups, and multiple
comparisons were performed using ANOVA and post hoc Tukey's test.
Spearman's and Pearson's correlation analyses were performed to
analyze the associations of LMNB2 and CDCA3. The experiments were
repeated at least three times.
Results
LMNB2 is upregulated in human bladder
cancer tissues and correlated with the prognosis and clinical
features of patients
According to the bioinformatics data from GEPIA
database, it was observed that LMNB2 was widely expressed in
multiple types of tumor tissues, such as thyroid cancer, glioma,
and colorectal cancer (Fig. 1A),
and associated with the prognosis of patients with several types of
cancers, including renal, prostate, liver, and stomach cancer
(Fig. 1B). These data suggested
LMNB2 was a potential tumor regulator.
To clarify whether LMNB2 could affect the
progression of bladder cancer, the expression levels of LMNB2 in
human bladder cancer tissues and normal tissues were first
investigated using GEPIA database. A total of 404 tumor tissues and
28 normal tissues were included in this comparison. It was observed
that the mRNA levels of LMNB2 in tumor tissues were obviously
higher than those in normal tissues (Fig. 1C). Subsequently, the effects of
LMNB2 on the prognosis of patients was further investigated using
GEPIA database, and it was revealed that LMNB2 was significantly
associated with the disease-free survival rates of patients in two
cohorts (19,20) (P=0.023 and P=0.045, respectively;
Fig. 1D), suggesting an
association between LMNB2 and prognosis. Therefore, it was
determined that LMNB2 was upregulated in human bladder cancer
tissues and associated with the prognosis of patients.
The expression levels of LMNB2 in 107 human bladder
cancer tissues and the corresponding adjacent tissues collected at
our hospital were then detected using IHC assays. Similar to the
results obtained in the GEPIA database, it was observed that LMNB2
expression was significantly higher in tumor tissues than that in
adjacent tissues (Fig. 2),
further confirming that LMNB2 was upregulated in human bladder
cancer tissues.
Subsequently, the patients were divided into two
groups, including LMNB2 low and high expression groups, according
to the expression levels of LMNB2 in tumor tissues. It was observed
that 32 (32/107; 29.9%) patients exhibited LMNB2 low expression,
whereas the remaining (75/107, 70.1%) exhibited high expression of
LMNB2 in tumor tissues (Table I).
To further clarify the associations between LMNB2 expression and
the clinical features of patients, the age, sex, tumor stage, tumor
grade, lymph node metastasis, and recurrence of disease in patients
were investigated. No evident associations between LMNB2 expression
and clinical features including age (P=0.364), sex (P=0.782), tumor
grade (P=0.269), and lymph node metastasis (P=0.274; Table I) were found. However, it was
determined that the expression of LMNB2 was significantly
associated with the tumor stage (P=0.001) and recurrence of disease
(P=0.006; Table I) of patients
with bladder cancer. Collectively, our data confirmed that LMNB2
was upregulated in human bladder cancer tissues and associated with
the prognosis and clinical features of patients.
LMNB2 depletion impairs the proliferation
of bladder cancer cells via stimulating cell cycle arrest and
apoptosis in vitro
Subsequently, it was determined that LMNB2 was
overexpressed in bladder cancer cells, T24 and 5637 cells, compared
with normal epithelial cell line, SV-HUC-1 (Fig. 3A). It was then investigated
whether LMNB2 could affect the proliferation of bladder cancer
cells in vitro. The shRNA plasmids of LMNB2 were constructed
and stably transfected into two types of human bladder cancer
cells, including T24 and 5637 cells, to deplete its expression.
Through RT-qPCR assays, it was demonstrated that the transfection
of LMNB2 shRNA plasmids could significantly decrease the mRNA
levels of LMNB2 in T24 and 5637 cells (Fig. 3B). Similarly, it was established
that the protein expression of LMNB2 was significantly decreased
upon the transfection of LMNB2 shRNA plasmids in both T24 and 5637
cells, as confirmed by western blot analysis (Fig. 3C).
Notably, using MTT and CCK-8 assays, it was
determined that the knockdown of LMNB2 in T24 and 5637 cells
decreased the OD value at 570 and 490 nm, respectively, suggesting
the impairment of bladder cancer cell proliferation (Fig. 3D and E). It was then detected
whether LMNB2 affected the proliferation of bladder cancer cells
via colony formation assays. The results revealed that LMNB2
ablation significantly suppressed the proliferation of T24 and 5637
cells, as observed by the decreased number of colonies (Fig. 3F). Furthermore, consistent with
the previous results found, through western blotting, it was
observed that the expression levels of two proliferation markers,
Ki67 and PCNA, were significantly decreased after LMNB2 depletion
in T24 and 5637 cells, suggesting the inhibition of cell
proliferation (Fig. 3G and
H).
Next, it was determined whether LMNB2 affected
bladder cancer cell proliferation via mediation of the cell cycle
or cell apoptosis through flow cytometric assays. According to the
results, it was revealed that LMNB2 depletion led to cell cycle
arrest with an increase in the number of cells at the G2 phase in
T24 and 5637 cells (Fig. 4A).
Additionally, through flow cytometric assays, it was determined
that the knockdown of LMNB2 promoted the apoptosis of bladder
cancer cells, with an increase in the number of early or late
apoptotic cells (Fig. 4B).
Furthermore, knockdown of LMNB2 in bladder cancer cells increased
the expression of caspase-3 and caspase-8 (Fig. 4C). Subsequently, overexpression of
LMNB2 in SV-HUC-1 cells demonstrated that LMNB2 promoted cell
proliferation and impaired cell apoptosis as observed by the
decrease in the expression of caspase-3 and caspase-8 (Fig. 4D-F). Therefore, these data
provided the evidence that LMNB2 depletion suppressed the
proliferation of bladder cancer cells via stimulating cell cycle
arrest and apoptosis.
LMNB2 promotes the tumor growth of
bladder cancer cells in mice
To further confirm the effects of LMNB2 on bladder
cancer progression, in vivo assays were performed. Nude mice
were subcutaneously injected with T24 cells stably-transfected with
control or LMNB2 shRNA plasmids. After 7 days, the tumor volume was
measured every 7 days. The representative images from each group
were captured and are presented in Fig. 5A. After 42 days, all the tumors
were isolated, and the growth curves were calculated and compared
between two groups. Consistent with the in vitro data, the
tumors from LMNB2-depleted cells were significantly smaller than
those in the control groups, according to the growth curves
(Fig. 5A). LMNB2 expression in
tumor tissues was then detected using western blotting. The results
revealed that the expression levels of LMNB2 in the LMNB2-depleted
tissues were significantly decreased compared with the control
(Fig. 5B). Therefore, these
results indicated that LMNB2 depletion suppressed tumor growth of
bladder cancer cells in mice.
LMNB2 contributes to bladder cancer
progression via promotion of CDCA3 expression
The molecular mechanisms underlying LMNB2 promotion
of bladder cancer progression were then investigated. Through
bioinformatics analysis, it was observed that LMNB2 expression was
positively correlated with the expression of CDCA3 in bladder
cancer tissues (Fig. 6A). As has
been reported, CDCA3 is widely involved in the regulation of
multiple types of cancers (21,22). In addition, through the GEPIA
database and IHC staining, it was determined that CDCA3 mRNA and
protein expression were upregulated in bladder cancer tissues
compared with normal and adjacent tissues, respectively (Fig. 6B and C).
The associations between CDCA3 expression and the
clinical features of these 107 bladder cancer patients were then
investigated. Similarly, it was revealed that CDCA3 expression was
also significantly associated with tumor stage (P=0.007) and
recurrence of disease (P=0.001) of bladder cancer patients
(Table II). It was therefore
theorized that LMNB2 promoted bladder cancer progression via the
promotion of CDCA3 expression.
Through RT-qPCR assays, it was determined that the
mRNA level of CDCA3 was increased in the LMNB2-overexpressed T24
cells, and decreased in the LMNB2-depleted T24 cells (Fig. 7A). Notably, the mRNA expression of
LMNB2 was not affected in the CDCA3-overexpressed or depleted T24
cells (Fig. 7B).
 | Figure 7LMNB2 promotes the proliferation of
bladder cancer cells via transcriptional activation of CDCA3. (A
and B) LMNB2 and CDCA3 expression levels in T24 cells upon the
transfection of the indicated plasmids were detected by RT-qPCR
assays. (C) Colony formation assays showed the proliferation
capacity of T24 cells upon the transfection of the indicated
plasmids. (D and E) MTT and CCK-8 assays showed the OD values at
570 and 490 nm wavelengths, respectively, of T24 cells with the
transfection of the indicated plasmids. (F) ChIP assays were
performed, and PCR amplification was used after the enrichment of
CDCA3 promoter fragment by the anti-IgG or anti-LMNB2 antibody in
T24 cells. (G) The luciferase activity of pGL3-Basic and pGL3-CDCA3
in T24 cells co-transfected with pcDNA3.1-LMNB2 or pcDNA3.1-vector
plasmids was measured by luciferase reporter assays. Results are
presented as the mean ± SEM. *P<0.05,
**P<0.01 and ***P<0.001. LMNB2, Lamin
B2; CDCA3, cell division cycle-associated protein 3; RT-qPCR,
reverse transcription-quantitative PCR; CCK-8, Cell Counting Kit-8;
sh, short hairpin; OE, overexpressed. |
To further clarify whether LMNB2 promoted bladder
cancer cell proliferation through the promotion of CDCA3
expression, rescue assays were performed. The colony formation
assays revealed that the cell proliferation caused by LMNB2
overexpression was markedly reversed by CDCA3 depletion (Fig. 7C). In addition, the cell
proliferation was confirmed by MTT and CCK-8 assays, revealing that
LMNB2 regulated cell proliferation through CDCA3 (Fig. 7D and E). These data confirmed that
LMNB2 promoted bladder cancer cell proliferation via CDCA3.
Through ChIP assays, it was revealed that the
antibody of LMNB2 could be specifically co-immunoprecipitated by
the promoter fragment of CDCA3 in T24 cells, whereas the IgG
antibody could not be co-immunoprecipitated, suggesting the
specific binding of LMNB2 with CDCA3 promoter (Fig. 7F). Furthermore, by performing
luciferase assays, the pGL-CDCA3 plasmids containing the promoter
region of CDCA3 were co-transfected with pEnter-vector or
pEnter-LMNB2 plasmids into T24 cells. It was determined that LMNB2
could promote the activation of CDCA3 promoter in T24 cells
(Fig. 7G). Therefore, it was
further revealed that LMNB2 promoted bladder cancer progression via
transcriptionally activating CDCA3.
LMNB2 cooperates with CDCA3 to promote
bladder cancer progression
The aforementioned data revealed that LMNB2 could
bind to CDCA3 promoter and transactivate CDCA3, and therefore
promoted bladder cancer progression. IHC assays were then
performed, and the expression levels of both LMNB2 and CDCA3 in 107
human bladder cancer tissues were detected and analyzed in
continuous slices of tumor tissues. Notably, it was revealed that
the expression of CDCA3 was markedly associated with the expression
levels of LMNB2 (P=0.003; Fig. 8
and Table III). It was
therefore concluded that LMNB2 promoted the progression of bladder
cancer through transcriptional activation of CDCA3. Collectively,
it was determined that LMNB2 promoted the progression of bladder
cancer through the promotion of CDCA3 expression.
 | Table IIIAssociations between LMNB2 and CDCA3
expression in 107 patients with bladder cancer. |
Table III
Associations between LMNB2 and CDCA3
expression in 107 patients with bladder cancer.
| Total no. of
patients (n=107) | LMNB2
| χ2 | P-value | Pearson | Spearman |
|---|
| Low (n=32) | High (n=75) |
|---|
| CDCA3 | | | 8.644 | 0.003 | 0.284 | 0.284 |
| Low (n=35) | 17 | 18 | | | | |
| High (n=72) | 15 | 57 | | | | |
Discussion
Early diagnosis and targeted therapy of bladder
cancer are very important to improve the prognosis of patients with
bladder cancer (23-25). The key of targeted therapy is to
elucidate the pathogenesis of bladder cancer and explore new
therapeutic targets (26). In the
present study, the high expression of LMNB2 in human bladder cancer
tissues was revealed. The expression of LMNB2 was associated with
the prognosis and clinical features of bladder cancer patients. Our
data further confirmed that LMNB2 affected the cell cycle and
apoptosis, and promoted the proliferation of bladder cancer cells
in vitro and in vivo via transcriptional activation
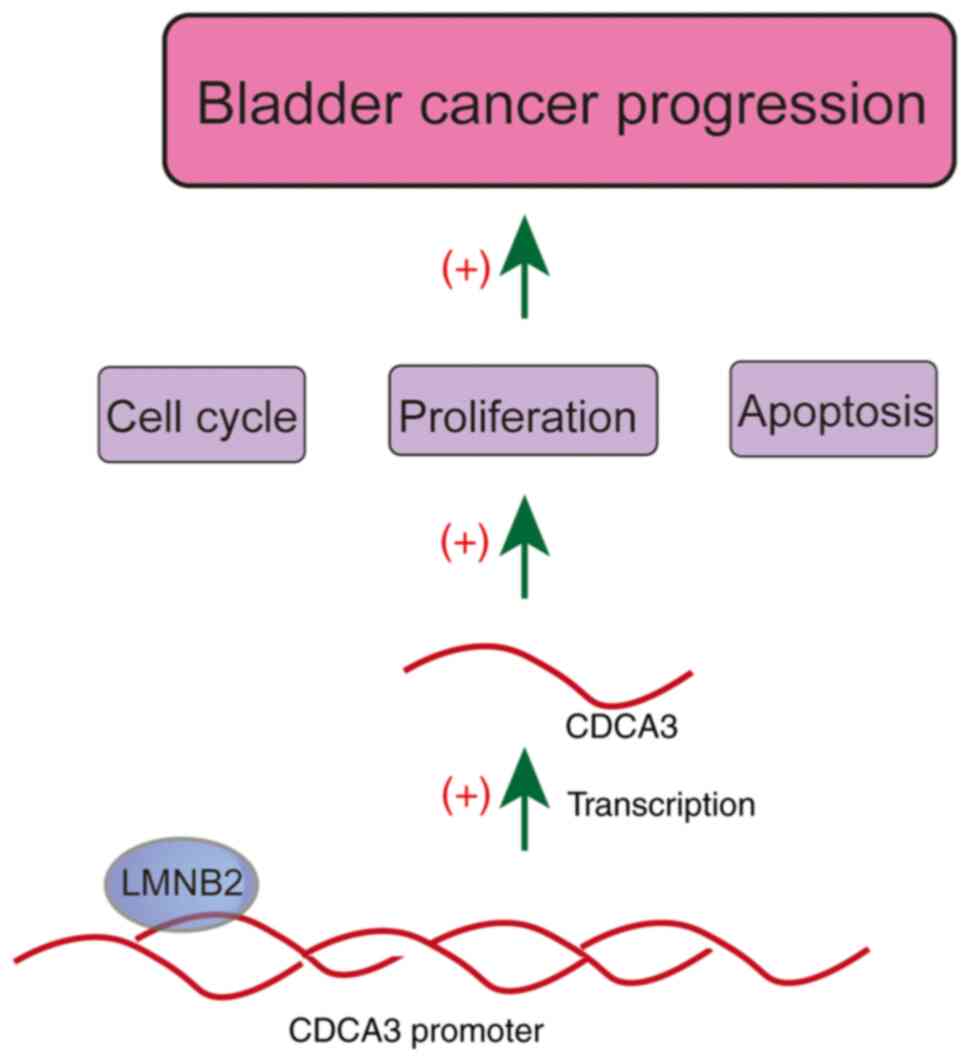
of CDCA3 (Fig. 9). The present
data therefore provided the evidence that LMNB2 could serve as a
promising molecular target for bladder cancer treatment.
As is known, LMNB2 is ubiquitously expressed in all
types of cells and developmental stages (27). The defects and mutations of LMNB2
have led to several types of diseases (28). A previous study indicated that the
mutations of LMNB2 led to acquired partial lipodystrophy (28). Another study provided evidence
that the mutation of LMNB2 stimulated progressive myoclonus
epilepsy with early ataxia (29).
These studies suggested the critical function of LMNB2 in
development. In the present study, the involvement of LMNB2 in the
progression of bladder cancer was demonstrated, however the precise
regulatory mechanism requires further study. LMNB2 has also been
shown to localize to mitochondria in axons, and therefore
suppressed axonal degeneration via maintaining mitochondrial
function and axonal integrity (30). In the future, whether LMNB2
regulated bladder cancer progression via regulating mitochondrial
function will be studied.
It has also been widely reported that LMNB2 affects
the progression and metastasis of multiple types of cancers
(15,16). LMNB2 was also aberrantly highly
expressed in several cancer tissues, including liver and lung
cancer (15,16). LMNB2 protein levels were
correlated with the prognosis and survival rates of patients with
lung cancer, similar to the results of our study (31). Additionally, LMNB2 also affected
the proliferation of lung cancer cells via binding MCM7 and
promoting MCM7 helicase activity (16). In the present study it was
determined that LMNB2 promoted the proliferation of bladder cancer
cells. It was also revealed that LMNB2 affected the cell cycle and
apoptosis of bladder cancer cells. The present data indicated that
LMNB2 affected the proliferation of bladder cancer cells via
regulation of CDCA3 expression. Therefore, it is suggested that
LMNB2 could act as a therapeutic target for bladder cancer
treatment.
CDCA3 is also known as a critical cancer regulator
in multiple types of cancers (21,22). It has been reported that CDCA3
affected tumor formation in pancreatic cancer, and promoted cell
proliferation in gastric and lung cancer (21,32,33). In colorectal cancer, CDCA3 could
mediate p21-dependent proliferation via promoting E2F1 expression
(22). In addition, in colorectal
cancer, CDCA3 promoted cell proliferation via activating the NF-κB
pathway (34). Notably, CDCA3 was
correlated with the TNM staging and prognosis of patients with
bladder cancer (35). Our
hypothesis that LMNB2 promoted bladder cancer progression via
regulation of CDCA3 expression is considered to be feasible.
Limitedly, the binding site of LMNB2 regulating CDCA3 mRNA
expression was not been revealed in the present study.
In summary, the aberrant expression of LMNB2 in
human bladder cancer tissues, and that its expression was
associated with the prognosis and clinical features of patients
with bladder cancer was revealed. LMNB2 depletion resulted in the
impairment of cell proliferation, the arrest of the cell cycle, and
the promotion of apoptosis in bladder cancer cells. It was further
confirmed that LMNB2 promoted the proliferation of bladder cancer
in vitro and in vivo via transcriptional activation
of CDCA3. It is therefore considered that LMNB2 could serve as a
promising therapeutic target for the treatment of bladder
cancer.
Availability of data and materials
The datasets supporting the conclusions of this
study are included within the manuscript.
Authors' contributions
JJ and WW conceived and designed the study, and
confirmed the authenticity of all the raw data. HL collected the
patient data. JJ and JC acquired the data. JC analyzed the data. JJ
wrote and reviewed the manuscript. WW supervised the study. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All applicable hospital guidelines for the care and
use of animals were followed. The use of animals was approved
(approval no. SYXK 2020-0127) by the Ethics Committee of the
College of Clinical Medicine of Henan University of Science and
Technology. All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
hospital research committee and with the 1964 Helsinki declaration
and its later amendments or comparable ethical standards. The
present study was approved (approval no. AMI-2020-073) by the
Ethics Committee of the College of Clinical Medicine of Henan
University of Science and Technology. Preoperative patients signed
informed consent for the use of postoperative specimens for
scientific research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang L, Liang J, Liu X, Wu J, Tan D and
Hu W: Aloperine exerts antitumor effects on bladder cancer in
vitro. Onco Targets Ther. 13:10351–10360. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jian Y, Xu Z, Xu C, Zhang L, Sun X, Yang D
and Wang S: The roles of glycans in bladder cancer. Front Oncol.
10:9572020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Hefnawy AS, Rizk E, Al Demerdash Khamis
NM, Barakat MAA, Khater SM and Shokeir AA: Urinary hyaluronic acid:
A versatile marker of bladder cancer. Int Urol Nephrol.
52:1691–1699. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dal Moro F, Valotto C, Guttilla A and
Zattoni F: Urinary markers in the everyday diagnosis of bladder
cancer. Urologia. 80:265–275. 2013. View Article : Google Scholar
|
|
6
|
Tao K, Liu S, Wang L, Qiu H, Li B, Zhang
M, Guo M, Liu H, Zhang X, Liu Y, et al: Targeted multifunctional
nanomaterials with MRI, chemotherapy and photothermal therapy for
the diagnosis and treatment of bladder cancer. Biomater Sci.
8:342–352. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Payumo AY and Huang GN: Lamin B2, guardian
of cardiomyocyte nuclear division. Dev Cell. 53:5–7. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ranade D, Koul S, Thompson J, Prasad KB
and Sengupta K: Chromosomal aneuploidies induced upon Lamin B2
depletion are mislocalized in the interphase nucleus. Chromosoma.
126:223–244. 2017. View Article : Google Scholar :
|
|
9
|
Kuga T, Nie H, Kazami T, Satoh M,
Matsushita K, Nomura F, Maeshima K, Nakayama Y, Tomonaga T and
Lamin B: 2 prevents chromosome instability by ensuring proper
mitotic chromosome segregation. Oncogenesis. 3:e942014. View Article : Google Scholar
|
|
10
|
Han L, Choudhury S, Mich-Basso JD,
Ammanamanchi N, Ganapathy B, Suresh S, Khaladkar M, Singh J, Maehr
R, Zuppo DA, et al: Lamin B2 levels regulate polyploidization of
cardiomyocyte nuclei and myocardial regeneration. Dev Cell.
53:42–59.e11. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Razafsky D, Ward C, Potter C, Zhu W, Xue
Y, Kefalov VJ, Fong LG, Young SG and Hodzic D: Lamin B1 and lamin
B2 are long-lived proteins with distinct functions in retinal
development. Mol Biol Cell. 27:1928–1937. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Parry DA, Martin CA, Greene P, Marsh JA;
Genomics England Research Consortium; Blyth M, Cox H, Donnelly D,
Greenhalgh L, Greville-Heygate S, et al: Heterozygous lamin B1 and
lamin B2 variants cause primary microcephaly and define a novel
laminopathy. Genet Med. 23:408–414. 2021. View Article : Google Scholar :
|
|
13
|
Galazis N, Olaleye O, Haoula Z, Layfield R
and Atiomo W: Proteomic biomarkers for ovarian cancer risk in women
with polycystic ovary syndrome: A systematic review and biomarker
database integration. Fertil Steril. 98:1590–1601.e1. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang MY, Han YC, Han Q, Liang Y, Luo Y,
Wei L, Yan T, Yang Y, Liu SL and Wang EH: Lamin B2 promotes the
malignant phenotype of non-small cell lung cancer cells by
upregulating dimethylation of histone 3 lysine 9. Exp Cell Res.
393:1120902020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kong W, Wu Z, Yang M, Zuo X, Yin G and
Chen W: LMNB2 is a prognostic biomarker and correlated with immune
infiltrates in hepatocellular carcinoma. IUBMB Life. 72:2672–2685.
2020. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma Y, Fei L, Zhang M, Zhang W, Liu X, Wang
C, Luo Y, Zhang H and Han Y: Lamin B2 binding to minichromosome
maintenance complex component 7 promotes non-small cell lung
carcinogenesis. Oncotarget. 8:104813–104830. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abdelbaqi K and Di Paola D: Ku protein
levels, localization and association to replication origins in
different stages of breast tumor progression. J Cancer. 4:358–370.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of clear cell renal cell
carcinoma. Nature. 499:43–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoadley KA, Yau C, Hinoue T, Wolf DM,
Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et
al: Laird, cell-of-origin patterns dominate the molecular
classification of 10,000 tumors from 33 types of cancer. Cell.
173:291–304.e6. 2018. View Article : Google Scholar
|
|
21
|
Zou RC, Guo ZT, Wei D, Shi ZT, Ye ZC, Zhai
G, Zhong C, Tang B, Wang L and Ge JY: Downregulation of CDCA3
expression inhibits tumor formation in pancreatic cancer.
Neoplasma. 67:1223–1232. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qian W, Zhang Z, Peng W, Li J, Gu Q, Ji D,
Wang Q, Zhang Y, Ji B, Wang S, et al: CDCA3 mediates p21-dependent
proliferation by regulating E2F1 expression in colorectal cancer.
Int J Oncol. 53:2021–2033. 2018.PubMed/NCBI
|
|
23
|
Nguyen EK, Yu H, Pond G, Shayegan B,
Pinthus JH, Kapoor A, Mukherjee SD, Neville A, Lalani AA, Hotte SJ,
et al: Outcomes of trimodality bladder-sparing therapy for
muscle-invasive bladder cancer. Can Urol Assoc J. 14:122–129.
2020.
|
|
24
|
Pearce S and Daneshmand S: Enhanced
endoscopy in bladder cancer. Curr Urol Rep. 19:842018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
De Keukeleire S, De Maeseneer D, Jacobs C
and Rottey S: Targeting FGFR in bladder cancer: Ready for clinical
practice? Acta Clin Belg. 75:49–56. 2020. View Article : Google Scholar
|
|
26
|
Vartolomei L, Ferro M, Mirone V, Shariat
SF and Vartolomei MD: Systematic review: Depression and anxiety
prevalence in bladder cancer patients. Bladder Cancer. 4:319–326.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Coffinier C, Fong LG and Young SG: LINCing
lamin B2 to neuronal migration: Growing evidence for cell-specific
roles of B-type lamins. Nucleus. 1:407–411. 2010. View Article : Google Scholar
|
|
28
|
Hegele RA, Cao H, Liu DM, Costain GA,
Charlton-Menys V, Rodger NW and Durrington PN: Sequencing of the
reannotated LMNB2 gene reveals novel mutations in patients with
acquired partial lipodystrophy. Am J Hum Genet. 79:383–389. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Damiano JA, Afawi Z, Bahlo M, Mauermann M,
Misk A, Arsov T, Oliver KL, Dahl HH, Shearer AE, Smith RJ, et al:
Mutation of the nuclear lamin gene LMNB2 in progressive myoclonus
epilepsy with early ataxia. Hum Mol Genet. 24:4483–4490. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Puzzi L, Marchetti L, Peverali FA,
Biamonti G and Giacca M: DNA-protein interaction dynamics at the
Lamin B2 replication origin. Cell Cycle. 14:64–73. 2015. View Article : Google Scholar :
|
|
31
|
Giusti L, Da Valle Y, Bonotti A, Donadio
E, Ciregia F, Ventroni T, Foddis R, Giannaccini G, Guglielmi G,
Cristaudo A and Lucacchini A: Comparative proteomic analysis of
malignant pleural mesothelioma evidences an altered expression of
nuclear lamin and filament-related proteins. Proteomics Clin Appl.
8:258–268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y, Yin W, Cao W, Chen P, Bian L and
Ni Q: CDCA3 is a potential prognostic marker that promotes cell
proliferation in gastric cancer. Oncol Rep. 41:2471–2481.
2019.PubMed/NCBI
|
|
33
|
Adams MN, Burgess JT, He Y, Gately K,
Snell C, Zhang SD, Hooper JD, Richard DJ and O'Byrne KJ: Expression
of CDCA3 is a prognostic biomarker and potential therapeutic target
in non-small cell lung cancer. J Thorac Oncol. 12:1071–1084. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang W, Lu Y and Li X, Zhang J, Zheng L,
Zhang W, Lin C, Lin W and Li X: CDCA3 promotes cell proliferation
by activating the NF-κB/cyclin D1 signaling pathway in colorectal
cancer. Biochem Biophys Res Commun. 500:196–203. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li S, Liu X, Liu T, Meng X, Yin X, Fang C,
Huang D, Cao Y, Weng H, Zeng X and Wang X: Identification of
biomarkers correlated with the TNM staging and overall survival of
patients with bladder cancer. Front Physiol. 8:9472017. View Article : Google Scholar : PubMed/NCBI
|