Introduction
Cancer is the most challenging disease worldwide due
to tumor heterogeneity among patients which makes difficult the
diagnostic and prognostic procedures, as well as the therapeutic
approaches (1). In particular,
the lack of therapeutic response or the onset of drug resistance is
widely associated with cancer heterogeneity, which often occurs
among cancer cells within the same tumor bulk (2,3).
Several studies have suggested that epigenetic aberrations along
with somatic mutations are involved in the early stage of tumor
development and cancer variability (4,5).
Among the epigenetic mechanisms, DNA methylation (methDNA) is
subjected to profound changes during the tumorigenesis and cancer
progression leading researchers to define specific cancer-related
epigenetic signatures (6).
Specifically, the methDNA consists of the addition of a methyl
group at the carbon-5 position of the cytosine within the CpG
dinucleotides of the DNA sequence forming a 5-methylcytosine (5mC).
Gene expression may be profoundly affected by 5mCs when they
localize within key regulatory elements, including transcriptional
factors (TFs), enhancers and silencer consensus sites.
Additionally, methDNA may drive chromatin remodeling to facilitate
or inhibit the access of TFs to gene locus (7,8).
Notably, the methDNA patterns are maintained through cancer cell
generations representing potential tumor epigenetic hallmarks,
which could be used as putative diagnostic or prognostic biomarkers
(9). Over the years, several
molecular approaches have been developed to evaluate the methDNA
status of cancer-related genes, including whole genome methylation
analysis through omics technologies (10,11). Currently, bisulfite conversion
represents the gold standard pre-processing method for the deep
analysis of methDNA status. The procedure is based on the
conversion of the unmethylated cytosine in uracil under sodium
bisulfite treatment, whereas 5mCs remain unchanged. Subsequently,
the amount of the converted cytosines is assessed by
methyl-sensitive PCR or DNA sequencing (for example, Next
Generation Sequencing) reflecting the CpGs methylation status.
However, the degradative processes mediated by bisulfite conversion
result in a consistent fragmentation of DNA affecting the
downstream analyses, especially for low-quality and/or -quantity
DNA samples (12-15). In this context, the
Methylation-Sensitive Restriction Enzymes (MSRE) method could be
valuable to overcome this limitation since the quality of DNA is
unaffected during MSRE steps (16). The MSRE assay is based on the use
of MSREs (such as HpaII, AatII and ClaI),
which can recognize and digest specific sequences depending on the
methylation status of the cytosine of CpG dinucleotides within the
restriction sites. In particular, the methylation of these
restriction sites inhibits the MSRE endonuclease activity on
methDNA targets, whereas the unmethylation results in their
complete digestion. The cleavage rate of methDNA target is assessed
by PCR whose amplification signal reflects the methylation status
of the target at the restriction site (17,18). Furthermore, the MSRE enzymatic
efficiency is evaluated by isoschizomers that are not
methyl-sensitive allowing the complete cleavage of methDNA targets
in the absence of inhibitors. Besides the presence of digestion
inhibitors, the diluted samples may be unsuitable for the MSRE
pre-processing due to the large quantity of the MSRE digestion mix
that should be loaded to reach the sensitive threshold of the
following PCR-based analyses (i.e. Real-Time PCR). As a
consequence, the amplification efficiency may be affected by the
presence of the MSRE buffer as well as the restriction enzymes
(19,20).
To overcome some limitations of the standard MSRE
and bisulfite conversion for the pre-processing of low-quality
and/or -quantity DNA samples, several studies have proposed novel
approaches based on the combination of MSRE and the high
sensitivity droplet digital PCR (ddPCR) (21-24). Notably, the ddPCR is a
high-throughput technology for the absolute quantification of low
copies of DNA or RNA targets derived from non-canonical biological
specimens, including cell-free DNA (cfDNA) (25-29). In the present study, the
MSRE-ddPCR assay was proposed as a one-tube method for the methDNA
analysis using a methylation control (spike-in template) for the
evaluation of the assay efficiency and data normalization. The
advantage of the proposed MSRE-ddPCR method is represented by the
capability to perform the MSRE reaction directly in the ddPCR mix
before amplification in a one-step protocol. Furthermore, the
introduction of a methylation control allows to avoid the use of a
restriction enzyme unaffected by methylation (e.g. MspI)
used in standard MSRE assays to test the digestion efficiency. This
method is suitable for the analysis of poor quality and/or low
concentrate DNA (up to 0.625 ng) samples obtained from different
biological matrices, including serum and formalin-fixed,
paraffin-embedded (FFPE) tissues. As a proof of concept, the
MSRE-ddPCR assay needs further validation studies on different
biological matrices before introducing this method in a routine
setting.
Materials and methods
Cell cultures and melanoma patient
specimens
The A375 (cat. no. CRL-1619) and A2058 (CRL-3601)
melanoma cell lines were obtained from the American Type Culture
Collection, whereas the SK-MEL-23 melanoma cell line was already
available at the National Cancer Institute Pascale of Naples. A375,
A2058 and SK-MEL-23 cells were cultured in a complete RPMI-1640
medium supplemented with 10% fetal bovine serum (FBS), 2 mmol/l of
L-glutamine, 100 UI of penicillin and 100 μg/ml streptomycin
(all provided from Gibco; Thermo Fisher Scientific, Inc.). Each
cell line was seeded in 100-mm cell-culture dishes (Qiagen GmbH) at
the density of 1×106 cells and cultured in a humidified
incubator (5% CO2) at 37°C for 48 h. Cell pellets were
collected by scraping cell cultures in cold PBS 1X (Gibco; Thermo
Fisher Scientific Inc.) and frozen at -80°C until analyses.
Consecutive cohorts of 10 FFPE melanoma tissues (Age range: 35-75
years) and 10 FFPE nevi without atypical histological features (Age
range: 18-55 years), as well as two serum samples from two melanoma
patients that also provided FFPE tissues, were obtained from the
National Cancer Institute 'Fondazione G. Pascale', Naples (Italy),
using standard procedures. The FFPE tissues and serum samples were
collected from January 2019 to May 2020 at the Melanoma Cancer
Immunotherapy and Innovative Therapy Unit of the National Cancer
Institute 'Fondazione 'G. Pascale' (Naples, Italy). The
histopathological features of melanoma samples and sociodemographic
characteristics of patients and healthy controls are reported in
Table SI. The present study was
conducted in accordance with the guidelines of the Declaration of
Helsinki (seventh revision, 2013) and approved by the Institutional
Review Board of the National Cancer Institute 'Fondazione 'G.
Pascale' (Naples, Italy) (approval no. 33/17oss, approved on 10
January 2018). Informed consent was obtained from all subjects
involved in the study. All experiments were conducted in
duplicate.
DNA extraction
Genomic DNA from A375, A2058 and SK-MEL-23 melanoma
cell lines was extracted by using the PureLink Genomic DNA Mini Kit
(cat. no. K1820-01; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The cfDNA from serum samples was
extracted according to a custom protocol, as previously described
(30). Briefly, 1 ml of serum
was treated with 240 μl of extraction solution (EDTA (250
mmol/l)/NaCl (750 mmol/l): 100 μl; sodium dodecyl sulfate
(100 g/l): 100 μl; proteinase K (stock solution 20 mg/ml):
40 μl) and incubated at 56°C for 2 h. Subsequently, 200
μl of saturated 6M NaCl was added to the mixture to
precipitate the proteins. The collected supernatant was mixed 1:1
with phenol-chloroform and incubated at RT for 5 min. The DNA
solution was treated with an equal volume of absolute ethanol at
−20°C overnight and centrifuged at 14,000 x g for 15 min at 4°C.
The DNA pellet was washed with 70% ethanol and finally resuspended
in 20 μl of RNAse/DNase-free water. Since the cfDNA amount
was undetectable by Nanodrop-1000 (Thermo Fisher Scientific, Inc.),
a fixed volume of 5 μl of cfDNA sample was used for
downstream analyses. Genomic DNA from FFPE tissues (four sections
with a thickness of 8 μm) was extracted using the QIAamp DNA
FFPE Tissue kit (cat. no. 56404; Qiagen GmbH) and the
deparaffinization solution (cat. no. 19093; Qiagen GmbH) according
to the manufacturer's protocols. Nanodrop-1000 was used to assess
the amount and quality of extracted DNA by evaluating the 260/280
nm ratio (~1.8 for pure DNA samples).
DNA bisulfite conversion and Sanger
sequencing
To perform bisulfite sequencing of SLC22A17
methDNA hotspot (chr14:23,821,229-23,821,230-Assembly: GRCh37/hg19)
(Fig. S1A), 1.2 μg of
genomic DNA from A2058, A375 and SK-MEL-23 cells were
bisulfite-converted by using the EpiTect Plus DNA Bisulfite kit
(cat. no. 59124; Qiagen GmbH) according to the manufacturer's
protocol. The amplification of bisulfite-converted SLC22A17
target (chr14:23,821,176-23,821,349-Assembly: GRCh37/hg19)
(Fig. S1A), whose unconverted
sequence contains 15 CpG dinucleotides and 1 CCGG restriction site,
was conducted preparing a reaction mix (20 μl) containing
100 ng of the bisulfite-converted DNA, 10 μl of the 2X ddPCR
Supermix for Probes (No dUTP) (cat. no. 1863024; Bio-Rad
Laboratories, Inc.), 10 μM (final concentration) of forward
and reverse primers. The Bisulfite Primer Seeker (https://www.zymore-search.eu/pages/bisulfite-primer-seeker-accessed
on 7th March 2022) was used to design the bisulfite primers. PCR
thermal conditions and primer sequences are reported in Table I. PCR product was cleanup using
the PureLink PCR Purification kit (cat. no. K310001; Thermo Fisher
Scientific, Inc.) and sequenced with the Mix2Seq kit (Eurofins
Genomics Germany GmbH) according to the manufacturer's protocols.
The analysis of DNA sequences was performed by Chromas Lite
software version 2.6.6 (https://technelysium.com.au/wp/chromas/) (accessed on
10th June 2022).
 | Table IPrimers and amplification
conditions. |
Table I
Primers and amplification
conditions.
| Name | Sequence
(5'-3') | Amplification
condition |
|---|
| SLC22A17
amplification from bisulfite-converted DNA |
|
| SLC22A17
Prom2 | F:
GTGAGTATAGGAAGGTTATTATAGTTTT
R: TAACTAAAAACAACCTCCCAATAC | 95°C for 10 min,
followed by 40 cycles of 94°C for 30 sec, 55°C for 1 min, and
finally 98°C for 10 min |
|
| Methylated and
unmethylated controls |
|
| SLC22A17
cloning | F:
TTGGTGGTGAGCACAGGAAG
R: GGTGCTCTTCGTGGCTCTGG | 94°C for 3 min,
followed by 30 cycles of 92°C for 1 min, 72°C for 40 sec, and
finally 72°C for 10 min |
|
| Methylation
internal control |
|
T7
EGFP-N bis reverse | F:
TAATACGACTCACTATAGGG
R: CTTGCCGTTGGTGGCATCGC | 98°C for 20 sec,
followed by 35 cycles of 98°C for 1 sec, 72°C for 15 sec, and
finally 72°C for 1 min |
|
|
Methylation-sensitive restriction
enzyme-droplet digital PCR |
|
| methCTRL | F:
CACTATAGGGAGACCCAAG
R: AACTTGTGGCCGTTTAC
Probe: [HEX]5'-CTGTTCACCGGGGTGG-3'
[IowaBlack] | |
|
SLC22A17 | F:
GAGGCAATGGTTGAAGTCCG
R: CTAATGCCTCTGGCTGGGAG
Probe: [FAM]5'-GCCGCTGCACGAGGGGTCGG-3' [BHQ1] | 95°C for 10 min,
followed by 40 cycles of 94°C for 30 sec, 55°C for 1 min, and
finally 98°C for 10 min (ramp rate 2°C/sec) |
DNA methylated and unmethylated
controls
A sequence (276 bp) of SLC22A17 gene
(chr14:23,821,170-23,821,445-Assembly: GRCh37/hg19), containing 3
CCGG restriction sites, was used to generate DNA unmethylated and
methylated controls (Fig. S1B).
Specifically, the SLC22A17 sequence was obtained by PCR
amplification of A375 gDNA using the Taq DNA Polymerase,
recombinant (5 U/μl) (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Primers and PCR
thermocycling conditions are included in Table I. The PCR product was purified
using the PureLink PCR Purification kit (cat. no. K310001; Thermo
Fisher Scientific Inc.) and quantified by NanoDrop-1000 (Thermo
Fisher Scientific, Inc.). To obtain methylated control (100% of
methylation), the amplified SLC22A17 sequence was treated
with CpG methyltransferase (M.SssI) kit (cat. no. EM0821; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Briefly, 200 ng of SLC22A17 PCR product was
added to a reaction mixture containing 1 μl of M.SssI
enzyme, 2 μl of M.SssI buffer 10X, 0.4 μl of SAM
(S-adenosylmethionine) 50X and nuclease-free water to obtain a
final volume of 20 μl. The unmethylated control (0% of
methylation) was generated from SLC22A17 PCR product using
the same methylation mixture without M.SssI enzyme. Both reactions
were incubated at 37°C for 30 min and then stopped by heating at
65°C for 20 min. Finally, the methylated and unmethylated controls
were purified by using the PureLink PCR Purification kit (cat. no.
K310001; Thermo Fisher Scientific, Inc.) and quantified with
Nanodrop-1000.
The SLC22A17 methylated (100%) and
unmethylated (0%) CTRLs were mixed in different ratios (100, 75,
50, 25 and 0%) maintaining a constant total DNA concentration of
1.25×10−6 ng/μl.
Custom methylation internal control
To assess the digestion efficiency of HpaII
and MspI in ddPCR-MSRE reaction and normalize the percentage
of methylation of each methDNA target, methCTRL was generated by
PCR amplification of a sequence of the fluorescent protein Clover,
which contains 1 CCGG restriction site (Fig. S1C). Briefly, 10 ng of
pcDNA3-Clover plasmid gently provided by Dr Michael Lin (Department
of Bioengineering, Stanford University, Stanford, USA) (Addgene
plasmid #40259; http://n2t.net/addgene:40259; RRID: Addgene_40259) was
amplified using the Phusion High-Fidelity DNA Polymerase (2
U/μl) kit (cat. no. F-530XL; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. PCR thermal conditions
and primer sequences are reported in Table I. The PCR product was
subsequently treated with 1 μl DpnI (cat. no. FD1703;
Thermo Fisher Scientific, Inc.) at 37°C for 15 min and then the
enzyme was inactivated at 80°C for 20 min. Finally, the PCR
reaction was purified using the PureLink PCR Purification kit (cat.
no. K310001; Thermo Fisher Scientific, Inc.) and quantified with
Nanodrop-1000.
Standard MSRE assay
MSRE digestion was performed on gDNA obtained from
A375, SK-MEL-23 and A2058 cells. For each sample, three different
reaction tubes (final volume 10 μl) were prepared by mixing
200 ng of gDNA, 10−6 ng/μl of methCTRL, 1X
CutSmart Buffer (cat. no. B7204), and 20 UI of HpaII (cat.
no. R0171S) for tube 1, 20 UI of MspI (cat. no. R0106S) for
tube 2, and no enzyme for tube 3 (all the reagents were purchased
from New England Biolabs). All the reaction tubes were incubated at
37°C for 1 h and stopped with Proteinase K (cat. no. EO0491; Thermo
Fisher Scientific, Inc.) (final concentration 1 mg/ml) incubating
the samples at 55°C for 30 min followed by an inactivation step at
95°C for 10 min. Following the standard MSRE digestion, 4 μl
of RNase/DNase-free water molecular biology-grade was added to 1
μl of each digested sample for the downstream ddPCR
amplification. In addition, to evaluate the effect of potential
inhibitors (melanin) on the MSRE digestion, 20 ng instead of 200 ng
of SK-MEL-23 gDNA were digested in 10 μl of final reaction
volume according to the aforementioned standard MSRE protocol. Of
note, 5 μl of these digested samples were directly added to
the ddPCR mix for the downstream analysis.
BamHI digestion for ddPCR interference
test
To further evaluate the ddPCR efficiency in the
amplification of targets digested by restriction enzymes and to
evaluate the highest amount of the MSRE digestion mix that can be
uploaded in the ddPCR mix, BamHI was used as a restriction
enzyme that does not digest the SLC22A17 methDNA target.
Briefly, 20 μl of digestion mix containing 20 UI of
BamHI (cat. no. R0136S; New England Biolabs, Inc.), 1X
CutSmart Buffer (cat. no. B7204; New England Biolabs, Inc.), 80 ng
of A375 gDNA, and 4×10−6 ng of methCTRL was incubated at
37°C for 30 min, stopped with 1 mg/ml of Proteinase K (cat. no.
EO0491; Thermo Fisher Scientific Inc.) at 55°C for 30 min followed
by an inactivation step (95°C for 10 min). A total of 5 μl
of the digested sample (20 ng) was used for the ddPCR
amplification. As a control, 20 ng of A375 gDNA and 10−6
ng methCTRL in 5 μl of molecular biology-grade water were
amplified in ddPCR.
Standard ddPCR mix preparation
The ddPCR amplification mix was prepared by mixing
11 μl of 2X ddPCR Supermix for Probes (no dUTP) (cat. no.
1863024; Bio-Rad Laboratories, Inc.), 5 μl of each sample,
450 nM of FAM probe (SLC22A17 target), 450 nM of HEX probe
(methCTRL), 900 nM of forward and reverse primers for each target
and RNase/DNase-free water molecular biology-grade up to a final
volume of 22 μl (probe and primer sequences are reported in
Table I).
MSRE-ddPCR mix preparation and digestion
protocol
Custom MSRE-ddPCR assay consists of one-tube
reactions in which methylation-sensitive restriction enzymes (i.e.
HpaII and MspI) directly digest the DNA targets in
the ddPCR reaction mix. In particular, three different
amplification mixes were prepared for each sample, containing
HpaII, MspI and no enzyme as undigested control,
respectively. The MspI mix was performed as an additional
control to assess the enzymatic digestion efficiency. However, this
optional control can be avoided because of the use of methCTRL.
Briefly, each amplification mix (22 μl) was prepared by
using 11 μl of 2X ddPCR Supermix for Probes (no dUTP) (cat.
no. 1863024; Bio-Rad Laboratories, Inc.), 900 nM of forward/reverse
primers and 450 nM of FAM/HEX probes for SLC22A17 target and
methCTRL (probe and primer sequences are reported in Table I). Up to 20 ng of DNA sample and
10−6 ng of methCTRL (final volume 5 μl) were
added to each MSRE-ddPCR mix along with 10 UI of restriction enzyme
in the HpaII and MspI mix. All amplification mixes
were incubated at 37°C for 30 min before droplet generation.
Droplet generation and analysis
Droplet generation was performed by loading 20
μl of ddPCR amplification mix in DG8 Cartridges along with
70 μl of Droplet Generation Oil (cat. no. 1863005; Bio-Rad
Laboratories, Inc.) within the sample and oil wells, respectively.
Then the cartridge was covered with Gasket (cat. no. 1863009;
Bio-Rad Laboratories, Inc.) and transferred into Droplet Generator
QX100 (Bio-Rad Laboratories, Inc.) for droplet generation according
to the manufacturer's instructions. The droplet mixture was
recovered from the cartridge and transferred into a 96-well plate
(cat. no. 12001925; Bio-Rad Laboratories, Inc.) to perform PCR
amplification by the C1000 Touch Thermal Cycler (Bio-Rad
Laboratories, Inc.) according to the manufacturer's protocol. The
ddPCR thermal conditions are reported in Table I. Finally, the QX200 Droplet
Reader (Bio-Rad Laboratories, Inc.) was used for droplet
quantification. Absolute quantification (copies/μl) of DNA
targets was retrieved using QuantaSoft software (version 1.7.4
(QuantaSoft). Amplitude thresholds were set manually by the
operator on the basis of positive and negative droplet amplitudes.
The fluorescence amplitude of droplets was also considered to
evaluate the efficiency of the amplification reaction.
Quantitative analysis of the methDNA
percentage
The methDNA percentage of the target genes was
retrieved considering the ratio between the ddPCR absolute
quantification of the methDNA target in HpaII and the undigested
control mix for each sample (first term of Formula 1 and 2).
Notably, this ratio is 1 when the methDNA target is fully
methylated (100% of methylation), while it is 0 when the methDNA
target is completely unmethylated (0% of methylation).
To overcome the bias in methDNA percentage
estimation due to the inhibition of the enzymatic digestion, data
normalization was performed by using an enzymatic digestion
coefficient computed by the reciprocal ratio between the ddPCR
absolute quantification of the methDNA target in MspI and
undigested control mix (second term of Formula 1). Similarly, the
enzymatic digestion coefficient was obtained by the reciprocal
ratio between the methCTRL absolute quantification in HpaII
and the undigested control mix avoiding the MspI mix (second
term of Formula 2). Notably, the enzymatic digestion coefficient
does not affect the methDNA percentage estimation when the
enzymatic digestion is completed (enzymatic digestion
coefficient=1). Conversely, the methDNA percentage is adjusted when
the enzymatic digestion is partially inhibited.
Statistical analysis
Linear regression analysis was computed to evaluate
the goodness of fit (R2) obtained from the MSRE-ddPCR
analysis on scalar dilution samples of SLC22A17 methylated
control. To test the sensitivity of MSRE-ddPCR, the horizontal
best-fit lines through the mean of all the methDNA values, obtained
from the serial dilutions of SK-MEL-23 and A375 gDNAs, were
calculated using Non-linear fit analysis. Differential analysis of
SLC22A17 methDNA in melanoma and nevi samples was evaluated
by Mann-Whitney test. The difference between the comparing groups
was reported as differences between the median levels of
SLC22A17 methDNA. Receiver Operating Characteristic (ROC)
curve analysis was performed to evaluate the performance and
accuracy of the diagnostic test. P≤0.05 was considered to indicate
a statistically significant difference. All analyses were executed
using GraphPad Prism software (version 8.0.2) (Dotmatics).
Results
MSRE-ddPCR efficiency test
To evaluate the potential interference between
restriction enzymes and ddPCR chemistry in enzymatic digestion and
amplification processes, MSRE-ddPCR protocol was performed
analyzing DNA methylated (100%) and unmethylated (0%) controls
obtained from the SLC22A17 sequence
(chr14:23,821,170-23,821,445-Assembly: GRCh37/hg19). Specifically,
5×10−6 ng of each DNA methylated and unmethylated
controls along with 10−6 ng of Methylation Internal
Control (methCTRL) were added to HpaII, MspI and
undigested mixes. methCTRL and SLC22A17 forward/reverse
primers and probes were used for the amplification step.
As expected, the results revealed that the number of
copies for the SLC22A17 methylated control within the
HpaII mix was similar to the undigested sample (45.4±2.38
copies/μl and 42.5±1.7 copies/μl, respectively).
Interestingly, the amplitude of SLC22A17 methylated control
(FAM fluorescence) in the HpaII mix was similar to the
undigested mix, demonstrating that the HpaII enzyme did not
affect the ddPCR amplification efficiency (Fig. 1A). Moreover, no amplification was
detected for SLC22A17 methylated control within the
MspI mix. Similarly, the SLC22A17 unmethylated
control showed no amplification within HpaII and MspI
reactions compared with the undigested sample, suggesting that the
enzymatic digestion efficiency was not affected by ddPCR chemistry
(Fig. 1A).
Regarding methCTRL (HEX fluorescence), a similar
amplification signal was observed in the undigested mix of
SLC22A17 methylated and unmethylated samples (35.4±0.45 and
34.00±0.84 copies/μl, respectively), while a weak
amplification signal was detected in both HpaII and
MspI reactions (from 0.26 to 1.70 copies/μl)
(Fig. 1B). The positive droplets
of methCTRL in HpaII and MspI reactions are due to a
not complete digestion that never reaches the theoretical 100%
efficiency of HpaII and MspI in ddPCR mix. This
background signal, detected in each MSRE-ddPCR mix, is removed by
normalization procedures during the estimation of methDNA
percentage.
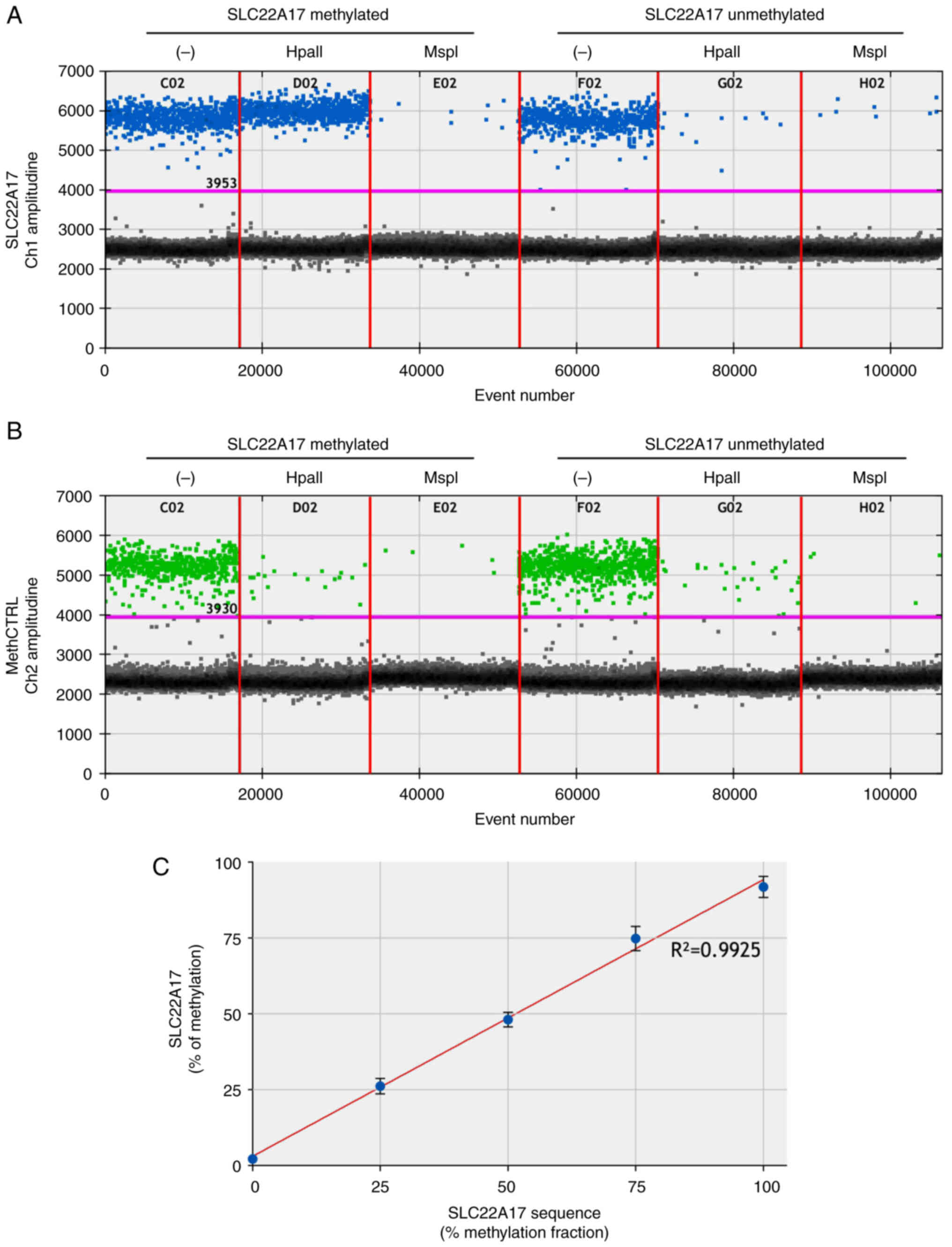
To evaluate the accuracy of the MSRE-ddPCR method,
linear regression analysis was performed comparing the expected and
measured methDNA levels of each SLC22A17
methylated/unmethylated sample (100, 75, 50, 25 and 0%). The
results demonstrated a high linear correlation of all
expected/measured couples (R2=0.9925) indicating the
reliability of the method at different percentages of methDNA
target (Fig. 1C).
MSRE-ddPCR analysis on cellular genomic
DNA
The MSRE-ddPCR assay was tested on gDNA extracted
from different melanoma cell lines evaluating the levels of
SLC22A17 methDNA hotspot (Fig. S1) selected in the present study.
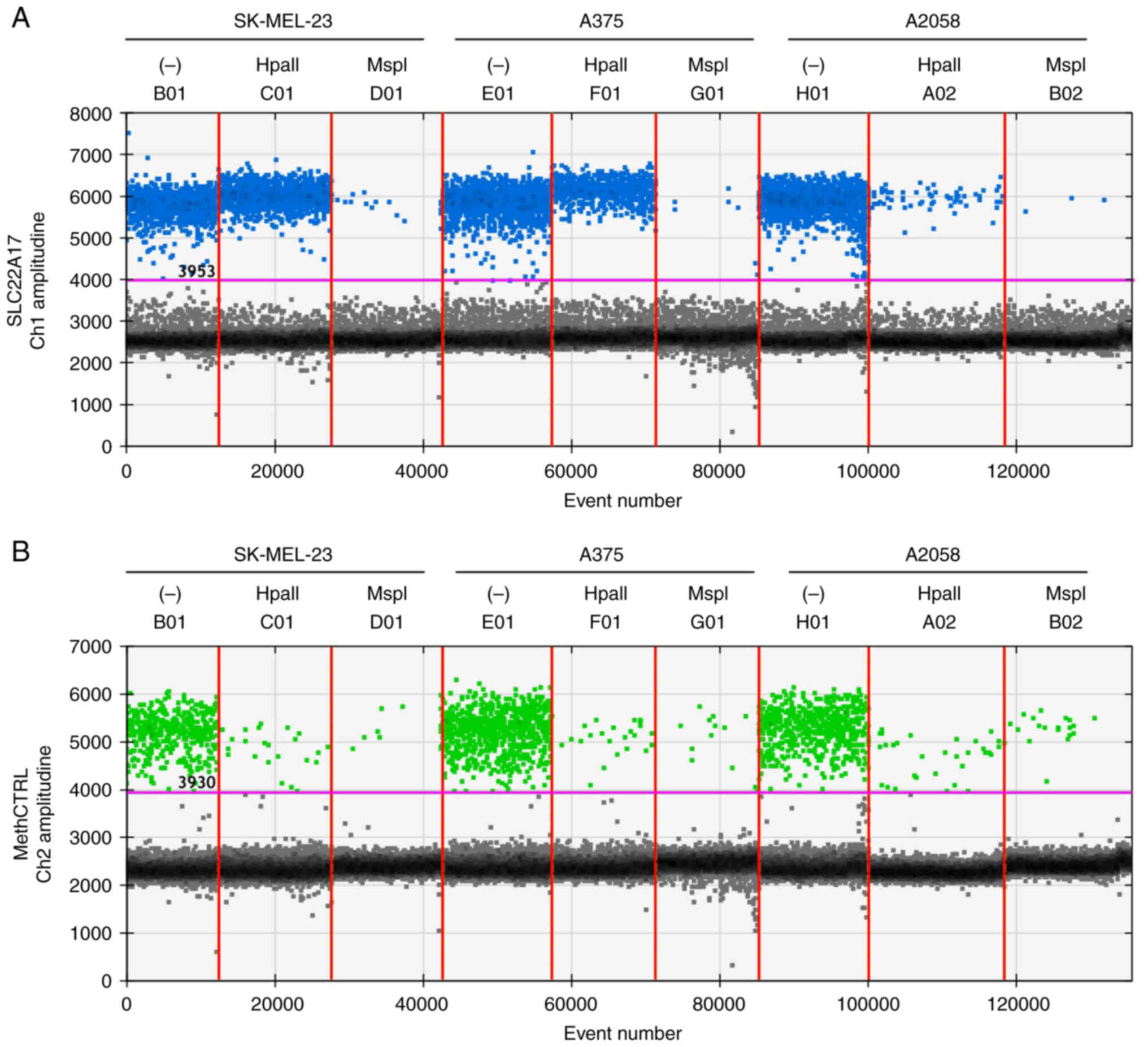
In particular, the SLC22A17 target was assessed on
SK-MEL-23, A375 and A2058 cells (Fig. 2A and B).
The methylation levels of the SLC22A17
hotspot were higher in SK-MEL-23 (98.18±0.15%) compared with A375
(39.7±0.60%) and A2058 cells (1.16±0.19%) (Fig. 2A and B). Notably, the amplitude
of FAM and HEX droplets was comparable to that observed in the
setup experiment reported in Fig.
1.
The MSRE-ddPCR results agreed with those obtained
from bisulfite sequencing of SLC22A17 hotspot in the same
cell lines. In particular, the internal cytosine of CCGG within the
SLC22A17 hotspot was unconverted in SK-MEL-23 (Fig. S2A), partially converted (~34% of
methDNA) in A375 (Fig. S2B) and
converted in A2058 (Fig. S2C),
indicating that this hotspot was highly hypermethylated in
SK-MEL-23, partially methylated in A375 and unmethylated in the
A2058 cells. The absence of amplification signal in the MspI
mix for the SLC22A17 target (Fig. 2A), as well as the slight signal
detected for methCTRL in HpaII and MspI mix (Fig. 2B), indicated that the restriction
enzymes were highly efficient in MSRE-ddPCR mix.
Comparison between MSRE-ddPCR and
standard MSRE assays
To assess the reliability of MSRE-ddPCR results, the
methylation levels of the SLC22A17 hotspot were also
evaluated by the standard MSRE procedure using HpaII and
MspI enzymes as aforementioned. The methylation percentage
computed through both methods was normalized by the methCTRL signal
in HpaII mix and the amplification signal of each target in
MspI mix (Table II).
 | Table IIComparison between MSRE-ddPCR and
standard MSRE assays performed on melanoma cell lines. |
Table II
Comparison between MSRE-ddPCR and
standard MSRE assays performed on melanoma cell lines.
| Cell line | Target | Normalization by
methCTRL
| Normalization by
MspI mix
|
|---|
| MSRE-ddPCR (% of
methyl ± DV) | MSRE (% of methyl ±
SD) | MSRE-ddPCR (% of
methyl ± SD) | MSRE (% of methyl ±
SD) |
|---|
| SK-MEL-23 |
SLC22A17 | 98.18±0.15 | NA | 98.26±0.79 | NA |
| A375 | | 39.7±0.60 | 40.95±0.64 | 39.95±0.35 | 41.40±0.42 |
| A2058 | | 1.16±0.19 | 2.09±0.11 | 1.29±0.05 | 2.16±0.08 |
The results indicated that the methylation levels of
SLC22A17 target obtained with both MSRE-ddPCR and standard
MSRE procedures were comparable showing only a slight reduction of
the methDNA percentage estimation (<0,5%) when methCTRL
normalization was applied (Table
II). Interestingly, the standard MSRE assay was unable to
quantify the SLC22A17 methylation hotspot in SK-MEL-23 cells
since the SLC22A17 signal was relevant in the MspI mix (~90
copies/μl), suggesting that the MspI enzymatic
digestion was inhibited by DNA sample contaminants (Fig. S3). It was probably due to the
presence of melanin in the gDNA sample obtained from the highly
melanin-pigmented SK-MEL-23 cells that appeared brown-pigmented
during the DNA extraction procedure (data not shown). To test this
hypothesis, the standard MSRE assay was performed on 20 ng of
SK-MEL-23 gDNA (instead of 200 ng of gDNA) in 10 μl of final
reaction volume, of which 5 μl were directly added to the
ddPCR mix for the amplification. The results revealed an efficient
digestion of MspI in SK-MEL-23 diluted gDNA, indicating that the
high melanin content of SK-MEL-23 may reduce the efficiency of
MspI (Fig. S3A and B).
However, the detected amplitude for each diluted sample was
significantly reduced compared with standard MSRE procedure due to
the amount of the MSRE digestion mix (5 μl of diluted gDNA)
used for the downstream analysis, demonstrating that MSRE mix
affected the ddPCR amplification (Fig. S3A and B). Notably, the amount of
SK-MEL-23 gDNA sample loaded in each standard MSRE mix (200 ng of
gDNA in 10 μl final reaction volume) was ~7.5% of the
reaction and ~0.75% for diluted gDNA sample (20 ng in 10 μl
final reaction volume), whereas it was ~0.34% in each MSRE-ddPCR
mix (20 ng of gDNA in 22 μl final reaction volume),
considering an initial DNA concentration of 266.7 ng/μl.
Therefore, using a lower quantity of DNA in each MSRE-ddPCR mix
compared with the standard MSRE mix, the enzymatic inhibition was
efficiently prevented.
Interference of restriction enzyme
digestion mix on ddPCR amplification
To further evaluate the interference of the standard
MSRE digestion mix on the downstream ddPCR amplification and to
assess the highest amount of the MSRE digestion mix that may be
processed by ddPCR, A375 gDNA was digested with BamHI using
its standard reagents. This restriction enzyme was selected since
it does not digest the SLC22A17 methDNA hotspot allowing to
simulate the inhibition of ddPCR amplification by digestion
components independently from the negative effect of MSRE digestion
on the number of positive droplets. Specifically, 5 μl of
BamHI reaction (1X CutSmart Buffer, 5 UI of BamHI)
containing 20 ng of A375 gDNA and 10−6 ng of methCTRL
were amplified in ddPCR to detect SLC22A17 target and
methCTRL. The same amplification conditions were applied to the
control sample consisting of 20 ng of A375 gDNA and 10−6
ng of methCTRL in 5 μl of molecular biology-grade water.
The results revealed that the SLC22A17
amplitude (FAM channel) was reduced by ~1,000 AU fluorescence in
the BamHI mix compared with the control mix (Fig. S4A and C). Similarly, a
significant reduction of methCTRL amplitude signal was observed in
the BamHI reaction compared with the control (~5561 AU
fluorescence for the control mix and ~4477 AU fluorescence for the
BamHI mix) (Fig. S4B and
D). The data confirmed the results obtained for SK-MEL-23
diluted MSRE mix (Fig. S3)
indicating that the restriction enzyme reaction, including the MSRE
reaction, consistently affected the ddPCR amplification. Therefore,
the standard MSRE digestion of low-concentrated DNA samples cannot
be analyzed by ddPCR because up to 5 μl of this reaction
should be added in each ddPCR mix to reach the sensitivity
threshold.
Linearity and sensitivity of MSRE-ddPCR
assay
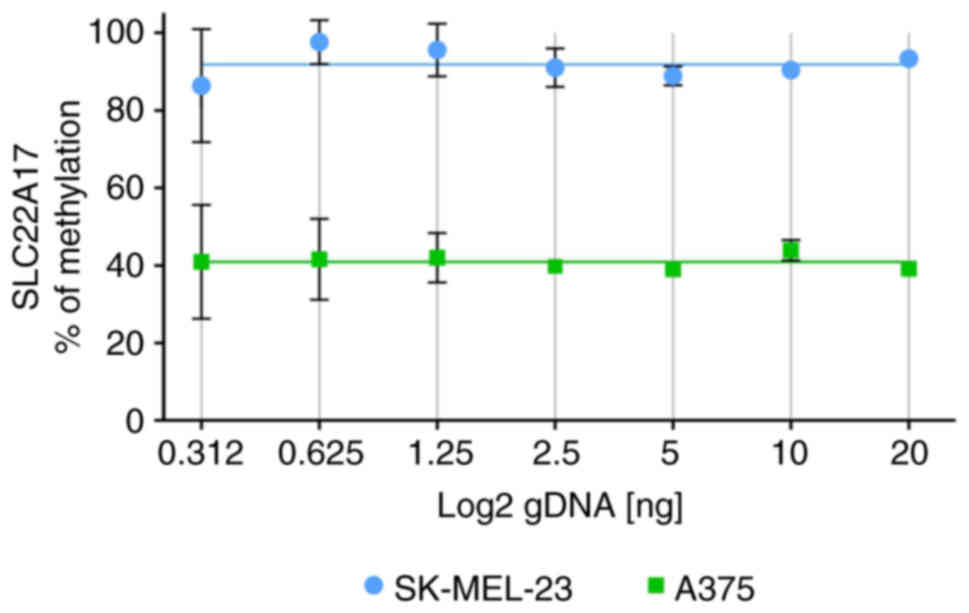
Linearity test of MSRE-ddPCR assay was performed
evaluating the SLC22A17 methDNA target using a 2-fold serial
dilution starting from 20 to 0.312 ng of gDNA obtained from
SK-MEL-23 and A375 cells. The results suggested that the
methylation percentage of the SLC22A17 hotspot, whose
average values ranged from 88.29 to 95.49% for SK-MEL-23 and 37.65
to 44.20% for A375, demonstrated a slight deviation (3.6% for
SK-MEL-23 and 3.27% for A375) from best-fit value (91.89% for
SK-MEL-23 and 40.92% for A375) in both the gDNA dilutions. However,
a significant increase in standard deviation (SD) was observed
starting from 1.25 ng of gDNA, with the highest SD observed for
samples diluted at 0.312 ng, suggesting that the accuracy of the
MSRE-ddPCR was suitable until the concentration of 0.625 ng
(Fig. 3).
Performance analysis of MSRE-ddPCR in
low-quality DNA samples
MSRE-ddPCR assay was designed for the methylation
analysis of low quantity and/or quality DNA samples, including gDNA
from FFPE samples and cfDNA from different body fluids. To test the
analytic performance of the MSRE-ddPCR, the SLC22A17 methDNA
hotspot (Fig. S1B) was analyzed
on a pilot cohort of FFPE melanoma tissues (n=10) and FFPE nevi
(n=10). Moreover, to evaluate the adaptability of the MSRE-ddPCR
for the analysis of cfDNA, two serum samples from melanoma patients
were analyzed as a pilot test.
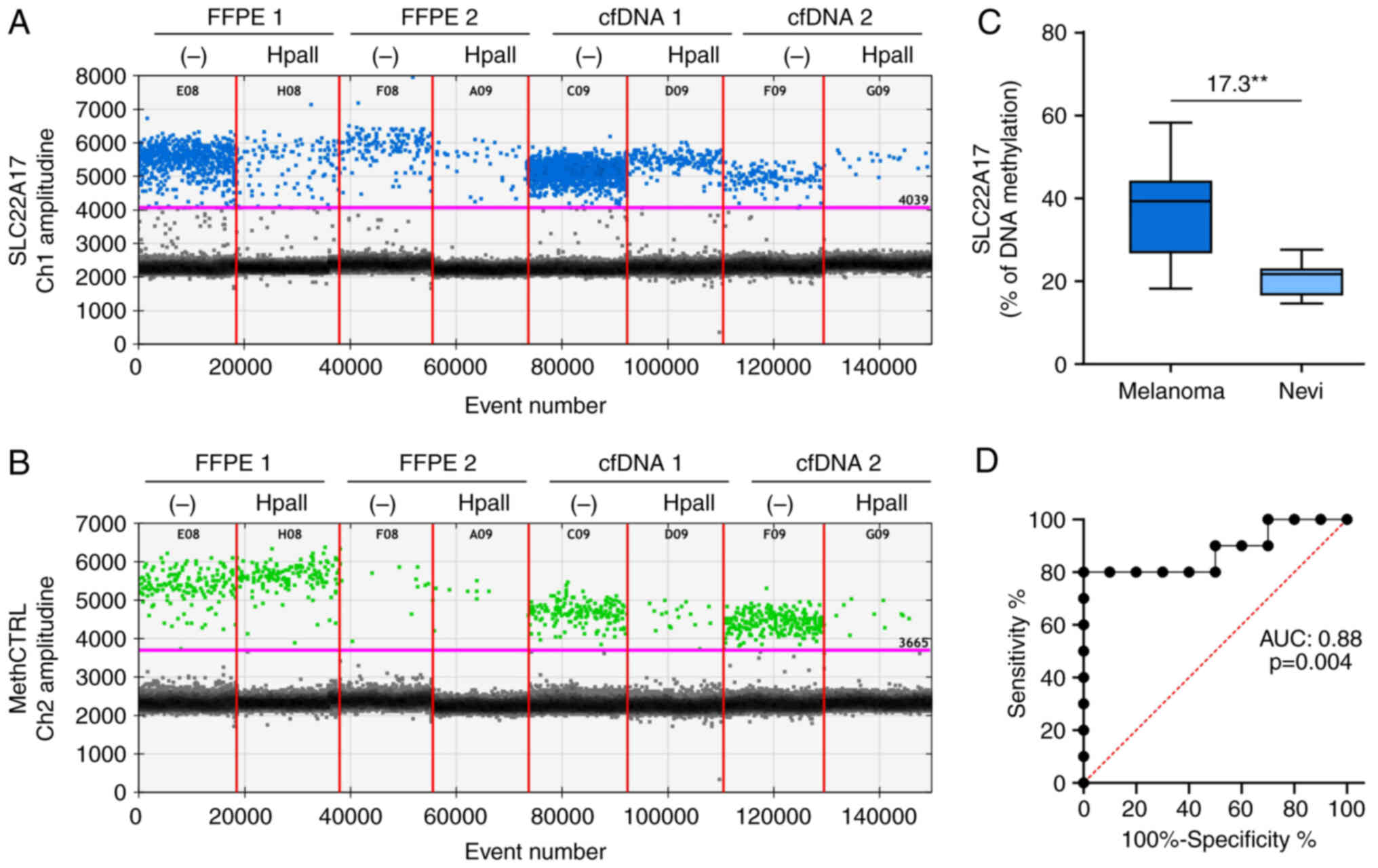
The plots indicated that the amplitude signal of
both FAM and HEX channels was similar to the signal detected for
melanoma cell lines (Fig. 4A and
B). However, the droplet rain between positive and negative
droplets was observed for SLC22A17 methDNA hotspot in the
FFPE samples, indicating the presence of PCR inhibitors and DNA
fragmentation (Fig. 4A and B).
Moreover, these inhibitors slightly reduced the amplitude of the
SLC22A17 methDNA hotspot and methCTRL in FFPE and cfDNA
samples than melanoma cell lines. However, the amplitude difference
did not influence the accuracy of target detection by ddPCR as
demonstrated in Fig. S4.
The MSRE-ddPCR analysis revealed that the
methylation levels of SLC22A17 were 21.04±0.90% for FFPE 1
and 33.43±1.88% for FFPE 2. Regarding the methylation analysis of
serum cfDNA, the SLC22A17 hotspot was 9.74±0.38% for cfDNA 1
and 12.69±0.53% for cfDNA 2 (Fig. 4A
and B). The results of SLC22A17 methDNA hotspot analysis
on the melanoma and nevi cohorts revealed that the median
percentage of methDNA was significantly (P≤0.01) higher in melanoma
(39.37%) compared with healthy samples (21.78%) (Fig. 4C). Of note, despite the small
number of analyzed samples, the ROC curve analysis demonstrated
high sensitivity and specificity [Area Under Curve (AUC): 0.88;
P≤0.01] of the SLC22A17 methDNA test as a diagnostic
biomarker for melanoma (Fig.
4D).
Discussion
In the last decades the development of
high-throughput technologies, including Next Generation Sequencing
(NGS) and microarray, has drastically enhanced the knowledge of
methDNA status in cancer patients. These technologies allow the
generation of large-scale data useful to define the whole methylome
and are often publicly available for in silico studies to
identify new methDNA targets involved in tumor development and
progression (31-33). In this context, the
locus-specific evaluation of methDNA is mandatory to validate the
in silico results, as well as to analyze low DNA templates
typically obtained from FFPE tissues, liquid biopsies and
nanovesicles (16,34). Actually, the gold standard
pre-processing method to analyze both the whole genome and the
single methDNA hotspots, especially for sequencing approaches (i.e.
NGS, pyrosequencing, Sanger sequencing) or microarray analyses
(i.e. Infinium HumanMethylation450 Bead Chip array by Illumina), is
based on the bisulfite conversion (35,36). However, during the bisulfite
conversion, the DNA samples are exposed to acidic conditions and
high temperatures leading to a consistent DNA fragmentation that
may reduce the DNA recovery during the cleanup, as well as the DNA
sequencing of long fragments (12,13,16,37,38). Furthermore, the conversion of
unmethylated cytosine in uracil (and then thymine during PCR)
considerably reduces the complexity of the genome resulting in low
specificity of the PCR amplification, as well as the loss of
accuracy in the NGS alignment. In addition, the complete conversion
of unmethylated cytosines could not be reached for all DNA samples
affecting the precise estimation of the methDNA levels (16). Despite the MSRE procedure does
not affect the quality or the quantity of DNA for downstream
analyses, this method can be only applied to evaluate the CpG
hotspots located in the restriction sites recognized by the
methyl-sensitive enzymes used in MSRE. Moreover, the MSRE reaction
may be inhibited by different DNA contaminants, especially when a
large quantity of low-concentrated samples is used for the analysis
(19,20). Similarly, a large amount of the
MSRE digested sample must be used to reach the PCR sensitivity
threshold inducing a significant reduction of the DNA amplification
efficiency (19,20).
In the last few years, the ddPCR technology has
emerged as a suitable tool for the analysis of low-quantity and/or
-quality DNA samples (25,39). Moreover, this technology has
recently been adopted for the analysis of methDNA by MSRE to
overcome the limitations of standard procedures. In particular,
several studies have demonstrated that the ddPCR amplification of
MSRE-digested samples enhanced the sensitivity of methDNA target
detection compared with previous generations of PCR (endpoint,
qPCR) (21,22,24,40). Interestingly, since the ddPCR
reaction mix is compatible with restriction enzyme activity, van
Zogchel et al (23)
evaluated the methylation status of Ras association domain
family member 1 (RASSF1A) in liquid biopsy samples
performing MSRE reaction directly within the ddPCR mix. In this
field, the MSRE-ddPCR assay was proposed by the authors, a custom
one-tube protocol based on the combination of the MSRE reaction and
the high-sensitive ddPCR amplification and the use of spike-in
methylation control, named methCTRL. In particular, the MSRE-ddPCR
approach is based on the use of the methyl-sensitive restriction
enzymes (i.e. HpaII and MspI), which are directly
loaded in the ddPCR mix to perform digestion of the DNA targets
before droplet generation and amplification steps. This allows us
to reduce the systematic errors related to the high number of
analytic steps and pipetting, which occur in the standard MSRE
procedure. Furthermore, up to 5 μl of DNA diluted sample can
be loaded in each MSRE-ddPCR mix to reach the ddPCR sensitive
threshold. Notably, the ddPCR amplification performed on diluted
DNA samples, pre-processed with standard MSRE digestion, needs a
large amount of the digested sample that could drastically inhibits
the amplification due to the presence of PCR inhibitors (digestion
buffer and enzymes) contained within the MSRE mix (Figs. S3 and S4).
The main significant advantage of the MSRE-ddPCR is
represented by the design of the methCTRL that consists of an
exogenous unmethylated DNA sequence containing one CCGG site
specific for HpaII and MspI. In particular, the
methCTRL is added to the sample to assess the efficiency of the
enzymatic digestion in each MSRE-ddPCR mix. Since the cleavage of
the methCTRL may be affected by inhibitors within the sample, no
amplification signal should be detected in each MSRE-ddPCR
digestion mix containing no inhibitors.
Of note, the methCTRL is useful to directly assess
the HpaII efficiency in each MSRE-ddPCR mix independently
from the MspI reaction. Since the assessment of the methDNA
levels depends on the HpaII activity on CCGG sites, the use
of methCTRL ensures the accurate estimation of the methylation
status of each methDNA hotspot and allows the evaluation of
specific HpaII inhibitors or other issues, including
inappropriate enzyme storage. Therefore, the use of the MspI
control mix is not required to normalize the methDNA
quantification.
To test the validity of the MSRE-ddPCR assay, setup
and validation experiments were performed analyzing a methDNA
hotspot within SLC22A17, a gene known to be widely involved
in the development, progression, and drug resistance of several
tumor types, including melanoma (41-46). Specifically, the gDNA extracted
from melanoma cell lines, melanoma and nevi FFPE tissues, and cfDNA
obtained from two melanoma patients was analyzed for
SLC22A17 methDNA hotspot. The results indicated that the
MSRE-ddPCR assessment of this methDNA hotspot was comparable to the
standard MSRE assay demonstrating that restriction enzyme activity
was not affected by ddPCR chemistry. This result was confirmed by
bisulfite sequencing of the SLC22A17 methDNA hotspot, which
shows a hypermethylation of the SLC22A17 hotspot in
SK-MEL-23, a partial methylation in A375 cells, and hypomethylation
in A2058 cells. Furthermore, the MSRE-ddPCR assay was suitable to
analyze critical DNA samples, including the low-concentrated DNA
and gDNA from high melanin-pigmented SK-MEL-23 cells. Notably, the
standard MSRE failed the evaluation of the SLC22A17 methDNA
hotspot since the PCR amplification signal was strongly detected in
the MspI mix. Although the issue was overcome by diluting
the gDNA sample, the ddPCR amplification was significantly affected
by the large amount of the MSRE mix required to reach the
sensitivity threshold (Fig.
S3). Conversely, the same methDNA target was efficiently
measured by ddPCR-MSRE as demonstrated by the absence of signal in
the MspI mix (Fig. 2A).
However, optimization experiments should be performed to test the
MSRE-ddPCR accuracy of each specific target considering the
pigmentation levels of melanoma specimens. Of note, the presence of
melanin, which is not removed during the purification steps, may
affect PCR-based assays even in low-concentrated DNA samples
(47,48).
Setup experiments demonstrated the sensitivity and
linearity of the MSRE-ddPCR for the methylation analysis of
low-concentrated DNA samples. In addition, the analytical potential
of this assay in the detection of methDNA hotspots was also
demonstrated in a cohort of melanoma and normal samples, indicating
the strength and reproducibility of the MSRE-ddPCR method in the
detection of epigenetic cancer-related biomarkers. Interestingly,
although the pilot test on cfDNA suggested the potential
extensibility of the MSRE-ddPCR for the analysis of liquid biopsy
samples, further investigations should be undertaken to validate
its application in routine setting.
The MSRE-ddPCR is a valuable method for methDNA
analysis, which not requires any DNA pre-processing steps (i.e.
standard MSRE and bisulfite conversion) affecting the DNA
quality/quantity for the downstream amplification. However, the
number and location of HpaII restriction sites may reduce
the coverage of methylation analysis compared with the bisulfite
sequencing that allows quantifying the methylation status with the
single-CpG resolution. This limitation may be partially overcome
using other methylation-sensitive enzymes (i.e. HhaI,
BstUI and AciI) for each of which optimization tests
could be required to assess the compatibility with ddPCR chemistry,
as well as the design of appropriate methCTRLs.
Another limitation of the MSRE-ddPCR assay may be
due to the presence of multiple enzymatic restriction sites within
the DNA target, which result in a misestimation of the methylation
levels. In particular, a single unmethylated restriction site is
responsible for the total digestion of the DNA target affecting the
evaluation of the methylation status for other CpG restriction
sites. Therefore, the design of gene-specific primers flanking a
single methylation-sensitive restriction site is mandatory to
perform an accurate analysis.
Overall, the ddPCR-MSRE assay represents an
efficient one-tube method for reproducible and highly sensitive
analyses of specific methDNA hotspots in several DNA samples,
including low quantity and/or quality DNA from canonical and
non-canonical biological matrices.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study
are included in this published article and its supplementary
information files. The sequencing data generated in this study were
deposited in the Zenodo platform (https://doi.org/10.5281/zenodo.7840070) (created and
accessed on 18 April 2023) and are also available from the
corresponding author on reasonable request.
Authors' contributions
GG, AL, LF and SC performed the experiments. GG,
AL, SC, RC, GM, MC, LF, PAA and ML interpreted and analyzed data
and drafted the manuscript. LF, ML, RC, GM, MC and PAA edited the
manuscript and provided critical revisions. SC, ML and LF designed
and supervised the study. LF, GM and PAA obtained resources and
funding for the study. SC and GG confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the guidelines of the Declaration of Helsinki and approved by the
Institutional Review Board of the Istituto Nazionale Tumori IRCCS
Fondazione 'G. Pascale' (Naples, Italy) (protocol code 33/17oss,
approved on 10 January 2018). Informed consent was obtained from
all subjects involved in the study.
Patient consent for publication
Not applicable.
Competing interests
PAA has/had consultant/advisory roles for Bristol
Myers Squibb, Roche-Genentech, Merck Sharp & Dohme, Novartis,
Merck Serono, Pierre-Fabre, AstraZeneca, Sun Pharma, Sanofi, Idera,
Sandoz, Immunocore, 4SC, Italfarmaco, Nektar, Boehringer-Ingelheim,
Eisai, Regeneron, Daiichi Sankyo, Pfizer, Oncosec, Nouscom,
Lunaphore, Seagen, iTeos, Medicenna, and Bio-Al Health. He also
received research funding from Bristol Myers Squibb,
Roche-Genentech, Pfizer, and Sanofi; all of the above took place
outside the submitted work. All other authors declare that they
have no competing interests.
Acknowledgments
The authors would like to thank the Italian League
against Cancer (LILT) for the support.
Funding
The present study was supported by the Italian Ministry of
Health (IT-MOH) through 'Ricerca Corrente' (grant no. L2-1), the
Italian League Against Cancer (LILT) and the PON AIM R&I (grant
no. 2014-2020-E66C18001250007.
References
|
1
|
Bonin S and Stanta G: Pre-analytics and
tumor heterogeneity. N Biotechnol. 55:30–35. 2020. View Article : Google Scholar
|
|
2
|
Dagogo-Jack I and Shaw AT: Tumour
heterogeneity and resistance to cancer therapies. Nat Rev Clin
Oncol. 15:81–94. 2018. View Article : Google Scholar
|
|
3
|
Falzone L, Bordonaro R and Libra M:
SnapShot: Cancer chemotherapy. Cell. 186:1816–1816.e1. 2023.
View Article : Google Scholar
|
|
4
|
Baylin SB and Jones PA: Epigenetic
determinants of cancer. Cold Spring Harb Perspect Biol.
8:a0195052016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ilango S, Paital B, Jayachandran P, Padma
PR and Nirmaladevi R: Epigenetic alterations in cancer. Front
Biosci (Landmark Ed). 25:1058–1109. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Klutstein M, Nejman D, Greenfield R and
Cedar H: DNA methylation in cancer and aging. Cancer Res.
76:3446–3450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar
|
|
8
|
Dhar GA, Saha S, Mitra P and Nag Chaudhuri
R: DNA methylation and regulation of gene expression: Guardian of
our health. Nucleus (Calcutta). 64:259–270. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hao X, Luo H, Krawczyk M, Wei W, Wang W,
Wang J, Flagg K, Hou J, Zhang H, Yi S, et al: DNA methylation
markers for diagnosis and prognosis of common cancers. Proc Natl
Acad Sci USA. 114:7414–7419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pettini F, Visibelli A, Cicaloni V,
Iovinelli D and Spiga O: Multi-omics model applied to cancer
genetics. Int J Mol Sci. 22:57512021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Papanicolau-Sengos A and Aldape K: DNA
methylation profiling: An emerging paradigm for cancer diagnosis.
Annu Rev Pathol. 17:295–321. 2022. View Article : Google Scholar
|
|
12
|
Grunau C, Clark SJ and Rosenthal A:
Bisulfite genomic sequencing: SYstematic investigation of critical
experimental parameters. Nucleic Acids Res. 29:E652001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tanaka K and Okamoto A: Degradation of DNA
by bisulfite treatment. Bioorg Med Chem Lett. 17:1912–1915. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Q, Hermanson PJ and Springer NM:
Detection of DNA methylation by whole-genome bisulfite sequencing.
Methods Mol Biol. 1676:185–196. 2018. View Article : Google Scholar
|
|
15
|
Mehrmohamadi M, Sepehri MH, Nazer N and
Norouzi MR: A comparative overview of epigenomic profiling methods.
Front Cell Dev Biol. 9:7146872021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kurdyukov S and Bullock M: DNA methylation
analysis: Choosing the right method. Biology (Basel).
5:32016.PubMed/NCBI
|
|
17
|
Šestáková Š, Šálek C and Remešová H: DNA
methylation validation methods: A coherent review with practical
comparison. Biol Proced Online. 21:192019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beikircher G, Pulverer W, Hofner M,
Noehammer C and Weinhaeusel A: Multiplexed and sensitive DNA
methylation testing using methylation-sensitive restriction enzymes
'MSRE-qPCR'. Methods Mol Biol. 1708:407–424. 2018. View Article : Google Scholar
|
|
19
|
Melnikov AA, Gartenhaus RB, Levenson AS,
Motchoulskaia NA and Levenson Chernokhvostov VV: MSRE-PCR for
analysis of gene-specific DNA methylation. Nucleic Acids Res.
33:e932005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chapman KB and Higgs BW: Selective
amplification of hypermethylated DNA from diverse tumor types via
MSRE-PCR. Oncotarget. 11:4387–4400. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nell RJ, van Steenderen D, Menger NV,
Weitering TJ, Versluis M and van der Velden PA: Quantification of
DNA methylation independent of sodium bisulfite conversion using
methylation-sensitive restriction enzymes and digital PCR. Hum
Mutat. 41:2205–2216. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang D, O'Rourke D, Sanchez-Garcia JF, Cai
T, Scheuenpflug J and Feng Z: Development of a liquid biopsy based
purely quantitative digital droplet PCR assay for detection of MLH1
promoter methylation in colorectal cancer patients. BMC Cancer.
21:7972021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van Zogchel LMJ, Lak NSM, Verhagen OJHM,
Tissoudali A, Gussmalla Nuru M, Gelineau NU, Zappeij-Kannengieter
L, Javadi A, Zijtregtop EAM, Merks JHM, et al: Novel circulating
hypermethylated RASSF1A ddPCR for liquid biopsies in patients with
pediatric solid tumors. JCO Precis Oncol.
5:PO.21.001302021.PubMed/NCBI
|
|
24
|
Metzenmacher M, Hegedüs B, Forster J,
Schramm A, Horn PA, Klein CA, Bielefeld N, Ploenes T, Aigner C,
Theegarten D, et al: Combined multimodal ctDNA analysis and
radiological imaging for tumor surveillance in Non-small cell lung
cancer. Transl Oncol. 15:1012792022. View Article : Google Scholar
|
|
25
|
Olmedillas-López S, Olivera-Salazar R,
García-Arranz M and García-Olmo D: Current and emerging
applications of droplet digital PCR in oncology: An updated review.
Mol Diagn Ther. 26:61–87. 2022. View Article : Google Scholar
|
|
26
|
Gattuso G, Falzone L, Costa C, Giambò F,
Teodoro M, Vivarelli S, Libra M and Fenga C: Chronic pesticide
exposure in farm workers is associated with the epigenetic
modulation of hsa-miR-199a-5p. Int J Environ Res Public Health.
19:70182022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Crimi S, Falzone L, Gattuso G, Grillo CM,
Candido S, Bianchi A and Libra M: Droplet digital PCR analysis of
liquid biopsy samples unveils the diagnostic Role of
hsa-miR-133a-3p and hsa-miR-375-3p in oral cancer. Biology (Basel).
9:3792020.PubMed/NCBI
|
|
28
|
Pharo HD, Andresen K, Berg KCG, Lothe RA,
Jeanmougin M and Lind GE: A robust internal control for
high-precision DNA methylation analyses by droplet digital PCR.
Clin Epigenetics. 10:242018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin WH, Xiao J, Ye ZY, Wei DL, Zhai XH, Xu
RH, Zeng ZL and Luo HY: Circulating tumor DNA methylation marker
MYO1-G for diagnosis and monitoring of colorectal cancer. Clin
Epigenetics. 13:2322021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Salemi R, Falzone L, Madonna G, Polesel J,
Cinà D, Mallardo D, Ascierto PA, Libra M and Candido S: MMP-9 as a
candidate marker of response to BRAF inhibitors in melanoma
patients with BRAFV600E mutation detected in
circulating-free DNA. Front Pharmacol. 9:8562018. View Article : Google Scholar
|
|
31
|
Barros-Silva D, Marques CJ, Henrique R and
Jerónimo C: Profiling DNA methylation based on next-generation
sequencing approaches: New insights and clinical applications.
Genes (Basel). 9:4292018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nikolouzakis TK, Falzone L, Lasithiotakis
K, Krüger-Krasagakis S, Kalogeraki A, Sifaki M, Spandidos DA,
Chrysos E, Tsatsakis A and Tsiaoussis J: Current and future trends
in molecular biomarkers for diagnostic, prognostic, and predictive
purposes in non-melanoma skin cancer. J Clin Med. 9:28682020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Merkel A and Esteller M: Experimental and
bioinformatic approaches to studying DNA methylation in cancer.
Cancers (Basel). 14:3492022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gai W and Sun K: Epigenetic biomarkers in
cell-free DNA and applications in liquid biopsy. Genes (Basel).
10:322019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Smith J, Day RC and Weeks RJ:
Next-generation bisulfite sequencing for targeted DNA methylation
analysis. Methods Mol Biol. 2458:47–62. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Leti F, Llaci L, Malenica I and Di Stefano
JK: Methods for CpG methylation array profiling via bisulfite
conversion. Methods Mol Biol. 1706:233–254. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kint S, De Spiegelaere W, De Kesel J,
Vandekerckhove L and Van Criekinge W: Evaluation of bisulfite kits
for DNA methylation profiling in terms of DNA fragmentation and DNA
recovery using digital PCR. PLoS One. 13:e01990912018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hong SR and Shin KJ: Bisulfite-converted
DNA quantity evaluation: A multiplex quantitative real-time PCR
system for evaluation of bisulfite conversion. Front Genet.
12:6189552021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lavoro A, Scalisi A, Candido S, Zanghì GN,
Rizzo R, Gattuso G, Caruso G, Libra M and Falzone L: Identification
of the most common BRCA alterations through analysis of germline
mutation databases: Is droplet digital PCR an additional strategy
for the assessment of such alterations in breast and ovarian cancer
families? Int J Oncol. 60:582022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Van Wesenbeeck L, Janssens L, Meeuws H,
Lagatie O and Stuyver L: Droplet digital PCR is an accurate method
to assess methylation status on FFPE samples. Epigenetics.
13:207–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Candido S, Tomasello B, Lavoro A, Falzone
L, Gattuso G, Russo A, Paratore S, McCubrey JA and Libra M:
Bioinformatic analysis of the LCN2-SLC22A17-MMP9 network in cancer:
The role of DNA methylation in the modulation of tumor
microenvironment. Front Cell Dev Biol. 10:9455862022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chi Y, Remsik J, Kiseliovas V, Derderian
C, Sener U, Alghader M, Saadeh F, Nikishina K, Bale T,
Iacobuzio-Donahue C, et al: Cancer cells deploy lipocalin-2 to
collect limiting iron in leptomeningeal metastasis. Science.
369:276–282. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu F, Li N, Yang W, Wang R, Yu J and Wang
X: The expression analysis of NGAL and NGALR in clear cell renal
cell carcinoma. Gene. 676:269–278. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gomez-Chou SB, Swidnicka-Siergiejko AK,
Badi N, Chavez-Tomar M, Lesinski GB, Bekaii-Saab T, Farren MR, Mace
TA, Schmidt C, Liu Y, et al: Lipocalin-2 promotes pancreatic ductal
adenocarcinoma by regulating inflammation in the tumor
microenvironment. Cancer Res. 77:2647–2660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miyamoto T, Kashima H, Yamada Y, Kobara H,
Asaka R, Ando H, Higuchi S, Ida K, Mvunta DH and Shiozawa T:
Lipocalin 2 enhances migration and resistance against cisplatin in
endometrial carcinoma cells. PLoS One. 11:e01552202016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wei J, Gao X, Qin Y, Liu T and Kang Y: An
iron metabolism-related SLC22A17 for the prognostic value of
gastric cancer. Onco Targets Ther. 13:12763–12775. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Frouin E, Maudelonde T, Senal R, Larrieux
M, Costes V, Godreuil S, Vendrell JA and Solassol J: Comparative
methods to improve the detection of BRAF V600 mutations in highly
pigmented melanoma specimens. PLoS One. 11:e01586982016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Vicente ALSA, Bianchini RA, Laus AC,
Macedo G, Reis RM and Vazquez VL: Comparison of protocols for
removal of melanin from genomic DNA to optimize PCR amplification
of DNA purified from highly pigmented lesions. Histol Histopathol.
34:1089–1096. 2019.PubMed/NCBI
|