Introduction
Breast cancer is one of the most common malignant
tumors and the most frequent cause of cancer-related mortality
among females worldwide, severely endangering the life and health
of women (1,2). Even though notable improvements have
been made in recent years, 30% of patients with breast cancer are
suffer from relapse and metastasis, and patients with metastasis
have a 5-year survival rate of only 26%, thus leading to a great
burden for families and society (3,4).
Breast cancer still remains a major health concern and represents a
top biomedical research priority. Epidemiological research has
revealed various pathogenic factors that can result in the
development of breast cancer, including estrogen, alcohol
consumption, obesity and progestin use, as well as genetic
mutations (5,6). Recently, advances in genetic testing
for individuals who are at a high risk of developing cancer and in
targeted gene therapy for breast cancer are rapidly emerging,
contributing to a new era in cancer treatment (7). Thus, seeking for novel potential
target genes for providing effective therapeutic strategies for
breast cancer is of utmost importance.
Magnesium transporter protein 1 (MAGT1), an
evolutionally conserved Mg2+-specific ion transport
facilitator, possesses five predicted transmembrane regions with a
putative signaling sequence and a number of COOH-terminal
phosphorylation consensus sites (8,9).
It has been demonstrated that MAGT1 is a critical regulator of
intracellular free Mg2+ levels, and plays a crucial role
in coordinating natural killer (NK) and CD8+ T-cell
activation (10). Notably, recent
studies have indicated that MAGT1 is overexpressed in
hepatocellular carcinoma, glioma and colorectal cancer,
demonstrating that it is also associated with the overall survival
time and chemotherapeutic efficacy; thus, it may be a novel
therapeutic target for cancer treatment (9,11,12). However, the role of MAGT1 has not
yet been reported in breast cancer, at least to the best of our
knowledge.
The Kruppel-like factors (KLFs), a type of
transcription factors, are characterized by three zinc finger DNA
binding domains near their C terminus (13). KLFs can bind to GC-rich DNA
elements where they can regulate transcription promoter-dependently
(14). As a critical member of
the KLF family, KLF16 has been reported to coordinate various
biological processes including cell growth, death and metabolism,
and to participate in multiple diseases (15-17). KLF16 has been reported to affect
tumorigenesis and the development of malignant tumors, such as
prostate and gastric cancer (14,18). Notably, recent evidence has
indicated that KLF16 has critical oncogenic functions in breast
cancer, as KLF6 is involved in the proliferation, migration and
invasion of breast cancer cells (19).
The present study first investigated the role of
MAGT1 in breast cancer. Subsequently, the potential mechanisms of
action of MAGT1 and KLF16, particularly their associated roles in
the regulation of breast cancer progression were explored. The
findings presented herein provide a novel potential biomarker for
predicting the development and progression of breast cancer.
Materials and methods
Bioinformatics analysis
The expression of MAGT1 and KLF16 in breast cancer
and normal breast samples was retrieved from the Encyclopedia of
RNA Interactomes (ENCORI; http://starbase.sysu.edu.cn/index.php), including
1,104 tumor and 113 normal samples. The Human Protein Atlas (HPA;
https://www.proteinatlas.org/) was also
applied for the analysis of MAGT1 and KLF16 expression by
immunohistochemical assay. The binding between MAGT1 promoter and
KLF6 was predicted using the JASPAR database (https://jaspar.genereg.net/).
Cells, cell culture and transfection
The MCF-10A (cat no. CRL-10317), MCF-7 (cat no.
HTB-22), MDA-MB-231 (cat no. HTB-26) and SK-BR-3 (cat no. HTB-30)
cell lines were obtained from the American Type Culture Collection
(ATCC). The SUM190PT (cat no. YS1334C) cell line was obtained from
Shanghai Yaji Biological Technology Co., Ltd. All cells were
cultured in DMEM supplemented with 10% FBS (Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.), and
maintained at 37°C in a humidified incubator containing 5%
CO2.
For transfection, short hairpin RNAs (shRNA; pGPU6)
targeting KLF6 (shRNA-KLF6-1/2) and MAGT1 (shRNA-MAGT1-1/2) and an
empty pGPU6 plasmid which was used as the negative control
(shRNA-NC) were obtained from Shanghai GenePharma Co., Ltd. In
addition, the pcDNA3.1(+) KLF16 overexpression vector (Oe-KLF16)
and empty vector NC (Oe-NC) were supplied by Shanghai GenePharma
Co., Ltd. The full-length sequence of MAGT1 was amplified and
cloned into the pcDNA3.1 plasmid (Shanghai GenePharma Co., Ltd.) to
generate pcDNA-MAGT1, and the pcDNA3.1 empty plasmid was used as
its negative control (pcDNA-NC). When the cells reached 70-80%
confluency, they were transfected with the corresponding
aforementioned plasmids (50 nM) using Lipofectamine
3000® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions at 37°C for 48 h.
Following 48 h of transfection, subsequent experiments were
conducted.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the cells using the
RNeasy Mini kit (Qiagen, Inc.) and then reverse transcribed into
complementary DNA (cDNA) using the Strand cDNA Synthesis kit
(Takara Bio, Inc.) using the following thermocycling conditions:
70°C for 5 min, 42°C for 1 h and 70°C for 15 min. qPCR was
subsequently carried out using a PrimeScript RT-PCR kit (Qiagen,
Inc.) in accordance with the manufacturer's instructions. The
thermocycling conditions were as follows: Initial denaturation at
94°C for 5 min; followed by 22 cycles of denaturation at 94°C for
30 sec, annealing at 55°C for 30 sec and extension at 72°C for 30
sec. The mRNA levels were analyzed using the 2−ΔΔCq
method (20) and normalized to
the reference gene, GAPDH. The primer sequences used were as
follows: MAGT1 forward, 5′-CTC AGC CTC TGC CCA AAG AA-3′ and
reverse, 5′-CAC AAG GCG ACG GAA CTT GT-3′; KLF16 forward, 5′-CAA
GTC CTC GCA CCT AAA GTC-3′ and reverse, 5′-AGC GGG CGA ACT TCT TGT
C-3′; GAPDH forward, 5′-CCA TGG GGA AGG TGA AGG TC-3′ and reverse,
5′-AGT GAT GGC ATG GAC TGT GG-3′.
Western blot analysis
The MCF-7 cells and mouse tissues were collected and
lysed with RIPA buffer on ice for 30 min. After determining the
protein concentration using a BCA kit (Beyotime Institute of
Biotechnology), the same amount of protein (30 µg/lane) was
separated on a 12% SDS-PAGE gel and transferred to polyvinylidene
fluoride (PVDF) membranes (MilliporeSigma). The membranes were
blocked with 5% (w/v) skimmed milk in TBST for 1 h at room
temperature, followed by probing with primary antibodies against
MAGT1 (dilution 1:600; cat no. 27994-1-AP, ProteinTech Group,
Inc.), Ki67 (dilution 1:1,000; cat no. ab16667, Abcam),
proliferating cell nuclear antigen (PCNA; dilution 1:1,000; cat no.
ab18197, Abcam), MMP2 (dilution 1:1,000; cat no. ab92536, Abcam),
MMP9 (dilution 1:1,000; cat no. ab283575, Abcam), KLF16 (dilution
1:1,000; cat no. orb39548, Biorbyt, Ltd.) and GAPDH (dilution
1:2,500; cat no. ab9485, Abcam) at 4°C overnight, followed by
incubation with goat anti-rabbit horseradish peroxidase-labeled
secondary antibody (dilution 1:2,000; cat no. ab6721, Abcam) at
room temperature for 2 h. The bands were visualized by enhanced
chemiluminescence (ECL; Amersham Pharmacia). ImageJ software
(version 1.8.0; National Institutes of Health) was used for
densitometry.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell viability was assessed using MTT assay. The
transfected cells were seeded in 96-well plates at 37°C for
incubation of 24, 48 and 72 h, respectively. MTT solution (5 mg/ml;
MilliporeSigma) was added to each well to incubate the cells at
37°C for a further 4 h. Dimethyl sulfoxide (DMSO) was used to
dissolve the formazan. The absorbance at 570 nm was measured using
a multi-well scanning spectrophotometer (Bio-Rad Model 2550 EIA
Reader).
Cell colony formation assay
The MCF-7 cells were plated in six-well plates in
DMEM containing 10% FBS at 37°C in a humidified incubator
containing 5% CO2. The medium was refreshed every 3
days. After 2 weeks, the cells were washed with PBS, fixed with 4%
formaldehyde (Sigma-Aldrich; Merck KGaA) for 30 min at room
temperature and stained with crystal violet (Sigma-Aldrich; Merck
KGaA) for 30 min at room temperature for observation.
Wound healing assay
The MCF-7 cells were plated in six-well plates in
DMEM containing 10% FBS at 37°C in a humidified incubator
containing 5% CO2. After 24 h, a straight line was drawn
using a 200 µl pipette tip. The cells were then washed with
PBS three times and incubated in serum-free medium for 48 h. The
healing of the scratches was observed under a light microscope
(Olympus Corporation) at 0 and 24 h. The gap distance was
quantitatively evaluated using ImageJ software (1.8.0 172 version;
National Institutes of Health). Migration (%)=[(0 h average scratch
distance-24 h average scratch distance)/0 h average scratch
distance] ×100.
Transwell assay
A total of 1×105 MCF-7 cells was
suspended in 200 µl DMEM without FBS and inoculated into the
upper chamber of 24-well Transwell inserts (Corning Falcon;
Corning, Inc.) pre-coated with Matrigel (BD Biosciences).
Subsequently, 500 µl DMEM containing 10% FBS were added to
the lower chamber. The Transwell was incubated at 37°C in a
humidified incubator containing 5% CO2. After 24 h, the
non-invaded cells were wiped out using a cotton swab, and the cells
that had invaded to the bottom of the insert was fixed with 4%
formaldehyde for 10 min and stained with crystal violet for 10 min
at room temperature. The stained cells were observed under a light
microscope (Olympus Corporation).
Dual luciferase reporter gene assay
KLF16 binding motif and MAGT1 promoter (full length,
FL) or serial truncations (E1 Del and E2 Del) were cloned into the
pGL3-basic vector (E1761; Promega Corporation). A total of
1×105 MCF-7 cells were seeded in 24-well plates. On the
following day, the cells were transfected with the pGL3-based
reporter constructs (2 µg) and pRL-SV40 vector (2 µg)
using Lipofectamine 3000® (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the Firefly luciferase activity was
measured using the Dual-Luciferase Reporter Assay System (Promega
Corporation), and normalized to Renilla activity.
Chromatin immunoprecipitation (ChIP)
assay
The MCF-7 cells were cross-linked with 1%
formaldehyde at room temperature for 10 min and quenched in 125 mM
glycine for 5 min. The cell lysates were then sonicated into
fragments. Subsequently, 100 µl lysates containing chromatin
fragments were immunoprecipitated with antibody against KLF16 (5
µg; cat. no. sc-377519; Santa Cruz Biotechnology, Inc.) or
IgG (5 µg; cat. no. 2729; Cell Signaling Technology, Inc.)
at 4°C overnight. The immune complexes were recovered by the
addition of A/G-agarose beads (Santa Cruz Biotechnology, Inc.). The
precipitated DNA was purified using the ChIP DNA Clean &
Concentrator kits (Zymo Research) and then analyzed using RT-qPCR
as described above.
In vivo experiments
A total of 36 male BALB/c nude mice weighing 18-22 g
were obtained from HFK Bioscience Co, Ltd. and housed in a standard
environment with a controlled temperature (20±2°C), humidity
(50±5%), a 12/12-h light/dark cycle and free access to water and
food. The mice were allowed to acclimatize to their environment for
1 week prior to the experiments. All animal experiment procedures
were performed in compliance with the Guide for the Care and Use of
Laboratory Animals of The Second Clinical Medical College of North
Sichuan Medical College and were approved by the Ethics Committee
of The Second Clinical Medical College of North Sichuan Medical
College (Nanchong, China; approval no. NSMC-2021-94). Mice (6 mice
per group) were randomly divided into three groups in the first
section as follows: The control group, shRNA-NC group and
shRNA-MAGT1 group. In subsequent experiments, mice (6 mice per
group) were randomly divided into three groups as follows: The
control group, sh-KLF16 group and sh-KLF16s+pcDNA-MAGT1 group.
Approximately 1×107 transfected MCF-7 cells were
subcutaneously injected into the right axillary of the mice. After
the tumor was formed, the body weight and tumor size of the mice
were recorded every 3 days, and the tumor size was calculated using
the following equation: 1/2 × length × width2. At the
end of the 21st day, the 36 mice were euthanized by an
intraperitoneal injection of 120 mg/kg sodium pentobarbital, and
the tumors were removed after the heartbeat cessation and
respiratory arrest of the nude mice were confirmed. The mice were
monitored every day, and the humane endpoints were the following: A
marked reduction in food or water intake, labored breathing,
inability to stand and no response to external stimuli. No abnormal
signs that signified the humane endpoints of the experiment were
observed in any of the mice during the experiment. The tumors were
frozen at −80°C for further analysis.
Statistical analysis
The mean ± standard deviation (SD) for each
independent assay was used to present the experimental data. The
association of survival with MAGT1 expression was estimated using
the Kaplan-Meier analysis with the log-rank test. An unpaired
student's t-test or one-way ANOVA assay followed by Tukey's post
hoc test were applied for comparisons between two groups or among
more than two groups, respectively. A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
MAGT1 is upregulated in tumor samples of
breast cancer
Firstly, bioinformatics analysis was conducted to
examine the expression level of MAGT1 in breast cancer. As shown in
Fig. 1A, the analysis of the
ENCORI database (http://starbase.sysu.edu.cn/index.php) revealed a
relatively high expression of MAGT1 in breast cancer samples in
comparison to normal samples (P<0.01). The immunohistochemical
data obtained from the HPA database (https://www.proteinatlas.org/) further verified the
upregulated expression level of MAGT1 in breast tumor tissues
(Fig. 1B). Further analysis
revealed that there was no obvious association between MAGT1
expression and the tumor stage of breast cancer (Fig. 1C). The overall survival assay
revealed that the high expression of MAGT1 was significantly
associated with a poor survival time (Fig. 1D). These data suggested that MAGT1
expression was upregulated in breast cancer, and the upregulated
expression of MAGT1 predicted a poor outcome of patients.
MAGT1 knockdown suppresses the
proliferation, migration and invasion of MCF-7 cells
To explore the specific role of MAGT1 in breast
cancer, the MCF-7 cells were selected for use in further
experiments, as these cells exhibited a significantly high
expression of MAGT1 in comparison to the MCF-10A cells, and
exhibited the highest expression level of MAGT1 among several
breast cancer cell lines (MCF-7, MDA-MB-231, SK-BR-3 and SUM190PT
cells) (Fig. 2A). Due to the high
expression level of MAGT1 in breast cancer, the MCF-7 cells were
transfected with shRNA-MAGT1-1 or shRNA-MAGT1-2 to achieve MAGT1
knockdown. As shown in Fig. 2B and
C, transfection with both shRNA-MAGT1-1 and shRNA-MAGT1-2
resulted in the significantly decreased mRNA and protein expression
of MAGT1; however, as shRNA-MAGT1-2 decreased MAGT1 to a greater
degree, it was thus applied for use in subsequent experiments. A
series of cellular biological experiments were then carried out to
evaluate the effects of MAGT1 on the biological behaviors of breast
cancer cells. MTT and colony formation assays revealed that MAGT1
knockdown markedly restricted cell viability and the formation of
cell colonies, suggesting that the cell proliferative ability was
suppressed upon MAGT1 knockdown in MCF-7 cells. This was also
verified by the downregulated protein expression of Ki67 and PCNA
in the shRNA-MAGT1 group, compared to the shRNA-NC group (Fig. 2D-F). In addition, the decreased
wound healing and invasive abilities of the cells in the
shRNA-MAGT1 group, accompanied by the decreased protein expression
of MMP2 and MMP9, demonstrated that MAGT1 knockdown also restricted
the cell migratory and invasive abilities of the MCF-7 cells
(Fig. 2G-J).
MAGT1 knockdown inhibits tumor growth in
vivo
To further validate the aforementioned findings
in vitro, and in vivo experiment was conducted. Male
BALB/c nude mice were subcutaneously injected with MCF-7 cells
transfected with shRNA-NC or shRNA-MAGT1. As shown in Fig. 3A-C, the tumor size and tumor
weight were markedly decreased when MAGT1 was knocked down. In
particular, the mouse body weight and tumor size were monitored
every 3 days before the mice were sacrificed. No evident
differences in body weight were observed; however, the tumor volume
was significantly decreased upon MAGT1 knockdown (Fig. 3D and E). Furthermore, the protein
expression level of Ki67, PCNA, MMP2 and MMP9 exhibited a notable
decrease in the tumor tissues of mice injected with cells in which
was MAGT1 knocked down (Fig. 3F).
This finding was consistent with the in vitro findings.
KLF16 is upregulated in tumor tissues of
breast cancer and regulates MAGT1 through transcriptional
activation
Subsequently, to identify the role of KLF16 in
breast cancer, its expression in breast cancer was examined using
bioinformatics analysis. As exhibited in Fig. 4A and B, the expression of KLF16 in
tumor samples was higher than that in normal samples, according to
the ENCORI and HPA databases. Even though KLF16 expression was not
associated with tumor stage (Fig.
4C), these results also indicated a potential involvement of
KLF16 in breast cancer. Of note, it was predicted using the JASPAR
database that there were two potential KLF16 response elements (E1
and E2) binding to the MAGT1 promoter (Fig. 5A). To further ensure the
association between KLF16 and MAGT1, the MCF-7 cells were first
transfected with sh-KLF16-1/2 and sh-NC, respectively. The results
revealed that the expression level of KLF16 was markedly decreased
by transfection with sh-KLF16-1/2 (Fig. 5B and C). Due to a higher
transfection efficacy, sh-KLF16-1 was selected for use in further
experiments. KLF16 knockdown was then found to exert an inhibitory
effect on the expression level of MAGT1 (Fig. 5D). Subsequently, luciferase
reporter assay demonstrated that KLF16 knockdown reduced the
transcriptional activity of MAGT1 (Fig. 5E). To determine which responsive
elements were mainly responsible for the regulatory effects of
KLF16 on MAGT1, the MCF-7 cells were co-transfected with serial
truncations (E1 Del and E2 Del) of the MAGT1 promoter and
Oe-NC/Oe-KLF16. As shown in in Fig.
5F, the transcriptional activity of MAGT1 was strictly limited
in the E1 Del group upon KLF16 overexpression, indicating that E1
in the MAGT1 promoter was the main response element for this
binding association between KLF16 and the MAGT1 promoter. This
result was then verified by ChIP assay, as KLF16 was enriched at
the ZNF217 promoter within the E2 region (Fig. 5G).
The inhibitory effects of KLF16 on the
cell proliferation, migration and invasion, and tumor growth are
diminished by MAGT1 overexpression
As was expected, KLF16 was also highly expressed in
breast cancer cell lines, particularly in the MCF-7 cells (Fig. 6A). Subsequently, through gain- and
loss-of-function experiments, the effects of KLF16 and MAGT1 on
breast cancer progression were investigated. Firstly, transfection
with pcDNA-MAGT1 successfully overexpressed MAGT1 expression at the
mRNA and protein level in MCF-7 cells (Fig. 6B and C). The MCF-7 cells were then
transfected with shRNA-NC or shRNA-KLF16 alone, or co-transfected
with shRNA-KLF16 and pcDNA-NC/pcDNA-MAGT1. The mRNA and protein
expression level of MAGT1 was decreased by KLF16 knockdown,
followed by a restoration by a simultaneous transfection with
pcDNA-MAGT1 (Fig. 6D and E).
 | Figure 6The inhibitory effects of KLF16 on
the cell proliferation, migration and invasion are diminished by
MAGT1 overexpression. (A) The expression level of KLF16 in multiple
breast cancer cell lines (MCF-7, MDA-MB-231, SK-BR-3, and SUM190PT
cells) and MCF-10A cells was determined using RT-qPCR.
***P<0.001 vs. MCF-10A cells. MCF-7 cells were
transfected with pcDNA-NC or pcDNA-MAGT1, and the expression level
of MAGT1 was measured using (B) RT-qPCR and (C) western blot
analysis. MCF-7 cells were transfected with shRNA-NC or shRNA-KLF16
alone, or co-transfected with shRNA-KLF16 and pcDNA-NC/pcDNA-MAGT1.
The expression level of MAGT1 was measured using (D) RT-qPCR and
(E) western blot analysis. (F) Cell viability was then evaluated
using MTT assay. (G) Cell colony formation assay was conducted. (H)
Protein expression of Ki67 and PCNA was detected using western blot
analysis. (I) Wound healing and Transwell assays were performed to
examine cell migration and invasion, respectively. (J)
Quantification of cell migration rate. (K) Quantification of cell
invasion rate. (L) Protein expression of MMP2 and MMP9 was
determined using western blot analysis. All experiments were
performed in triplicate. **P<0.01 and
***P<0.001 vs. shRNA-NC; #P<0.05,
##P<0.01 and ###P<0.001 vs. sh-KLF16 +
pcDNA-NC. Kruppel-like factor 16; MAGT1, magnesium transporter
protein 1; RT-qPCR, reverse transcription-quantitative PCR; PCNA,
proliferation cell nuclear antigen. |
In addition, a series of cellular biological
behaviors were assessed using MTT, colony formation, wound healing
and Transwell assays, as aforementioned. The results revealed that
KLF16 knockdown markedly reduced cell viability and cell colonies,
and also decreased the protein expression of Ki67 and PCNA,
suggesting that KLF16 knockdown suppressed MCF-7 cell proliferation
(Fig. 6F-H). In addition, the
suppressive effects of KLF16 knockdown on cell migration and
invasion were also evidenced by the hindered wound healing and
decreased number of invasive cells, accompanied by the
downregulated protein expression of MMP2 and MMP9 (Fig. 6I-L). Nevertheless, simultaneous
transfection with pcDNA-MAGT1 and sh-KLF16 partly abolished these
suppressive effects of KLF16 knockdown on cell proliferation,
migration and invasion compared with transfection with sh-KLF16
alone (Fig. 6F-L). Eventually,
these in vitro findings were also verified in vivo.
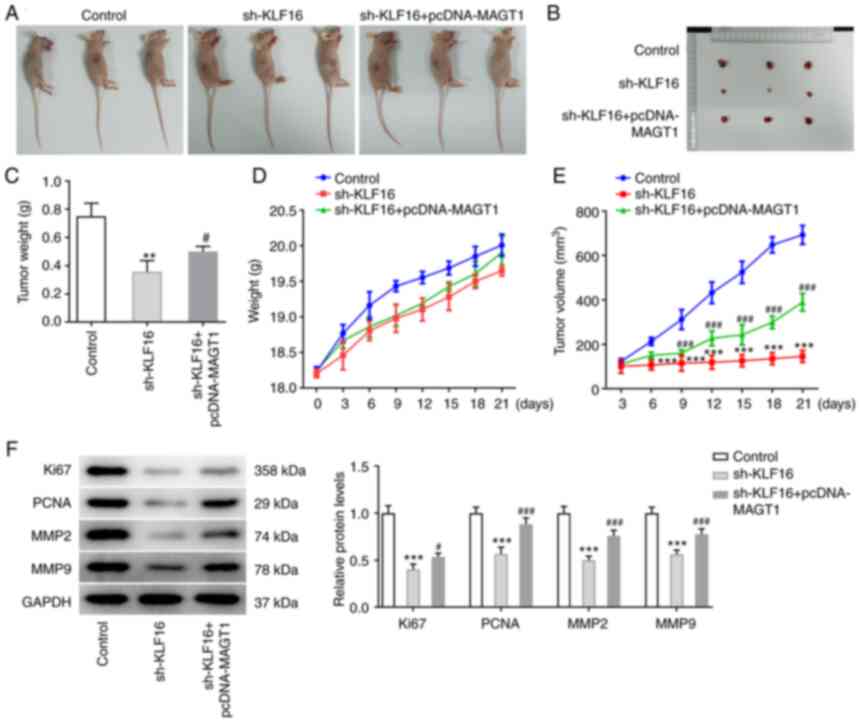
Male BALB/c nude mice were subcutaneously injected with MCF-7 cells
transfected with shKLF16 or co-transfected with sh-KLF16 and
pcDNA-MAGT1. As illustrated in Fig.
7A-C, the reduced tumor size and tumor weight induced by KLF16
knockdown were partly reversed by MAGT1 overexpression. The mouse
body weight continued to increase with time prolonging before,
without notable differences among the groups (Fig. 7D). The speed of tumor growth
during this period was hindered by KLF16 knockdown, which was
partly abolished by MAGT1 overexpression (Fig. 7E). Furthermore, the reduced
protein expression of Ki67, PCNA, MMP2 and MMP9 by KLF16 knockdown
was markedly elevated by MAGT1 overexpression (Fig. 7F).
Discussion
Breast cancer has continued to be the leading cause
of cancer-related mortality among females worldwide for years
(21). Despite the fact that the
current therapeutic management of breast cancer can control primary
tumor growth, the high invasiveness of breast cancer cells
predisposes the tumor to metastasis, resulting in relapses and
deterioration (22). Thus, it is
necessary to expand the knowledge of the pathogenesis of breast
cancer, and to identify strategies with which to prevent or
attenuate the metastasis of breast cancer.
The mechanisms of invasion and metastasis of breast
cancer cells are complex and involve the abnormal expression of
various genes (23). It has been
reported that gremlin-1 (GREM1) expression is significantly higher
in breast carcinoma tissues than that in corresponding normal
tissues. GREM1 contributes to the proliferation, migration and
invasion of breast cancer cells (24). Additionally, Aldo-keto reductase
family 1, member B10 has also been found to be overexpressed in
breast cancer tissues, which was then demonstrated to promote
breast cancer cell migration and invasion (25). Thus, targeting an effective gene
functioning on cell proliferation, migration and invasion is an
alternative option to develop therapies for breast cancer. MAGT1 is
a chromosome X-linked gene encoding a highly selective
Mg2+ transporter, and its critical role in temporally
coordinating NK and CD8+ T-cell activation has been
widely recognized (10,26). Recently, the importance of MAGT1
in tumor progression has been focused on by scholars and
illustrated in multiple studies. For instance, Li et al
(27) disclosed that MAGT1
functioned as a crucial targeted gene for miR-628-5p, which was
responsible for sevoflurane-mediated glioma progression. Bi et
al (28) demonstrated that
MAGT1 was indispensable for cervical cancer cell proliferation and
cell cycle progression by modulating the ERK/p38 MAPK signaling
pathway. In addition, the overexpression of MAGT1 has been linked
to tumor metastasis and anticancer drug resistance in colorectal
cancer (12). Nevertheless, the
role of MAGT1 in breast cancer has not been addressed to date, at
least to the best of our knowledge. The present study was the first
time to demonstrate that MAGT1 was abnormally upregulated in tissue
samples of breast cancer patients and breast cancer cell lines. The
oncogenic activity of MAGT1, as evidenced by the restricted cell
proliferative, migratory and invasive abilities of MCF-7 cells upon
MAGT1 knockdown was first demonstrated in breast cancer. Moreover,
MAGT1 knockdown attenuated tumor growth in vivo, further
verifying the oncogenic role of MAGT1. These novel findings
manifest that targeting MAGT1 may be a promising strategy for the
treatment of breast cancer.
Transcription factors drive cell fate transitions by
determining global transcriptional, epigenetic and topological
alterations (29). Indeed,
transcription factors were not originally considered ideal targets
for drug development; however, the advanced understanding of these
transcription factors, in terms of the their structure, interaction
with proteins and the dynamic mode of binding to DNA, provide
immense potential for novel therapeutic strategies targeted against
transcription factors (30). At
present, numerous transcription factors have been reported to be
associated with multiple tumor biomarkers, such as NF-κB, p53,
forkhead box O and others (31-33). The KLF family (KLF1-KLF17), a type
of zinc finger-containing transcription factor, has been found to
play a crucial role in tumorigenesis and development by modulating
cancer-promoting or cancer-suppressive genes via binding to the
GC-rich DNA sequence in primer regions of these genes (34,35). A previous study indicated that
KLF16 could transcriptionally repress the expression of
mitochondrial transcription factor A, which plays an oncogene role
in cancer, by interacting with its promoter, thereby suppressing
human glioma cell proliferation and tumorigenicity (36). Consistently, a potential binding
association between transcription factor KLF16 and the MAGT1
promoter was also found through the JASPAR database (https://jaspar.genereg.net/) in the present study, and
this connection was subsequently verified by luciferase reporter
and ChIP assays. In addition, the dysregulation of KLF16 in breast
cancer positively influenced MAGT1 expression. Further in
vitro and in vivo experiments not only revealed the
oncogenic role of KLF16 in breast cancer due to the suppressive
effects on cell proliferation, migration and invasion, and tumor
growth upon KLF16 knockdown, but also revealed a rescue of its
antitumor activity by simultaneous transfection with pcDNA-MAGT1,
suggesting that the anti-cancer effects of KLF16 knockdown may be
dependent on its inhibitory effect on MAGT1 expression.
In conclusion, the present study demonstrated that
MAGT1 and KLF16 were upregulated in tumor tissues of breast cancer
patients, as demonstrated from bioinformatics data and in breast
cancer cell lines. The knockdown of MAGT1 or KLF16 hindered the
development of breast cancer via restricting cell proliferation,
migration and invasion, and tumor growth. Mechanistically, KLF16
could directly bind to the MAGT1 promoter and transcriptionally
activated MAGT1 expression, thus regulating the oncogenic role of
MAGT1 and influencing the progression of breast cancer. The present
study provides a novel target for the treatment of breast cancer
and discloses its potential regulatory mechanisms.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XiaofenZ designed the study. LL, XiZ, YL, BX, SP and
HJ conducted the experiments and collected the data. LL, XiZ, YL
and BX analyzed and interpreted the data. LL and XiZ wrote the
manuscript. XiaofenZ revised the manuscript. All authors have read
and approved the final manuscript. XiaofenZ, LL and XiZ confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
All animal experiment procedures were performed in
compliance with the Guide for the Care and Use of Laboratory
Animals of The Second Clinical Medical College of North Sichuan
Medical College and were approved by the Ethics Committee of The
Second Clinical Medical College of North Sichuan Medical College
(Nanchong, China; approval no. NSMC-2021-94).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Science and Technology
Research Special Project of Sichuan Administration of Traditional
Chinese Medicine (grant no. 2020JC0082).
References
|
1
|
Khan F, Rahman A and Carrier M: Occult
cancer detection in venous thromboembolism: The past, the present,
and the future. Res Pract Thromb Haemost. 1:9–13. 2017. View Article : Google Scholar
|
|
2
|
Davari M, Amani B, Mokarian F, Hoseini M,
Akbarzadeh A and Heidarzadeh Khoramabadi N: Effectiveness of
trastuzumab as adjuvant therapy in patients with early stage breast
cancer: A systematic review and meta-analysis. Med J Islam Repub
Iran. 31:882017. View Article : Google Scholar
|
|
3
|
Peart O: Metastatic breast cancer. Radiol
Technol. 88:519M–539M. 2017.PubMed/NCBI
|
|
4
|
Tian T, Wang M, Lin S, Guo Y, Dai Z, Liu
K, Yang P, Dai C, Zhu Y, Zheng Y, et al: The impact of lncRNA
dysregulation on clinicopathology and survival of breast cancer: A
systematic review and meta-analysis. Mol Ther Nucleic Acids.
12:359–369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shamsi M and Pirayesh Islamian J: Breast
cancer: Early diagnosis and effective treatment by drug delivery
tracing. Nucl Med Rev Cent East Eur. 20:45–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hammerl D, Smid M, Timmermans AM, Sleijfer
S, Martens JWM and Debets R: Breast cancer genomics and
immuno-oncological markers to guide immune therapies. Semin Cancer
Biol. 52:178–188. 2018. View Article : Google Scholar
|
|
7
|
Walker-Smith TL and Peck J: Genetic and
genomic advances in breast cancer diagnosis and treatment. Nurs
Womens Health. 23:518–525. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng JM, Kong YY, Li YY and Zhang W:
MagT1 regulated the odontogenic differentiation of BMMSCs induced
byTGC-CM via ERK signaling pathway. Stem Cell Res Ther. 10:482019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goytain A and Quamme GA: Identification
and characterization of a novel mammalian Mg2+
transporter with channel-like properties. BMC Genomics. 6:482005.
View Article : Google Scholar
|
|
10
|
Chaigne-Delalande B, Li FY, O'Connor GM,
Lukacs MJ, Jiang P, Zheng L, Shatzer A, Biancalana M, Pittaluga S,
Matthews HF, et al: Mg2+ regulates cytotoxic functions
of NK and CD8 T cells in chronic EBV infection through NKG2D.
Science. 341:186–191. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang G, Li Y, Li J, Zhang D, Luo C, Zhang
B and Sun X: microRNA-199a-5p suppresses glioma progression by
inhibiting MAGT1. J Cell Biochem. 120:15248–15254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng K, Yang Q, Xie L, Qiu Z, Huang Y,
Lin Y, Tu L and Cui C: Overexpression of MAGT1 is associated with
aggressiveness and poor prognosis of colorectal cancer. Oncol Lett.
18:3857–3862. 2019.PubMed/NCBI
|
|
13
|
Philipsen S and Suske G: A tale of three
fingers: The family of mammalian Sp/XKLF transcription factors.
Nucleic Acids Res. 27:2991–3000. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaczynski J, Cook T and Urrutia R: Sp1-
and Kruppel-like transcription factors. Genome Biol. 4:2062003.
View Article : Google Scholar
|
|
15
|
Sun N, Shen C, Zhang L, Wu X, Yu Y, Yang
X, Yang C, Zhong C, Gao Z, Miao W, et al: Hepatic Krüppel-like
factor 16 (KLF16) targets PPARα to improve steatohepatitis and
insulin resistance. Gut. 70:2183–2195. 2021. View Article : Google Scholar
|
|
16
|
Yang X, Chen Q, Sun L, Zhang H, Yao L, Cui
X, Gao Y, Fang F and Chang Y: KLF10 transcription factor regulates
hepatic glucose metabolism in mice. Diabetologia. 60:2443–2452.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui A, Fan H, Zhang Y, Zhang Y, Niu D, Liu
S, Liu Q, Ma W, Shen Z, Shen L, et al: Dexamethasone-induced
Krüppel-like factor 9 expression promotes hepatic gluconeogenesis
and hyperglycemia. J Clin Invest. 129:2266–2278. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma P, Sun CQ, Wang YF, Pan YT, Chen QN,
Liu WT, Liu J, Zhao CH, Shu YQ and Li W: KLF16 promotes
proliferation in gastric cancer cells via regulating p21 and CDK4.
Am J Transl Res. 9:3027–3036. 2017.PubMed/NCBI
|
|
19
|
Bang S, Li J, Zhang M, Cui R, Wu X, Xin Z,
Ma D, Zhang J and Zhang H: The clinical relevance and function of
Krüppel-like factor 16 in breast cancer. Cancer Manag Res.
12:6373–6383. 2020. View Article : Google Scholar :
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar
|
|
22
|
Kozlowski J, Kozłowska A and Kocki J:
Breast cancer metastasis-insight into selected molecular mechanisms
of the phenomenon. Postepy Hig Med Dosw (Online). 69:447–451. 2015.
View Article : Google Scholar
|
|
23
|
Osborne C, Wilson P and Tripathy D:
Oncogenes and tumor suppressor genes in breast cancer: Potential
diagnostic and therapeutic applications. Oncologist. 9:361–377.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sung NJ, Kim NH, Surh YJ and Park SA:
Gremlin-1 promotes metastasis of breast cancer cells by activating
STAT3-MMP13 signaling pathway. Int J Mol Sci. 21:92272020.
View Article : Google Scholar :
|
|
25
|
Li J, Guo Y, Duan L, Hu X, Zhang X, Hu J,
Huang L, He R, Hu Z, Luo W, et al: AKR1B10 promotes breast cancer
cell migration and invasion via activation of ERK signaling.
Oncotarget. 8:33694–33703. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Baaij JH, Hoenderop JG and Bindels RJ:
Magnesium in man: Implications for health and disease. Physiol Rev.
95:1–46. 2015. View Article : Google Scholar
|
|
27
|
Li H, Xia T, Guan Y and Yu Y: Sevoflurane
regulates glioma progression by Circ_0002755/miR-628-5p/MAGT1 axis.
Cancer Manag Res. 12:5085–5098. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bi C, Zhang X, Chen Y, Dong Y, Shi Y, Lei
Y, Lv D, Cao X, Li W and Shi H: MAGT1 is required for HeLa cell
proliferation through regulating p21 expression, S-phase progress
and ERK/p38 MAPK MYC axis. Cell Cycle. 20:2233–2247. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Di Giammartino DC: Kloetgen A, Polyzos A,
Liu Y, Kim D, Murphy D, Abuhashem A, Cavaliere P, Aronson B, Shah
V, et al KLF4 is involved in the organization and regulation of
pluripotency-associated three-dimensional enhancer networks. Nat
Cell Biol. 21:1179–1190. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lambert M, Jambon S, Depauw S and
David-Cordonnier MH: Targeting transcription factors for cancer
treatment. Molecules. 23:14792018. View Article : Google Scholar :
|
|
31
|
Yadav RK, Chauhan AS, Zhuang L and Gan B:
FoxO transcription factors in cancer metabolism. Semin Cancer Biol.
50:65–76. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yeung SJ, Pan J and Lee MH: Roles of p53,
MYC and HIF-1 in regulating glycolysis-the seventh hallmark of
cancer. Cell Mol Life Sci. 65:3981–3999. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Viatour P, Merville MP, Bours V and
Chariot A: Phosphorylation of NF-kappaB and IkappaB proteins:
Implications in cancer and inflammation. Trends Biochem Sci.
30:43–52. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shields JM and Yang VW: Identification of
the DNA sequence that interacts with the gut-enriched Krüppel-like
factor. Nucleic Acids Res. 26:796–802. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miller IJ and Bieker JJ: A novel,
erythroid cell-specific murine transcription factor that binds to
the CACCC element and is related to the Krüppel family of nuclear
proteins. Mol Cell Biol. 13:2776–2786. 1993.PubMed/NCBI
|
|
36
|
Chen X, Li S, Ke Y, Wu S, Huang T, Hu W,
Fu H and Guo X: KLF16 suppresses human glioma cell proliferation
and tumourigenicity by targeting TFAM. Artif Cells Nanomed
Biotechnol. 46(Suppl 1): S608–S615. 2018. View Article : Google Scholar
|