Gynecological cancers are common, and ovarian cancer
(OC) is the deadliest due to difficulties diagnosing this disease
in the early stages. Specifically, 70–90% of patients with OC are
diagnosed at an advanced stage, indicating that the cancer has
already spread (1). In 2018,
there were ~300,000 reported cases of OC worldwide, with ~18,000
fatalities (2). Epithelial
ovarian tumors have the highest incidence, whereas germ cell and
sex cord-stromal OC account for ~5% of cases (3). At present, ovarian epithelial tumors
are classified into four subtypes with identical pathologies:
Serous, endometrioid, mucinous and clear cell (3). These tumors can be classified as
either type I or II based on the malignancy. Type II tumors are
more aggressive and have poorer survival outcomes compared with
type I (4). High-grade serous
cancers, a subtype of type II, are the most frequent and account
for 80% of OC-related deaths (5,6).
Platinum-based chemotherapy and cytoreductive surgery are the
primary clinical treatments for OC. However, this treatment
strategy is less effective for advanced OC as the lesions are
difficult to remove completely, and advanced OC tends to be
drug-resistant. Therefore, early diagnosis and targeted medicine
are crucial for the effective treatment of OC.
At present, tissue biopsy is the most common method
for cancer diagnosis, but it is invasive and time-consuming.
Additionally, biopsies carry risks such as tumor implantation and
metastasis, making them unsuitable for long-term monitoring
(7,8). Thus, novel and sensitive biomarkers
are needed for the early and quick diagnosis of cancer. All cell
types secrete extracellular vesicles (EVs), particularly exosomes,
which transport various cargoes such as lipids, proteins, DNA and
most non-coding RNAs (ncRNAs), including circular RNA (circRNA),
long ncRNA (lncRNA) and microRNA (miRNA). Exosomes can exchange
signals and materials between different cells by carrying and
transporting these cargoes (9).
Given that exosomes collect, transport and release cargo, they can
be considered new targets in OC diagnosis and treatment.
Cancer is a genetic disease primarily caused by
mutations in the non-coding genome (10). Previous genomic studies have
indicated that the non-coding regions of RNA are extensively
transcribed (11). The Human
Genome Project has demonstrated that 1.5% of the human genome
consists of genes that encode proteins; the rest are transcribed
without translation into proteins (12,13). The two subtypes of ncRNAs are
distinguished based on their size: lncRNA and small ncRNA (sncRNA).
sncRNAs can be further classified into miRNA, transfer RNA,
PIWI-interacting RNA and small nucleolar RNA (14). Particularly, miRNAs inhibit
protein translation and promote messenger RNA (mRNA) cleavage to
suppress gene expression (15).
Furthermore, miRNAs can migrate into biological fluids (16) and ~10% of circulating miRNAs are
secreted via exosomes (17,18). lncRNAs, which are transcripts of
>200 nucleotides with potential non-coding functions, exhibit
tissue- or cancer-type specificity (19). Serving as readouts of running
cellular programs or signals of certain cellular states, lncRNAs
can be used to distinguish various cellular pathologies, provide a
predictive value or even used to choose appropriate therapeutics
for patients with cancer (20).
For instance, the circulating lncRNA-GC1, derived from EVs, serves
as an early indicator of neoadjuvant chemotherapy (neoCT)
effectiveness and is correlated with improved survival outcomes in
patients with gastric cancer undergoing neoCT treatment (21). Serum exosomal lncRNA-UCA1,
initially discovered in human bladder cancer, can serve as a
potential non-invasive biomarker for the diagnosis of bladder
cancer. circRNAs are closed RNAs without 5′ or 3′ ends and are
classified into natural or synthetic circRNAs (22). In a number of tissues, endogenous
circRNAs are involved in various biological functions such as gene
transcription, protein translation, cell division, immune system
dysregulation and tumorigenesis (23).
In summary, ncRNAs in exosomes are involved in
numerous biological functions, including gene expression and
disease development, making them valuable diagnostic tools in
clinical settings. The present review summarizes the features,
isolation techniques, functions and mechanisms of exosomes and
exosomal ncRNAs and predicts their potential in diagnosing and
prognosing OC.
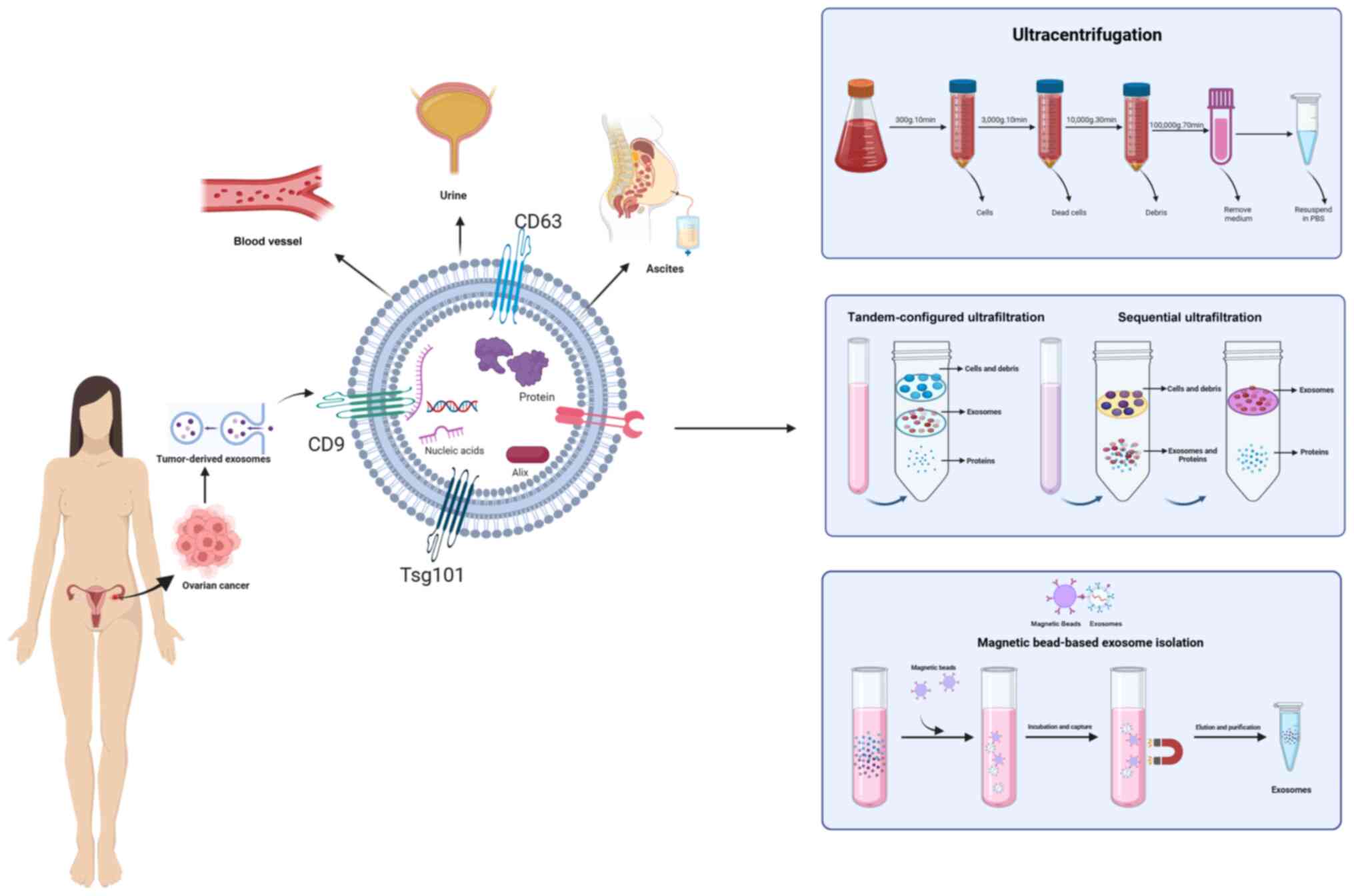
Exosomes can be isolated using several methods, such
as ultrafiltration, size exclusion chromatography and density
gradient ultracentrifugation. Most cells secrete EVs, first
reported in 1985 (24). Exosomes
are nanoparticles that facilitate cellular waste disposal into the
extracellular environment. Increasing evidence suggests that
exosomes participate in various physiological and disease-related
processes, serving as biomarkers for several diseases (25–27). Extracting exosomes is crucial for
understanding their characteristics and functions. Numerous
techniques have been developed to separate exosomes based on their
main features, such as surface proteins, size, shape and density
(Fig. 1 and Table I).
Tissue biopsy is currently the gold standard for
cancer diagnosis, but it has limitations. First, tissue biopsy only
provides a snapshot of the tumor tissue, possibly neglecting tumor
heterogeneity and the different processes and changes that occur
during interactions with the immune system and other systems.
Additionally, tissue biopsy requires time to detect cancer, thus
delaying further therapeutic decisions. Tissue biopsy also cannot
show whether a patient responds favorably to current treatment and
is impractical for frequent sample collection (28). With the development of science and
technology, liquid biopsy is regarded as a non-invasive, real-time,
tumor-specific method that can track the advancement and recurrence
of cancer and the response to treatment interventions (29). Assessing serum from patients for
human epididymis protein 4, carbohydrate antigen-125 (CA-125) and
p53 levels is one way to detect and monitor OC progression or the
response to treatment (30).
However, these methods lacked specificity and sensitivity.
Therefore, a novel approach is required to accurately predict,
diagnose and determine the prognosis of OC (14). Exosomes are among the most
important biological components of liquid biopsy and possess a
number of advantages over tumor-educated platelets (TEPs),
circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs).
For instance, exosomes display exceptional stability within
biological fluids, including plasma and urine (31). The immune conditions of the
patient and different treatments can affect the RNA content of
TEPs. Additionally, both CTCs and ctDNA have brief half-lives and
are unstable, making sample collection challenging (32,33).
The term ‘liquid biopsy’ was first proposed in 2010
when researchers used CTCs to enhance breast cancer treatment and
prognosis (34). Initially, the
use of liquid biopsies focused on identifying CTCs and ctDNA in
patients with cancer. However, components of CTCs and ctDNA are
scarce in circulating biofluids, making them inappropriate for use
as clinical diagnostic biomarkers. Conversely, other molecules
secreted from cancer cells (such as ncRNAs) are more abundant than
CTCs or ctDNA and are relatively stable in circulating biofluids
(35). In recent decades,
exosomes have been used to develop novel biomarkers, a primary goal
of translational research (26,27). Exosomes are abundant and widely
distributed in human bodily fluids, such as serum, saliva and
urine. Exosomes carry various biomolecules, including proteins,
lipids and nucleic acids, from their parent cells, making them
accessible and potential biomarkers for tumor diagnosis, prognosis
and monitoring (36).
Additionally, exosomes are crucial for tumor development, spread
and therapy resistance, particularly chemotherapy, by transferring
their contents (37). ncRNAs are
pivotal exosome components; exosomal ncRNAs, including miRNAs,
lncRNAs and circRNAs are specific biomarkers for cancer diagnosis,
prognosis, prediction and monitoring (38). With the advancements in
transcriptomic analysis, ncRNAs have been shown to affect and
participate in various aspects of OC (39), including cancer progression,
metastasis and drug resistance (40).
In general, miRISCs induce translational inhibition
by binding to target mRNA. Then, P-body (GW182) family proteins
binding to AGO are recruited, providing a scaffold to recruit
downstream effector proteins such as poly(A)-deadenylases, PAN2/3,
and the glucose-repressible alcohol dehydrogenase transcriptional
effector, carbon catabolite repression 4-negative on TATA-less
(52). Ultimately, the
chromatin-binding exonuclease, 5′-3′ exoribonuclease 1, degrades
the decapped mRNA (53,54). While most research has focused on
miRNAs that prevent target gene expression, other studies have
revealed that certain miRNAs can increase gene expression. For
instance, AGO2 and FMR1 autosomal homolog 1 are linked to certain
miRNAs, such as let-7, which stimulate translation during cell
cycle arrest (55).
miRNAs are divided into two main categories based on
their target genes: Tumor suppressor miRNAs and oncogenic miRNAs
(56). Tumor suppressor miRNAs
silence uncontrolled genetic proliferation, thereby preventing
cancer progression; tumorigenesis may occur when these miRNAs are
downregulated in tumor cells (57). Oncogenic miRNAs increase oncogene
expression and promote tumor development; tumorigenesis may occur
when these miRNAs are upregulated in tumor cells (58).
Various miRNAs are associated with OC progression,
diagnosis, prognosis and monitoring, which is summarized in
Table II. miR-34a, which
inhibits nearly 700 target genes, is a classical tumor suppressor
(58). miR-34a can also encode a
microprotein, termed miRPEP133, through an open reading frame (ORF)
in the pri-miRNA-34a. A study overexpressed miRPEP133 in the SKOV3
OC cell line, finding that it may increase apoptosis and limit cell
survival (59). Numerous studies
have assessed the expression of miR-1246 in various cancer cell
lines using various miRNA profiling techniques. The upregulation of
miR-1246 has been noted in lung and liver tissues. Compared with
normal colonic mucosa, the marked upregulation of miR-1246 in
colorectal cancer tissues bolsters its status as a promising
biomarker and suggests a role in the pathogenic processes of
colorectal cancer (60–62). OC exosomes show significant levels
of oncogenic miR-1246 expression (63). Based on data from The Cancer
Genome Atlas, OC cells can be sensitized to paclitaxel in patients
with OC receiving anti-miR-1246 therapy and exhibiting caveolin 1
(Cav1) upregulation. Moreover, miR-1246 inhibits Cav1 by acting on
platelet derived growth factor receptor β receptor, preventing cell
proliferation (64). miR-221
plays both oncogenic and tumor-suppressive roles in human cancer.
miR-221 is capable of suppressing the proliferation of lung cancer
cells, potentially through S phase arrest (65). miR-221 is upregulated in OC
tissues compared with matched normal tissues. Furthermore, patients
with high miR-221 expression have shorter overall survival (OS) and
disease-free survival times than those with low expression. miR-221
targets protease activating factor 1 (APAF1), which induces
apoptosis via gene inhibitors (66). Expressing higher levels of APAF1
by transfecting OC cell lines with miR-221 inhibitors prevents the
proliferation, migration and invasion of cancer cells in
vitro (66). According to a
previous study, miR-27b-5p expression is lower in OC cell lines and
tissues than in surrounding tissues. Furthermore, the OS rate was
lower in patients with low miRNA-27b-5p expression. This study also
verified that C-X-C motif chemokine ligand 1 (CXCL1), which is
involved in OC malignancy, is a target gene of miR-27b-5p.
Additionally, reverse transcription-quantitative PCR and
immunoblotting analyses showed that the CXCL1 mRNA and protein
levels were decreased in A2780 and OVCAR3 cells following
miR-27b-5p overexpression (67).
In total, four molecular subtypes of OC,
characterized by low overall survival rates, have been identified
through extensive transcriptional profiling of cancer tissues:
Immunoreactive, differentiated, proliferative and mesenchymal
(68,69). To further investigate the
different patterns of immune cell infiltration in OC, Liu et
al (70) utilized the
expression deconvolution algorithm, CIBERSORT, to confirm the
distinct infiltration of immune cells in the four subtypes.
Concentrated in plasma cells, the mesenchymal subtype of high-grade
serous ovarian cancer (HGSOC) can induce the mesenchymal features
of OC by releasing exosomes. Exosomal miRNA profiling revealed that
plasma cell-derived miR-330-3p is an essential modulator of
mesenchymal identity in OC. miR-330-3p increases the transcription
of junctional adhesion molecule 2 through enhancer-induced gene
activation pathways (71).
The promoter regions of lncRNA genes are conserved,
similar to those of protein-coding genes, and the production of
lncRNAs controlled by these genes is tissue-specific (77). At present, different methods are
used to classify lncRNAs, mainly depending on their function,
interactions with protein-coding genes, structure, sequence and
metabolism (78). lncRNAs can be
categorized into four functional groups: Skeleton, guide, signal
and bait molecules. In terms of interactions with protein-coding
genes, lncRNAs fall into four categories: Overlapping,
bidirectional, intron and cis-antisense (79). lncRNAs are crucial for various
biological processes, including: i) Engaging with chromatin
complexes to support the regulation of epigenetic genes; ii) acting
as proteins or multiprotein compound modulators; iii) influencing
transcriptional expression by binding proteins linked to DNA/RNA;
iv) controlling the stability of DNA by forming triple helices and
R-loops; and v) assisting in the creation of higher-order chromatin
structures (80).
At present, RNAs are the main targets for
therapeutic and diagnostic strategies. ncRNAs, for instance, show
notable variety in nucleotide length and can form different
structures when interacting with proteins, DNA and other
substances. Notably, lncRNAs are more likely to form intricate
secondary or tertiary structures due to their long nucleotide
sequences. These structures, similar to enzymes, fulfill biological
functions through their unique domains that show reproducibility
and conservatism (81). The
function of a number of proteins is determined by their distinct
structural domains. However, current understanding of the
structural domains that determine their functionality and define
their interactome is lacking. Recently, various methods have been
developed to assess the secondary structure of lncRNAs (82). These techniques are roughly
divided into two types: Experimental and computational methods.
Experimental techniques include enzymatic footprinting, chemical
probing, nuclear magnetic resonance, small-angle scattering, atomic
force microscopy and cryo-electron microscopy (81).
Various mechanisms have demonstrated that lncRNAs
are involved in cancer progression. In the clinic, most OC cases
have abdominal cavity metastases. Thus, the
epithelial-to-mesenchymal transition (EMT) pathway serves a crucial
role in tumor cell dissemination. The mechanisms by which lncRNAs
mediate EMT in OC are diverse (83). For instance, through
bioinformatics analysis, a previous study identified lncRNA
pro-transition associated RNA as a lncRNA-mediated competing
endogenous RNA (ceRNA) that competitively inhibits miR-101 to
modulate the expression of zinc finger E-box binding homeobox 1
(84). A number of patients with
OC develop carboplatin resistance. Chemotherapy-resistant
strategies often involve multipotent mesenchymal stem cells or an
increased EMT phenotype in cancer (85,86). In a clinical study, which included
134 primary OC cases (63 treated with carboplatin, 55 treated with
cisplatin and 16 without therapy), expression of the lncRNA HOX
transcript antisense RNA (HOTAIR) and the corresponding DNA
methylation were detected. The research showed that patients with
high HOTAIR expression receiving carboplatin had significantly
lower survival rates, whereas this effect was not observed in
patients who did not receive carboplatin (87). To reduce transcription, HOTAIR
attracts polycomb repressive complex 2 (PRC2) to certain polycomb
group target (PCGT) genes, particularly in embryonic fibroblasts
(88). During differentiation,
PCGTs essential for the identity of specialized cells are
derepressed (89,90). However, promoters of these stem
cell PCGTs are methylated and repressed in cancer (91). In several cancer types, high
HOTAIR expression levels are strongly associated with metastasis,
cancer invasiveness and poor prognosis (88,92).
The OC process is aided by ferroptosis, an
iron-dependent type of cell death. One of the predominant methods
used in cancer therapy involves triggering apoptosis to eradicate
malignant cells. lncRNAs can regulate ferroptosis, a process that
can trigger apoptosis and may therefore be beneficial in cancer
treatment (93). A recent study
found that lncRNA CACNA1G antisense RNA 1 promotes ferritin heavy
chain 1 synthesis by reducing ferroptosis through insulin like
growth factor 2 mRNA binding protein 1-mediated N6-methyladenosine
(m6A) modification, which enhances the migration and division of OC
cells (94). Additionally,
another study showed that lncRNA ADAMTS9 antisense RNA 1 sponges
miR-587 in OC, preventing ferroptosis and enhancing the expression
of solute carrier family 7 member 11 (95).
Numerous studies have shown that lncRNAs can control
the expression of tumor suppressor genes to either promote or
prevent new tumor growth. Leukemia inhibitory factor receptor
antisense RNA1 (LIFR-AS1), a tumor suppressor gene in colorectal
cancer, was found to be downregulated in serous ovarian carcinoma
(SOC) in a previous study. Generally, patients with SOC and low
LIFR-AS1 expression had a poor prognosis. Low LIFR-AS1 expression
was also associated with tumor size, clinical stage, lymph node
metastasis and distant metastasis (96). LIFR-AS1 upregulation enhances the
production of cleaved caspase-3 and E-cadherin, suppressing the
malignant activities of SOC cells, including their migration,
invasion and proliferation (96).
lncRNAs can also repress tumor progression by regulating oncogene
expression. For instance, long intergenic non-protein-coding RNA
857 (LINC00857), which inactivates the Hippo signaling pathway, can
accelerate OC progression by regulating yes-associated protein 1
(YAP1), which acts as an oncogene in OC. LINC00857 competitively
binds to miR-486-5p to regulate YAP1 expression, which elevates OC
progression (97). A summary of
the oncogenic lncRNAs in critical signaling pathways in OC is
detailed in Table III.
When a tumor suppressor gene exon is spliced,
circRNAs join in a different order from their genomic sequences.
circRNAs were first described in 1976 (98). In 2015, high-throughput
technologies were used to ascertain that circRNAs were two times
more abundant in EVs than in parental cells (99). Premature mRNAs, which often do not
encode proteins, can be alternatively spliced to become circRNAs,
which are ncRNAs. circRNAs are more stable than other linear RNAs
as they form covalently closed loops without 5′- or 3′-end features
(100). This structure makes
circRNAs resistant to degradation by classical RNA pathways,
leading to their exocytosis from cells (101–103). circRNAs were initially thought
to be non-functional byproducts of splicing mistakes. However, as
high-throughput technologies and bioinformatics research progress,
there is a growing body of evidence indicating that they exist
widely in eukaryotic cells, where they regulate miRNAs and proteins
to participate in various biological processes (104,105). Moreover, these circRNAs have
been found to be stable and extremely conservative (99,106). circRNA expression in tumor
tissues differs from that in normal tissues, indicating their
involvement in tumorigenesis (107). circRNAs originate from various
genomic regions, including intergenic, intronic, antisense and
untranslated regions, and are categorized into three groups based
on the region from which they are derived: Intronic, exon-intron
and exonic (108). circRNAs have
three major biological functions (109), including: i) Controlling the
expression of target genes by competitively binding to miRNAs,
similar to lncRNAs, acting as sponges for miRNAs or ceRNAs
(110); ii) controlling the
transcription, splicing, translation and interaction of genes with
RNA-binding proteins to control the expression of genes (111); and iii) serving a role in
polysomes, initiating putative ORFs and the AUG codon, and encoding
regulatory peptides (112).
circRNAs are involved in the development, metastasis
and carcinogenesis of numerous human disorders (113,114). For instance, in
platinum-resistant OC, the expression of circPLPP4 is notably
elevated. Functionally, circPLPP4 operates as a molecular sponge
for miR-136, effectively sequestering it. By doing so, circPLPP4
competitively increases the expression of PIK3R1, thereby
contributing to resistance against cisplatin (115). Hsa_circ_0004712 exhibits
abundant regulation in both OC tissues and cellular models, and
functions by specifically targeting and reducing the expression of
miR-331-3p. FZD4 was identified as a downstream target of
miR-331-3p, experiencing repression when miR-331-3p levels were
high, yet its expression was notably enhanced in the presence of
hsa_circ_0004712, illustrating a regulatory axis between the
circular RNA, microRNA and target gene (116). A summary of the oncogenic
circRNAs in critical signaling pathways in OC is detailed in
Table IV.
The majority of the tumor mass consists of the TME,
which is mostly composed of tumor stroma (117). Stem cells, fibroblasts,
endothelial cells, immune cells and numerous other cells that
produce growth factors and cytokines are found in the TME (118). EVs are essential for
communication between stromal and tumor cells in both nearby and
distant microenvironments. EVs support tumor growth and establish a
complex microenvironment called the pre-metastatic niche (PMN). The
PMN has been primed by the primary tumor in distant organs or
regions free of tumors, anticipating metastasis spread (119). The characteristic features of
the PMN include extracellular matrix remodeling and deposition,
lymphangiogenesis, vascular permeability, angiogenesis and
immunological suppression (120).
Among the deadliest gynecological cancers, OC
frequently results in peritoneal metastases, leading to higher
recurrence rates and resistance to standard platinum-based
treatments, which can result in a poor prognosis (121,122). Metastatic cancer cells spread to
secondary sites, where they find a supportive TME owing to the
biological components of the host. For instance, OC-generated EVs
target YAP1, a key effector of the Hippo pathway. This interaction
reduces the nuclear YAP1/transcriptional co-activator with
PDZ-binding motif protein ratio and increases CXCL1 production by
stromal fibroblasts. Pro-inflammatory cytokines, such as CXCL1,
play a crucial role in promoting tumor development and metastasis
(123). This finding indicates
that pro-inflammatory CXCL1 is extensively expressed in the TME of
ovarian malignancies and has oncogenic-promoting properties
(123). Exosomes derived from OC
that contain activating transcription factor 2 and
metastasis-associated protein 1 may enhance angiogenesis (124). Tumor-secreted EVs (TEVs) are
taken up by macrophages and lymphocytes in animal models,
significantly decreasing the cross-presentation of tumor antigens
from dendritic cells, which in turn affects CD8+ T cell
activity (125). Another study
demonstrated that TEVs expressing programmed cell death ligand 1
reduced the number of CD8+ T cells that invaded the
lymph nodes (126).
Phosphatidylserine, which impedes T-cell activation by obstructing
intracellular signaling cascades, is present in some exosomes from
OC (127). To counteract the
immunosuppressive effects, normal cells, particularly immune cells,
also produce EVs. These findings highlight the critical role of EVs
in controlling the immune system. For instance, exosomes release
cytokines, respond to specific antigens and mitogens, stimulate or
inhibit B cell antibody production and directly kill target cells
(128). Cytotoxic EVs are also
released by natural killer (NK) cells. Fais et al (129) provided the first evidence that
NK-derived EVs are lethal to cancer cells. It was shown that NK
cells obtained from healthy donors, both in the resting and active
states, were capable of killing a range of cancer cell lines and
extending their cytotoxicity to resting immune cells but not to
activated cells. Another study has reported that NK-derived EVs
exhibit characteristic protein markers that are typical of NK
cells, showing a propensity to be selectively internalized by SKOV3
cells, and demonstrating cytotoxic effects against OC cells
(130). FasL and perforin are
expressed in NK-derived EVs. These proteins can induce target cell
demise via two pathways: i) A conventional ligand/receptor
engagement between the membrane-bound FasL of NK-derived EVs and
the Fas receptor on the surface of the target cell and; ii) an
atypical cytotoxic mechanism initiated upon the uptake of exosomes
by the target cells, potentially involving the intracellular
release of effector molecules, such as perforin (129).
EVs have become important mediators of immunological
evasion and repression of complex interactions between tumors and
normal cells. TEVs disrupt immunological functions and diminish the
effectiveness of immunotherapy. Normal cells, particularly immune
cells, release EVs to counteract these effects.
Current clinical screening methods for OC include
imaging and blood testing. Imaging techniques comprise positron
emission tomography, magnetic resonance imaging, computed
tomography and ultrasound screening. Serum tests primarily measure
CA-125 and cancer antigen 19-9 (131). Common diagnostic methods for OC,
such as pelvic examination, ultrasound screening and CA-125
measurement, lack sensitivity and specificity (132), and only 50–60% of individuals
diagnosed with stage I or II OC exhibit elevated CA-125 levels
(133). Therefore, the discovery
of new biomarkers is urgently needed to improve detection rates.
Patients with OC show specific miRNA expression patterns in
exosomes (134). Thus, these
patterns may serve as potential biomarkers for various cancer
types, including OC. For instance, ~95 exosomal miRNAs are
differentially expressed in patients with epithelial ovarian cancer
(EOC) and healthy donors (135).
Individuals with EOC exhibit significantly lower levels of
miR-320d, miR-4479 and miR-6763-5p than healthy donors. According
to a study that used receiver operating characteristic curves to
assess the diagnostic efficiency of exosomal miRNAs, these three
exosomal miRNAs have diagnostic potential (135). A study also reported that
exosomal miR-200b and CA-125, which are used for EOC screening, are
associated with OS. According to this study, miR-200b influences
cell proliferation and apoptosis. Furthermore, exosomal miR-200b
can distinguish patients with EOC from healthy individuals with a
64% sensitivity and 86% specificity (136). Additionally, the level of serum
exosomal miR-34a can be used to differentiate patients with
early-stage and advanced-stage OC, suggesting that miR-34 is a
potential biomarker for OC (137).
Exosomal ncRNAs show promise as potential novel
diagnostic biomarkers for OC and are currently in the preclinical
stage of development. Numerous prior studies have demonstrated that
exosomal ncRNAs can function as prognostic biomarkers for OC. For
instance, exosomal metastasis-associated lung adenocarcinoma
transcript 1 (MALAT1) has been linked to more malignant tendencies
and a late International Federation of Gynecology and Obstetrics
stage of EOC. Based on nomogram modeling and multivariate survival
analysis, serum exosomal MALAT1 may be a potential predictive
biomarker for EOC (138). Zhang
et al (139) demonstrated
that OS was associated with high levels of exosomal miR-200b.
Additionally, circFoxp1 is identified as an independent factor that
can predict survival and disease recurrence in individuals with
EOC, according to the International Union of Obstetrics and
Gynecology (140). Unlike other
human cancer types, OC preferably invades the peritoneal cavity,
with a particular potency at interacting with different viscera
inside the compartment (121).
The exosomes of the primary ovarian tumor could prime the distant
TME for an expedited metastatic invasion. Within the TME, exosomes
control intercellular communication between tumor cells and the
normal stroma, cancer-associated fibroblasts and local immune
cells.
Globally, epithelial ovarian cancer leads to
>185,000 fatalities annually. HGSOC, the predominant variant,
contributes to ~60% of these deaths. Despite advancements in
surgical techniques and chemotherapy protocols, the mortality rate
for HGSOC has remained high over the past several decades (141). The primary cause is the lack of
reliable biomarkers for early diagnosis. Exosomes, which contain
numerous ncRNAs, have increasingly been recognized as key players
in intercellular communication. ncRNAs, including circRNAs, lncRNAs
and miRNAs, are crucial for tumor development. Exosomal ncRNAs can
be packaged, secreted and transported by tumor and normal cells to
influence each other and complete the establishment of OC. Exosomes
from OC could stimulate the establishment of PMNs by
immunosuppression, angiogenesis, stromal cell modification and
oncogenic reprogramming to promote tumor metastasis and growth.
Although exosomes and exosomal ncRNAs are innovative indicators for
the prognosis and diagnosis of OC, several issues hinder their use
in clinical settings. First, only a limited quantity of exosomes is
present in biological fluids. Second, there is limited knowledge
about the biological functions of ncRNAs. Despite these challenges,
exosomes and ncRNAs are promising biomarkers for the identification
and management of OC.
Not applicable.
Funding: No funding was received.
Not applicable.
XW was responsible for writing and revising the
manuscript. MY participated in the literature review and provided
feedback. JZ and YZ provided substantial contributions to
conception and design. GL was responsible for revising the
manuscript and providing financial support. Data authentication is
not applicable. All the authors read and approved the final version
of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Dr Gencui Li ORCID: 0009-0004-1397-3204.
|
1
|
Yang C, Kim HS, Song G and Lim W: The
potential role of exosomes derived from ovarian cancer cells for
diagnostic and therapeutic approaches. J Cell Physiol.
234:21493–21503. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stewart C, Ralyea C and Lockwood S:
Ovarian cancer: An integrated review. Semin Oncol Nurs. 35:151–156.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koshiyama M, Matsumura N and Konishi I:
Subtypes of ovarian cancer and ovarian cancer screening.
Diagnostics (Basel). 7:122017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lheureux S, Braunstein M and Oza AM:
Epithelial ovarian cancer: Evolution of management in the era of
precision medicine. CA Cancer J Clin. 69:280–304. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lisio MA, Fu L, Goyeneche A, Gao ZH and
Telleria C: High-grade serous ovarian cancer: Basic sciences,
clinical and therapeutic standpoints. Int J Mol Sci. 20:9522019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sierra J, Marrugo-Ramirez J,
Rodríguez-Trujillo R, Mir M and Samitier J: Sensor-integrated
microfluidic approaches for liquid biopsies applications in early
detection of cancer. Sensors (Basel). 20:13172020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bellassai N, D'Agata R, Jungbluth V and
Spoto G: Surface plasmon resonance for biomarker detection:
Advances in non-invasive cancer diagnosis. Front Chem. 7:5702019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miao M, Miao Y, Zhu Y, Wang J and Zhou H:
Advances in exosomes as diagnostic and therapeutic biomarkers for
gynaecological malignancies. Cancers (Basel). 14:47432022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maurano MT, Humbert R, Rynes E, Thurman
RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et
al: Systematic localization of common disease-associated variation
in regulatory DNA. Science. 337:1190–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morris KV and Mattick JS: The rise of
regulatory RNA. Nat Rev Genet. 15:423–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Beg A, Parveen R, Fouad H, Yahia ME and
Hassanein AS: Role of different non-coding RNAs as ovarian cancer
biomarkers. J Ovarian Res. 15:722022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu G, Brkić J, Hayder H and Peng C:
MicroRNAs in human placental development and pregnancy
complications. Int J Mol Sci. 14:5519–5544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Théry C: Exosomes: Secreted vesicles and
intercellular communications. F1000 Biol Rep. 3:152011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo X, Gao Y, Song Q, Wei J, Wu J, Dong J,
Chen L, Xu S, Wu D, Yang X, et al: Early assessment of circulating
exosomal lncRNA-GC1 for monitoring neoadjuvant chemotherapy
response in gastric cancer. Int J Surg. 109:1094–1104. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Obi P and Chen YG: The design and
synthesis of circular RNAs. Methods. 196:85–103. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan BT, Teng K, Wu C, Adam M and Johnstone
RM: Electron microscopic evidence for externalization of the
transferrin receptor in vesicular form in sheep reticulocytes. J
Cell Biol. 101:942–948. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Isaac R, Reis FCG, Ying W and Olefsky JM:
Exosomes as mediators of intercellular crosstalk in metabolism.
Cell Metab. 33:1744–1762. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qi Y, Xu R, Song C, Hao M, Gao Y, Xin M,
Liu Q, Chen H, Wu X, Sun R, et al: A comprehensive database of
exosome molecular biomarkers and disease-gene associations. Sci
Data. 11:2102024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kalluri R and McAndrews KM: The role of
extracellular vesicles in cancer. Cell. 186:1610–1626. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mannelli C: Tissue vs liquid biopsies for
cancer detection: Ethical issues. J Bioeth Inq. 16:551–557. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie H and Kim RD: The application of
circulating tumor DNA in the screening, surveillance, and treatment
monitoring of colorectal cancer. Ann Surg Oncol. 28:1845–1858.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang WL, Lu Z, Guo J, Fellman BM, Ning J,
Lu KH, Menon U, Kobayashi M, Hanash SM, Celestino J, et al: Human
epididymis protein 4 antigen-autoantibody complexes complement
cancer antigen 125 for detecting early-stage ovarian cancer.
Cancer. 126:725–736. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu W, Hurley J, Roberts D, Chakrabortty
SK, Enderle D, Noerholm M, Breakefield XO and Skog JK:
Exosome-based liquid biopsies in cancer: Opportunities and
challenges. Ann Oncol. 32:466–477. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kalluri R: The biology and function of
exosomes in cancer. J Clin Invest. 126:1208–1215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Spina V and Rossi D: Liquid biopsy in
tissue-born lymphomas. Swiss Med Wkly. 149:w147092019.PubMed/NCBI
|
|
34
|
Lianidou ES, Mavroudis D, Sotiropoulou G,
Agelaki S and Pantel K: What's new on circulating tumor cells? A
meeting report. Breast Cancer Res. 12:3072010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shigeyasu K, Toden S, Zumwalt TJ, Okugawa
Y and Goel A: Emerging role of MicroRNAs as liquid biopsy
biomarkers in gastrointestinal cancers. Clin Cancer Res.
23:2391–2399. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ebrahimi N, Faghihkhorasani F, Fakhr SS,
Moghaddam PR, Yazdani E, Kheradmand Z, Rezaei-Tazangi F, Adelian S,
Mobarak H, Hamblin MR and Aref AR: Tumor-derived exosomal
non-coding RNAs as diagnostic biomarkers in cancer. Cell Mol Life
Sci. 79:5722022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li C, Ni YQ, Xu H, Xiang QY, Zhao Y, Zhan
JK, He JY, Li S and Liu YS: Roles and mechanisms of exosomal
non-coding RNAs in human health and diseases. Signal Transduct
Target Ther. 6:3832021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj
A, Burnell M, Kalsi JK, Amso NN, Apostolidou S, Benjamin E,
Cruickshank D, et al: Ovarian cancer screening and mortality in the
UK collaborative trial of ovarian cancer screening (UKCTOCS): A
randomised controlled trial. Lancet. 387:945–956. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gareev I, Beylerli O, Yang G, Sun J,
Pavlov V, Izmailov A, Shi H and Zhao S: The current state of MiRNAs
as biomarkers and therapeutic tools. Clin Exp Med. 20:349–359.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Toden S and Goel A: Non-coding RNAs as
liquid biopsy biomarkers in cancer. Br J Cancer. 126:351–360. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Z, Qin YW, Brewer G and Jing Q:
MicroRNA degradation and turnover: Regulating the regulators. Wiley
Interdiscip Rev RNA. 3:593–600. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Denli AM, Tops BB, Plasterk RHA, Ketting
RF and Hannon GJ: Processing of primary microRNAs by the
Microprocessor complex. Nature. 432:231–235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Han J, Lee Y, Yeom KH, Kim YK, Jin H and
Kim VN: The Drosha-DGCR8 complex in primary microRNA processing.
Genes Dev. 18:3016–3027. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki
S, Liu Q and Tomari Y: ATP-dependent human RISC assembly pathways.
Nat Struct Mol Biol. 17:17–23. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Broughton JP, Lovci MT, Huang JL, Yeo GW
and Pasquinelli AE: Pairing beyond the seed supports MicroRNA
targeting specificity. Mol Cell. 64:320–333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Braun JE, Huntzinger E and Izaurralde E:
The role of GW182 proteins in miRNA-mediated gene silencing. Adv
Exp Med Biol. 768:147–163. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hayder H, O'Brien J, Nadeem U and Peng C:
MicroRNAs: Crucial regulators of placental development.
Reproduction. 155:R259–R271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Christie M, Boland A, Huntzinger E,
Weichenrieder O and Izaurralde E: Structure of the PAN3
pseudokinase reveals the basis for interactions with the PAN2
deadenylase and the GW182 proteins. Mol Cell. 51:360–373. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vasudevan S and Steitz JA:
AU-rich-element-mediated upregulation of translation by FXR1 and
argonaute 2. Cell. 128:1105–1118. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kang M, Tang B, Li J, Zhou Z, Liu K, Wang
R, Jiang Z, Bi F, Patrick D, Kim D, et al: Identification of
miPEP133 as a novel tumor-suppressor microprotein encoded by
miR-34a pri-miRNA. Mol Cancer. 19:1432020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang WC, Chin TM, Yang H, Nga ME, Lunny
DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, et al:
Tumour-initiating cell-specific miR-1246 and miR-1290 expression
converge to promote non-small cell lung cancer progression. Nat
Commun. 7:117022016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chai S, Ng KY, Tong M, Lau EY, Lee TK,
Chan KW, Yuan YF, Cheung TT, Cheung ST, Wang XQ, et al: Octamer 4/
microRNA-1246 signaling axis drives Wnt/β-catenin activation in
liver cancer stem cells. Hepatology. 64:2062–2076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cooks T, Pateras IS, Jenkins LM, Patel KM,
Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG and Harris
CC: Mutant p53 cancers reprogram macrophages to tumor supporting
macrophages via exosomal miR-1246. Nat Commun. 9:7712018.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang W, Jo H, Park S, Kim H, Kim SI, Han
Y, Lee J, Seol A, Kim J, Lee M, et al: Integrated analysis of
ascites and plasma extracellular vesicles identifies a miRNA-based
diagnostic signature in ovarian cancer. Cancer Lett.
542:2157352022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kanlikilicer P, Bayraktar R, Denizli M,
Rashed MH, Ivan C, Aslan B, Mitra R, Karagoz K, Bayraktar E, Zhang
X, et al: Exosomal miRNA confers chemo resistance via targeting
Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine.
38:100–112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yamashita R, Sato M, Kakumu T, Hase T,
Yogo N, Maruyama E, Sekido Y, Kondo M and Hasegawa Y: Growth
inhibitory effects of miR-221 and miR-222 in non-small cell lung
cancer cells. Cancer Med. 4:551–564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li J, Li Q, Huang H, Li Y, Li L, Hou W and
You Z: Overexpression of miRNA-221 promotes cell proliferation by
targeting the apoptotic protease activating factor-1 and indicates
a poor prognosis in ovarian cancer. Int J Oncol. 50:1087–1096.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu CH, Jing XN, Liu XL, Qin SY, Liu MW
and Hou CH: Tumor-suppressor miRNA-27b-5p regulates the growth and
metastatic behaviors of ovarian carcinoma cells by targeting CXCL1.
J Ovarian Res. 13:922020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tothill RW, Tinker AV, George J, Brown R,
Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro
B, et al: Novel molecular subtypes of serous and endometrioid
ovarian cancer linked to clinical outcome. Clin Cancer Res.
14:5198–5208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu R, Hu R, Zeng Y, Zhang W and Zhou HH:
Tumour immune cell infiltration and survival after platinum-based
chemotherapy in high-grade serous ovarian cancer subtypes: A gene
expression-based computational study. EBioMedicine. 51:1026022020.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yang Z, Wang W, Zhao L, Wang X, Gimple RC,
Xu L, Wang Y, Rich JN and Zhou S: Plasma cells shape the
mesenchymal identity of ovarian cancers through transfer of
exosome-derived microRNAs. Sci Adv. 7:eabb07372021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Brannan CI, Dees EC, Ingram RS and
Tilghman SM: The product of the H19 gene may function as an RNA.
Mol Cell Biol. 10:28–36. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhao L, Sun W, Zheng A, Zhang Y, Fang C
and Zhang P: Ginsenoside Rg3 suppresses ovarian cancer cell
proliferation and invasion by inhibiting the expression of lncRNA
H19. Acta Biochim Pol. 68:575–582. 2021.PubMed/NCBI
|
|
74
|
Hartford CCR and Lal A: When long
noncoding becomes protein coding. Mol Cell Biol. 40:e00528–19.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mattick JS, Amaral PP, Carninci P,
Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME,
Fitzgerald KA, et al: Long non-coding RNAs: Definitions, functions,
challenges and recommendations. Nat Rev Mol Cell Biol. 24:430–447.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang H, Meng Q, Qian J, Li M, Gu C and
Yang Y: Review: RNA-based diagnostic markers discovery and
therapeutic targets development in cancer. Pharmacol Ther.
234:1081232022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Graf J and Kretz M: From structure to
function: Route to understanding lncRNA mechanism. Bioessays.
42:e20000272020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Seetin MG and Mathews DH: RNA structure
prediction: An overview of methods. Methods Mol Biol. 905:99–122.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lampropoulou DI, Papadimitriou M,
Papadimitriou C, Filippou D, Kourlaba G, Aravantinos G and Gazouli
M: The role of EMT-related lncRNAs in ovarian cancer. Int J Mol
Sci. 24:100792023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liang H, Yu T, Han Y, Jiang H, Wang C, You
T, Zhao X, Shan H, Yang R, Yang L, et al: LncRNA PTAR promotes EMT
and invasion-metastasis in serous ovarian cancer by competitively
binding miR-101-3p to regulate ZEB1 expression. Mol Cancer.
17:1192018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Leung D, Price ZK, Lokman NA, Wang W,
Goonetilleke L, Kadife E, Oehler MK, Ricciardelli C, Kannourakis G
and Ahmed N: Platinum-resistance in epithelial ovarian cancer: An
interplay of epithelial-mesenchymal transition interlinked with
reprogrammed metabolism. J Transl Med. 20:5562022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kralj J, Pernar Kovač M, Dabelić S,
Polančec DS, Wachtmeister T, Köhrer K and Brozovic A: Transcriptome
analysis of newly established carboplatin-resistant ovarian cancer
cell model reveals genes shared by drug resistance and drug-induced
EMT. Br J Cancer. 128:1344–1359. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Teschendorff AE, Lee SH, Jones A, Fiegl H,
Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, et al:
HOTAIR and its surrogate DNA methylation signature indicate
carboplatin resistance in ovarian cancer. Genome Med. 7:1082015.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Bernstein BE, Mikkelsen TS, Xie X, Kamal
M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al:
A bivalent chromatin structure marks key developmental genes in
embryonic stem cells. Cell. 125:315–326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Lee TI, Jenner RG, Boyer LA, Guenther MG,
Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K,
et al: Control of developmental regulators by polycomb in human
embryonic stem cells. Cell. 125:301–313. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Teschendorff AE, Menon U, Gentry-Maharaj
A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell
CG, Maxwell AP, et al: Age-dependent DNA methylation of genes that
are suppressed in stem cells is a hallmark of cancer. Genome Res.
20:440–446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW,
Jin HY, Zhang Y, Li Q and Hua KQ: Overexpression of long non-coding
RNA HOTAIR predicts poor patient prognosis and promotes tumor
metastasis in epithelial ovarian cancer. Gynecol Oncol.
134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yang S, Ji J, Wang M, Nie J and Wang S:
Construction of ovarian cancer prognostic model based on the
investigation of ferroptosis-related lncRNA. Biomolecules.
13:3062023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jin Y, Qiu J, Lu X, Ma Y and Li G: LncRNA
CACNA1G-AS1 up-regulates FTH1 to inhibit ferroptosis and promote
malignant phenotypes in ovarian cancer cells. Oncol Res.
31:169–179. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Cai L, Hu X, Ye L, Bai P, Jie Y and Shu K:
Long non-coding RNA ADAMTS9-AS1 attenuates ferroptosis by Targeting
microRNA-587/solute carrier family 7 member 11 axis in epithelial
ovarian cancer. Bioengineered. 13:8226–8239. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Liu F, Cao L, Zhang Y, Xia X and Ji Y:
LncRNA LIFR-AS1 overexpression suppressed the progression of serous
ovarian carcinoma. J Clin Lab Anal. 36:e254702022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lin X, Feng D, Li P and Lv Y: LncRNA
LINC00857 regulates the progression and glycolysis in ovarian
cancer by modulating the Hippo signaling pathway. Cancer Med.
9:8122–8132. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.PubMed/NCBI
|
|
101
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Yao T, Chen Q, Fu L and Guo J: Circular
RNAs: Biogenesis, properties, roles, and their relationships with
liver diseases. Hepatol Res. 47:497–504. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extracellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ngo LH, Bert AG, Dredge BK, Williams T,
Murphy V, Li W, Hamilton WB, Carey KT, Toubia J, Pillman KA, et al:
Nuclear export of circular RNA. Nature. 627:212–220. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Pisignano G, Michael DC, Visal TH, Pirlog
R, Ladomery M and Calin GA: Going circular: History, present, and
future of circRNAs in cancer. Oncogene. 42:2783–2800. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Shang Q, Yang Z, Jia R and Ge S: The novel
roles of circRNAs in human cancer. Mol Cancer. 18:62019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ye D, Gong M, Deng Y, Fang S, Cao Y, Xiang
Y and Shen Z: Roles and clinical application of exosomal circRNAs
in the diagnosis and treatment of malignant tumors. J Transl Med.
20:1612022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang
Y, Li X, Wu Z, Yang D, Zhou Y, et al: Circular RNAs in cancer:
Emerging functions in hallmarks, stemness, resistance and roles as
potential biomarkers. Mol Cancer. 18:902019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Cortes R and Forner MJ: Circular RNAS:
Novel biomarkers of disease activity in systemic lupus
erythematosus? Clin Sci (Lond). 133:1049–1052. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Liu CX and Chen LL: Circular RNAs:
Characterization, cellular roles, and applications. Cell.
185:2016–2034. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhao Q, Liu J, Deng H, Ma R, Liao JY,
Liang H, Hu J, Li J, Guo Z, Cai J, et al: Targeting
mitochondria-located circRNA SCAR alleviates NASH via reducing mROS
output. Cell. 183:76–93.e22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Huang D, Zhu X, Ye S, Zhang J, Liao J,
Zhang N, Zeng X, Wang J, Yang B, Zhang Y, et al: Tumour circular
RNAs elicit anti-tumour immunity by encoding cryptic peptides.
Nature. 625:593–602. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li H, Lin R, Zhang Y, Zhu Y, Huang S, Lan
J, Lu N, Xie C, He S and Zhang W: N6-methyladenosine-modified
circPLPP4 sustains cisplatin resistance in ovarian cancer cells via
PIK3R1 upregulation. Mol Cancer. 23:52024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhou X, Jiang J and Guo S:
Hsa_circ_0004712 downregulation attenuates ovarian cancer malignant
development by targeting the miR-331-3p/FZD4 pathway. J Ovarian
Res. 14:1182021. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Camuzard O, Santucci-Darmanin S, Carle GF
and Pierrefite-Carle V: Autophagy in the crosstalk between tumor
and microenvironment. Cancer Lett. 490:143–153. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ye J, Wu D, Wu P, Chen Z and Huang J: The
cancer stem cell niche: Cross talk between cancer stem cells and
their microenvironment. Tumour Biol. 35:3945–3951. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Kaplan RN, Riba RD, Zacharoulis S, Bramley
AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et
al: VEGFR1-positive haematopoietic bone marrow progenitors initiate
the pre-metastatic niche. Nature. 438:820–827. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Yue M, Hu S, Sun H, Tuo B, Jia B, Chen C,
Wang W, Liu J, Liu Y, Sun Z and Hu J: Extracellular vesicles
remodel tumor environment for cancer immunotherapy. Mol Cancer.
22:2032023. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Mei S, Chen X, Wang K and Chen Y: Tumor
microenvironment in ovarian cancer peritoneal metastasis. Cancer
Cell Int. 23:112023. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lorusso D, Ceni V, Daniele G, Salutari V,
Pietragalla A, Muratore M, Nero C, Ciccarone F and Scambia G: Newly
diagnosed ovarian cancer: Which first-line treatment? Cancer Treat
Rev. 91:1021112020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Mo Y, Leung LL, Mak CSL, Wang X, Chan WS,
Hui LMN, Tang HWM, Siu MKY, Sharma R, Xu D, et al: Tumor-secreted
exosomal miR-141 activates tumor-stroma interactions and controls
premetastatic niche formation in ovarian cancer metastasis. Mol
Cancer. 22:42023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Yi H, Ye J, Yang XM, Zhang LW, Zhang ZG
and Chen YP: High-grade ovarian cancer secreting effective exosomes
in tumor angiogenesis. Int J Clin Exp Pathol. 8:5062–5070.
2015.PubMed/NCBI
|
|
125
|
Leary N, Walser S, He Y, Cousin N, Pereira
P, Gallo A, Collado-Diaz V, Halin C, Garcia-Silva S, Peinado H and
Dieterich LC: Melanoma-derived extracellular vesicles mediate
lymphatic remodelling and impair tumour immunity in draining lymph
nodes. J Extracell Vesicles. 11:e121972022. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Chen G, Huang AC, Zhang W, Zhang G, Wu M,
Xu W, Yu Z, Yang J, Wang B, Sun H, et al: Exosomal PD-L1
contributes to immunosuppression and is associated with anti-PD-1
response. Nature. 560:382–386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Kelleher RJ Jr, Balu-Iyer S, Loyall J,
Sacca AJ, Shenoy GN, Peng P, Iyer V, Fathallah AM, Berenson CS,
Wallace PK, et al: Extracellular vesicles present in human ovarian
tumor microenvironments induce a phosphatidylserine-dependent
arrest in the T-cell signaling cascade. Cancer Immunol Res.
3:1269–1278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Arenaccio C, Chiozzini C, Columba-Cabezas
S, Manfredi F, Affabris E, Baur A and Federico M: Exosomes from
human immunodeficiency virus type 1 (HIV-1)-infected cells license
quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and
ADAM17-dependent mechanism. J Virol. 88:11529–11539. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lugini L, Cecchetti S, Huber V, Luciani F,
Macchia G, Spadaro F, Paris L, Abalsamo L, Colone M, Molinari A, et
al: Immune surveillance properties of human NK cell-derived
exosomes. J Immunol. 189:2833–2842. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Luo H, Zhou Y, Zhang J, Zhang Y, Long S,
Lin X, Yang A, Duan J, Yang N, Yang Z, et al: NK cell-derived
exosomes enhance the anti-tumor effects against ovarian cancer by
delivering cisplatin and reactivating NK cell functions. Front
Immunol. 13:10876892023. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Shiao MS, Chang JM, Lertkhachonsuk AA,
Rermluk N and Jinawath N: Circulating exosomal miRNAs as biomarkers
in epithelial ovarian cancer. Biomedicines. 9:14332021. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Buys SS, Partridge E, Black A, Johnson CC,
Lamerato L, Isaacs C, Reding DJ, Greenlee RT, Yokochi LA, Kessel B,
et al: Effect of screening on ovarian cancer mortality: The
prostate, lung, colorectal and ovarian (PLCO) cancer screening
randomized controlled trial. JAMA. 305:2295–2303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Moss EL, Hollingworth J and Reynolds TM:
The role of CA125 in clinical practice. J Clin Pathol. 58:308–312.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Yokoi A, Yoshioka Y, Hirakawa A, Yamamoto
Y, Ishikawa M, Ikeda SI, Kato T, Niimi K, Kajiyama H, Kikkawa F and
Ochiya T: A combination of circulating miRNAs for the early
detection of ovarian cancer. Oncotarget. 8:89811–89823. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Wang S and Song X, Wang K, Zheng B, Lin Q,
Yu M, Xie L, Chen L and Song X: Plasma exosomal miR-320d, miR-4479,
and miR-6763-5p as diagnostic biomarkers in epithelial ovarian
cancer. Front Oncol. 12:9863432022. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Pan C, Stevic I, Müller V, Ni Q,
Oliveira-Ferrer L, Pantel K and Schwarzenbach H: Exosomal microRNAs
as tumor markers in epithelial ovarian cancer. Mol Oncol.
12:1935–1948. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Maeda K, Sasaki H, Ueda S, Miyamoto S,
Terada S, Konishi H, Kogata Y, Ashihara K, Fujiwara S, Tanaka Y, et
al: Serum exosomal microRNA-34a as a potential biomarker in
epithelial ovarian cancer. J Ovarian Res. 13:472020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY
and Hua KQ: Exosomal metastasis-associated lung adenocarcinoma
transcript 1 promotes angiogenesis and predicts poor prognosis in
epithelial ovarian cancer. Int J Biol Sci. 14:1960–1973. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zhang W, Su X, Li S, Liu Z, Wang Q and
Zeng H: Low serum exosomal miR-484 expression predicts unfavorable
prognosis in ovarian cancer. Cancer Biomark. 27:485–491. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Luo Y and Gui R: Circulating exosomal
circFoxp1 confers cisplatin resistance in epithelial ovarian cancer
cells. J Gynecol Oncol. 31:e752020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Chowdhury S, Kennedy JJ, Ivey RG, Murillo
OD, Hosseini N, Song X, Petralia F, Calinawan A, Savage SR, Berry
AB, et al: Proteogenomic analysis of chemo-refractory high-grade
serous ovarian cancer. Cell. 186:3476–3498.e35. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Martínez-Greene JA, Gómez-Chavarín M,
Ramos-Godínez MDP and Martínez-Martínez E: Isolation of hepatic and
adipose-tissue-derived extracellular vesicles using density
gradient separation and size exclusion chromatography. Int J Mol
Sci. 24:127042023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Bai HH, Wang XF, Zhang BY and Liu W: A
comparison of size exclusion chromatography-based tandem strategies
for plasma exosome enrichment and proteomic analysis. Anal Methods.
15:6245–6251. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Takov K, Yellon DM and Davidson SM:
Comparison of small extracellular vesicles isolated from plasma by
ultracentrifugation or size-exclusion chromatography: Yield, purity
and functional potential. J Extracell Vesicles. 8:15608092018.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Tian Y, Gong M, Hu Y, Liu H, Zhang W,
Zhang M, Hu X, Aubert D, Zhu S, Wu L and Yan X: Quality and
efficiency assessment of six extracellular vesicle isolation
methods by nano-flow cytometry. J Extracell Vesicles.
9:16970282019. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Aliakbari F, Stocek NB, Cole-André M,
Gomes J, Fanchini G, Pasternak SH, Christiansen G, Morshedi D,
Volkening K and Strong MJ: A methodological primer of extracellular
vesicles isolation and characterization via different techniques.
Biol Methods Protoc. 9:bpae0092024. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Knarr M, Avelar RA, Sekhar SC, Kwiatkowski
LJ, Dziubinski ML, McAnulty J, Skala S, Avril S, Drapkin R and
DiFeo A: miR-181a initiates and perpetuates oncogenic
transformation through the regulation of innate immune signaling.
Nat Commun. 11:32312020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wu M, Qiu Q, Zhou Q, Li J, Yang J, Zheng
C, Luo A, Li X, Zhang H, Cheng X, et al: circFBXO7/miR-96-5p/MTSS1
axis is an important regulator in the Wnt signaling pathway in
ovarian cancer. Mol Cancer. 21:1372022. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Luo L, Gao YQ and Sun XF: Circular RNA
ITCH suppresses proliferation and promotes apoptosis in human
epithelial ovarian cancer cells by sponging miR-10a-α. Eur Rev Med
Pharmacol Sci. 22:8119–8126. 2018.PubMed/NCBI
|
|
150
|
Ying X, Wu Q, Wu X, Zhu Q and Wang X,
Jiang L, Chen X and Wang X: Epithelial ovarian cancer-secreted
exosomal miR-222-3p induces polarization of tumor-associated
macrophages. Oncotarget. 7:43076–43087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Cappellesso R, Tinazzi A, Giurici T,
Simonato F, Guzzardo V, Ventura L, Crescenzi M, Chiarelli S and
Fassina A: Programmed cell death 4 and microRNA 21 inverse
expression is maintained in cells and exosomes from ovarian serous
carcinoma effusions. Cancer Cytopathol. 122:685–693. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Meng X, Müller V, Milde-Langosch K,
Trillsch F, Pantel K and Schwarzenbach H: Diagnostic and prognostic
relevance of circulating exosomal miR-373, miR-200a, miR-200b and
miR-200c in patients with epithelial ovarian cancer. Oncotarget.
7:16923–16935. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zavesky L, Jandakova E, Turyna R,
Langmeierova L, Weinberger V and Minar L: Supernatant versus
exosomal urinary microRNAs. Two fractions with different outcomes
in gynaecological cancers. Neoplasma. 63:121–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen
Y, Wang J, Liu Y, Chen P, Wu X and Wen J: Urinary microRNA-30a-5p
is a potential biomarker for ovarian serous adenocarcinoma. Oncol
Rep. 33:2915–2923. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Hong JS, Son T, Castro CM and Im H:
CRISPR/Cas13a-based MicroRNA detection in tumor-derived
extracellular vesicles. Adv Sci (Weinh). 10:e23017662023.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J,
Li X, Lu B, Cheng X, Liu P, et al: CircPLEKHM3 acts as a tumor
suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1
axis in ovarian cancer. Mol Cancer. 18:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Li L, Zhang F, Zhang J, Shi X, Wu H, Chao
X, Ma S, Lang J, Wu M, Zhang D and Liang Z: Identifying serum small
extracellular vesicle MicroRNA as a noninvasive diagnostic and
prognostic biomarker for ovarian cancer. ACS Nano. 17:19197–19210.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Yu CC, Liao YW, Hsieh PL and Chang YC:
Targeting lncRNA H19/miR-29b/COL1A1 axis impedes myofibroblast
activities of precancerous oral submucous fibrosis. Int J Mol Sci.
22:22162021. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Záveský L, Jandáková E, Turyna R,
Langmeierová L, Weinberger V, Záveská Drábková L, Hůlková M,
Hořínek A, Dušková D, Feyereisl J, et al: Evaluation of cell-free
urine microRNAs expression for the use in diagnosis of ovarian and
endometrial cancers. A pilot study. Pathol Oncol Res. 21:1027–1035.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Wang LL, Sun KX, Wu DD, Xiu YL, Chen X,
Chen S, Zong ZH, Sang XB, Liu Y and Zhao Y: DLEU1 contributes to
ovarian carcinoma tumourigenesis and development by interacting
with miR-490-3p and altering CDK1 expression. J Cell Mol Med.
21:3055–3065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Dong L and Hui L: HOTAIR promotes
proliferation, migration, and invasion of ovarian cancer SKOV3
cells through regulating PIK3R3. Med Sci Monit. 22:325–331. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Zhang Y, Dun Y, Zhou S and Huang XH:
LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells
proliferation and invasion by targeting miR-133a-3p and activating
Wnt/β-catenin signaling pathway. Biomed Pharmacother. 96:1216–1221.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Wang J, Tian Y, Zheng H, Ding Y and Wang
X: An integrated analysis reveals the oncogenic function of lncRNA
LINC00511 in human ovarian cancer. Cancer Med. 8:3026–3035. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
An J, Lv W and Zhang Y: LncRNA NEAT1
contributes to paclitaxel resistance of ovarian cancer cells by
regulating ZEB1 expression via miR-194. Onco Targets Ther.
10:5377–5390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Cao Y, Shi H, Ren F, Jia Y and Zhang R:
Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in
epithelial ovarian cancer. Exp Cell Res. 359:185–194. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Yiwei T, Hua H, Hui G, Mao M and Xiang L:
HOTAIR interacting with MAPK1 regulates ovarian cancer skov3 cell
proliferation, migration, and invasion. Med Sci Monit.
21:1856–1863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Yan H, Silva MA, Li H, Zhu L, Li P, Li X,
Wang X, Gao J, Wang P and Zhang Z: Long noncoding RNA DQ786243
interacts with miR-506 and promotes progression of ovarian cancer
through targeting cAMP responsive element binding protein 1. J Cell
Biochem. 119:9764–9780. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Duan M, Fang M, Wang C, Wang H and Li M:
LncRNA EMX2OS induces proliferation, invasion and sphere formation
of ovarian cancer cells via regulating the miR-654-3p/AKT3/PD-L1
axis. Cancer Manag Res. 12:2141–2154. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Yang M, Zhai Z, Zhang Y and Wang Y:
Clinical significance and oncogene function of long noncoding RNA
HAGLROS overexpression in ovarian cancer. Arch Gynecol Obstet.
300:703–710. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Wang Y, Zhang W, Wang Y and Wang S:
HOXD-AS1 promotes cell proliferation, migration and invasion
through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res.
8:170–182. 2018.PubMed/NCBI
|
|
171
|
Jin Y, Feng SJ, Qiu S, Shao N and Zheng
JH: LncRNA MALAT1 promotes proliferation and metastasis in
epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med
Pharmacol Sci. 21:3176–3184. 2017.PubMed/NCBI
|
|
172
|
Liu Y, Wang Y, Fu X and Lu Z: Long
non-coding RNA NEAT1 promoted ovarian cancer cells' metastasis
through regulation of miR-382-3p/ROCK1 axial. Cancer Sci.
109:2188–2198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Ding C, Wei R, Rodríguez RA and Del Mar
Requena Mullor M: LncRNA PCAT-1 plays an oncogenic role in
epithelial ovarian cancer by modulating cyclinD1/CDK4 expression.
Int J Clin Exp Pathol. 12:2148–2156. 2019.PubMed/NCBI
|
|
174
|
Özeş AR, Miller DF, Özeş ON, Fang F, Liu
Y, Matei D, Huang T and Nephew KP: NF-κB-HOTAIR axis links DNA
damage response, chemoresistance and cellular senescence in ovarian
cancer. Oncogene. 35:5350–5361. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Song R, Liu Z, Lu L, Liu F and Zhang B:
Long noncoding RNA SCAMP1 targets miR-137/CXCL12 axis to boost cell
invasion and angiogenesis in ovarian cancer. DNA Cell Biol.
39:1041–1050. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Chen S, Wang LL, Sun KX, Xiu YL, Zong ZH,
Chen X and Zhao Y: The role of the long non-coding RNA TDRG1 in
epithelial ovarian carcinoma tumorigenesis and progression through
miR-93/RhoC pathway. Mol Carcinog. 57:225–234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Mu Y, Li N and Cui YL: The lncRNA CCAT1
upregulates TGFβR1 via sponging miR-490-3p to promote TGFβ1-induced
EMT of ovarian cancer cells. Cancer Cell Int. 18:1452018.
View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Li J, Li J, Huang Y, Deng X, Luo M, Wang
X, Hu H, Liu C and Zhong M: Long noncoding RNA H19 promotes
transforming growth factor-β-induced epithelial-mesenchymal
transition by acting as a competing endogenous RNA of miR-370-3p in
ovarian cancer cells. Onco Targets Ther. 11:427–440. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Xu M, Zhou K, Wu Y, Wang L and Lu S:
Linc00161 regulated the drug resistance of ovarian cancer by
sponging microRNA-128 and modulating MAPK1. Mol Carcinog.
58:577–587. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Pan L, Meng Q, Li H, Liang K and Li B:
LINC00339 promotes cell proliferation, migration, and invasion of
ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomed
Pharmacother. 120:1094232019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Zou A, Liu R and Wu X: Long non-coding RNA
MALAT1 is up-regulated in ovarian cancer tissue and promotes
SK-OV-3 cell proliferation and invasion. Neoplasma. 63:865–872.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Chang L, Guo R, Yuan Z, Shi H and Zhang D:
LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging
miR-206 in ovarian cancer. Cell Physiol Biochem. 49:1289–1303.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Sun Q, Li Q and Xie F: LncRNA-MALAT1
regulates proliferation and apoptosis of ovarian cancer cells by
targeting miR-503-5p. Onco Targets Ther. 12:6297–6307. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
An Q, Liu T, Wang MY, Yang YJ, Zhang ZD,
Lin ZJ and Yang B: circKRT7-miR-29a-3p-COL1A1 axis promotes ovarian
cancer cell progression. Onco Targets Ther. 13:8963–8976. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Wang F, Niu Y, Chen K, Yuan X, Qin Y,
Zheng F, Cui Z, Lu W, Wu Y and Xia D: Extracellular
vesicle-packaged circATP2B4 mediates M2 macrophage polarization via
miR-532-3p/SREBF1 axis to promote epithelial ovarian cancer
metastasis. Cancer Immunol Res. 11:199–216. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Wu Y, Xu M, Feng Z, Wu H, Wu J, Ha X, Wu
Y, Chen S, Xu F, Wen H, et al: AUF1-induced circular RNA
hsa_circ_0010467 promotes platinum resistance of ovarian cancer
through miR-637/LIF/STAT3 axis. Cell Mol Life Sci. 80:2562023.
View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Li H, Luo F, Jiang X, Zhang W, Xiang T,
Pan Q, Cai L, Zhao J, Weng D, Li Y, et al: CircITGB6 promotes
ovarian cancer cisplatin resistance by resetting tumor-associated
macrophage polarization toward the M2 phenotype. J Immunother
Cancer. 10:e0040292022. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Zou T, Wang PL, Gao Y and Liang WT:
Circular RNA_LARP4 is lower expressed and serves as a potential
biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci.
22:7178–7182. 2018.PubMed/NCBI
|