Introduction
According to the World Health Organization
classification (WHO), adult diffuse gliomas include astrocytomas,
glioblastomas, oligodendrogliomas and oligoastrocytomas (1). Despite therapeutic advances, these
tumors remain incurable and the prognosis for patients afflicted
with anaplastic tumors and glioblastomas is still very poor
(2).
The epidermal growth factor receptor (EGFR) belongs
to HER family, a group of four receptors. Many reports have shown
that EGFR contribute to diffuse glioma oncogenesis (3–8). The
most prevalent EGFR pathway modifications involved are EGFR
gene amplification and receptor overexpression, the latter of which
remains controversial (9–15).
The EGFR gene is located in 7p12 and
generates a first type of mRNA transcript, referred to as EGFR
variant 1 (EGFRv1), which encodes the full-length receptor, known
as isoform a, EGFR or HER1. Isoform a is a transmembrane protein
with an intracellular tyrosine kinase domain. Ligand binding on the
extracellular domain leads to stimulation of cellular proliferation
and cell survival pathways. Glioblastomas harboring EGFR
gene amplification frequently contain a genetic variant, EGFRvIII,
which encodes a mutant receptor with an altered extracellular
domain that renders it constitutively active (16–18).
In addition to variant v1, the EGFR gene
generates three other alternatively spliced transcripts, referred
to as variants 2–4 (EGFRv2-v4). The corresponding mRNAs are shorter
than the EGFRv1 transcript and, respectively, encode truncated
forms of the receptor (isoforms b, c and d), which lack the
cytoplasmic tyrosine kinase domain. Soluble isoforms have been
reported (19–21). The role of these truncated EGFR
isoforms remains poorly known. In vitro, they have been
shown to decrease cellular proliferation (22,23).
The hypotheses for this cellular growth inhibition include the
competitive binding of EGFR ligands by the truncated isoforms
(24,25) and formation of catalytically
inactive heterodimers of different isoforms, which interfere with
the formation of functional holoreceptor dimers. This consequently
prevents intracellular kinase activity (26). To our knowledge, expressions of
truncated EGFR isoforms and their transcripts have never been
studied in gliomas.
To assess whether EGFR protein isoforms and their
corresponding transcripts are expressed in diffuse gliomas, we
performed an immunohistochemical analysis and determined the
expression patterns of EGFRv1, -v2, -v3, -v4 and mutant EGFRvIII
mRNAs. Results were analyzed with respect to the clinical data,
patient outcome, histological tumor type and presence or absence of
EGFR gene amplification.
Patients and methods
Patients and tissue samples
Tumors were obtained from 47 adult patients
diagnosed with infiltrating glioma who were undergoing surgery at
Limoges Dupuytren University Hospital between 1995 and 2011.
Clinical and survival data were obtained by a retrospective query
and all samples were used in accordance with French bioethics laws
regarding patient information and con-sent. No patients received
EGFR-targeted therapeutics. Tumor samples were fixed in 4% formalin
at the time of resection. They were then embedded in paraffin and
tumor sections were stained with hemalum phloxine saffron. Part of
the surgical specimen was snap-frozen and conserved at −80°C. The
histological type and grade of gliomas were determined according to
the WHO classification (1).
Non-tumor tissue was used as a control.
Immunohistochemistry
Sections (5 μm) were cut from paraffin-embedded
tumors and stained with two different anti-EGFR antibodies
(Fig. 1). One antibody targeted
the extracellular domain (Ext-Ab) and the other intracellular
domain (Int-Ab). Sample slides were processed automatically
(BenchMark XT ICH/ISH, Ventana Medical Systems, Tucson, AZ, USA)
according to protocols supplied by the manufacturers. EGFR staining
was quantified according to a semiquantitative method proposed by
Hirsch et al (27), as
previously described (28).
Staining was scored for labeling intensity (1, negative or trace;
2, weak; 3, moderate; 4, intense), percentage of positive cells per
slide (0%–100%) and for the Hirsch score resulting from
multiplication of these two parameters (absolute values ranging
from 0 to 400). For certain analyses, the level of expression in
samples was scored as: negative or low, intermediate, and high,
which corresponded to Hirsch values of 0–200, 201–300, and 301–400,
respectively.
We also tested the following antibodies: Ki67 (clone
MiB-1, DakoCytomation, Glostrup, Denmark; 1/200e), p53
(Dako Cytomation; 1/50e), Olig2 (Immuno-Biological
Laboratories, Gunma, Japan; 1/200e), and glial fibrillar
acidic protein (Dako Cytomation; 1/1600e). The
percentages of cells labeled with these antibodies were determined
by studying at least five hundred cells for each antibody in tumor
areas of highest positivity.
Total RNA extraction
Tumor tissue (30 mg) was incubated with 1 ml
Qiazol® solution (Qiagen, Courtaboeuf, France) and CK14
ceramic beads (Ozyme) and then pulverized two times for 40 sec each
at 6500 rpm in a Precellys 24 homogenizer (Bertin Technologies).
Homogenized tissues were then lysed and RNA purification was
performed according to the manufacturer’s protocol (RNeasy Lipid
Tissue Mini kit, Qiagen). RNA concentration and purity was
estimated by spectrophotometry (NanoDrop ND1000, Labtech, France).
RNA quality was assessed by capillary electrophoresis (Bioanalyzer
2100, Agilent Technologies) and only RNA with an Integrity Number
(R.I.N) >6 was used for analysis.
Quantitative and qualitative RT-PCR
Complementary DNA (cDNA) was synthesized from 2 μg
of total RNA using the Transcriptor First Strand cDNA
Synthesis® kit (Roche) and hexamer primers, according to
the manufacturer’s protocol. For PCR, primers were designed using
Amplify 1.2 or Primer 3 (http://fokker.wi.mit.edu/primer3/input.html/) software
and their specificity was determined by BLASTn in the NCBI database
(http://www.ncbi.nlm.nih.gov/). Primer
characteristics are listed in Table
I. Amplicon size and specificity were initially determined by
4% NuSieve agarose gel electrophoresis. Quantitative PCR [EGFRv1,
-v2, -v3, -v4 and hypoxanthine phosphoribosyl transferase (HPRT)]
and qualitative PCR (EGFRvIII) were performed on a Rotor Gene
thermocycler (Corbett Research) using the Light Cycler Fast Start
DNA Master SYBR Green I kit (Roche). All targets were amplified
twice in duplicate in the presence of 3 mM MgCl2 and 0.5
μM primers. Relative quantification of mRNA content was performed
using the ΔΔCt method
[(Ctsample-Ctcalibrator)interest
gene -
(Ctsample-Ctcalibrator)reference
gene], modified according to Pfaffl (29), with efficiency correction by Rotor
Gene software. mRNA content was expressed in relative arbitrary
units (R.A.U.).
 | Table ICharacteristics of the primer used
for quantitative and qualitative RT-PCR. |
Table I
Characteristics of the primer used
for quantitative and qualitative RT-PCR.
| Target | Primer | Location | Sequence 5′-3′ | Amplicon size
(bp) | Primer temperature
(°C) |
|---|
| EGFR variant 1 | Forward | Exon 29–30 |
CTCCCAGTGCCTGAATACATA | 83 | 58 |
| Reverse | Exon 30 |
GGCTGATTGTGATAGACAGGA | | |
| EGFR variant 2 | Forward | Exon 15 |
TGCACCTACGGGTCCTAAT | 97 | 58 |
| Reverse | Exon 16 |
TGAAGCAAAGGGAGAAATTG | | |
| EGFR variant 3 | Forward | Exon 10 |
AAGGAAATCACAGGTTTGAGC | 99 | 58 |
| Reverse | Exon 10bis |
TCCAAGGGAACAGGAAATATG | | |
| EGFR variant 4 | Forward | Exon 15 |
CTACGGGCCAGGAAATGAG | 86 | 62 |
| Reverse | Exon 17 |
CGCTGCCATCATTACTTTGA | | |
| HPRT | Forward | Exon 6 |
CTTTCCTTGGTCAGGCAGTA | 90 | 58 |
| Reverse | Exon 7 |
TGGCTTATATCCAACACTTCG | | |
| EGFRvIII | Forward | Exon 1 |
GCTCTGGAGGAAAAGAAAGGTAAT | 90 | 62 |
| Reverse | Exon 8 |
TCCTCCATCTCATAGCTGTCG | | |
Fluorescent in situ hybridization
EGFR gene amplification and 1p36 and 19q13
losses were analyzed by double fluorescent in situ
hybridization with the ‘LSI EGFR SpectrumOrange/CEP 7
SpectrumGreen Probe’, or with the ‘LSI 1p36 spectrum orange/LSI
1q25 spectrum green probe’ and the ‘LSI 19q13 spectrum orange/LSI
19p13 spectrum green probe’ (Abbott Molecular Inc., IL, USA) kits,
respectively. They were investigated on consecutive paraffin
sections from the same blocks used in immunohistochemical analyses.
This technique was a modification of the method previously
described (28): briefly, 4 μm
paraffin sections were incubated 16 h at 56°C, submitted to
deparaffinising, digested with pepsin (Abbott Molecular Inc.) at
37°C during 45 min and dehydrated in successive ethanol baths.
Slides were incubated with 10 μl of each probe for 5 min at 73°C to
denature DNA and 16 h at 37°C to ensure hybridization. Sections
were washed in 2X SSC/0.3% NP40 solution, once for 1 min at room
temperature, once for 2 min at 73°C, and dehydrated in successive
ethanol baths. Counterstaining and microscopic observation of
EGFR amplification were performed as previously described
(28). EGFR gene
amplification was considered to have occurred if more than 10% of
the cells analyzed produced a red (corresponding to the
EGFR-specific probe) to green (centromeric region of
chromosome 7) signal ratio ≥2, as recommended previously (30,31).
Eight sequential focus stacks with 0.3 μm were acquired and then
integrated into a single image in order to reduce thickness related
artifacts. Preparations were considered as valid when more of 80%
of the cells showed bright signals.
Statistical analyses
StatView® 5.0 software (SAS Institute,
Inc., Cary, NC, USA) was used for statistical analyses. Fisher’s
exact test was used to assess differences between nominal
variables. Means were compared with the non-parametric Mann-Whitney
test for pairs of variables and with the Kruskall-Wallis tests for
comparisons of more than two variables. Correlation Spearman test
was used to compare quantitative variables. Overall survival (OS)
and progression-free survival (PFS) were analyzed by Kaplan-Meier
and median OS or PFS medians were compared with the non-parametric
log-rank or Breslow tests. Results for which p<0.05 were
considered to be statistically significant.
Results
Patient and tumor characteristics
Patient characteristics are summarized in Table II. For the series, median
follow-up was 23.3 (0.5–240) months. Ki67 labeling index, Olig2 and
p53 protein expression, and presence or absence of a 1p36–19q13
loss were consistent with histological typing. Thus,
oligodendrogliomas were characterized by a 1p36/19q13 deletion,
stronger olig2 and weaker p53 expressions compared to other tumor
types (Table III).
 | Table IIDemographic, pathological and
clinical features. |
Table II
Demographic, pathological and
clinical features.
| No. | Grade | Sex
(male/female) | Age (median) | Tumor status
(primary/recurrent) | Radiotherapy
(yes/no) | Chemotherapy
(yes/no) |
|---|
| All | 47 | | 18/19 | 50.6 | 36/11 | 42/5 | 37/10 |
| A | 3 | 1 II, 2 III | 1/2 | 51.3 | 2/1 | 2/1 | 2/1 |
| GBM | 21 | 21 IV | 10/11 | 59.7 | 19/2 | 20/1 | 18/3 |
| O | 14 | 5 II, 9 III | 3/11 | 48.1 | 10/4 | 13/1 | 13/1 |
| OA | 9 | 6 II, 3 III | 4/5 | 43.2 | 5/4 | 7/2 | 4/5 |
 | Table IIIMolecular characterization of glioma
histological types. |
Table III
Molecular characterization of glioma
histological types.
| Ki67 (n=45) | p53 (n=45) | Olig2 (n=45) | 1p36-19q13 loss
(n=46) |
|---|
|
|
|
|
|
|---|
| % Mean ± SD | p-value | % Mean ± SD | p-value | % Mean ± SD | p-value | Yes | No | p-value |
|---|
| All patients | 19±14 | | 46.3±31.1 | | 58.6±23.9 | | 17 | 29 | |
| Histological
type |
| A | 13.7±10 | 0.06 | 80±10 | 0.0004 | 51.7±27.5 | 0.04 | 0 | 3 | <0.0001 |
| GBM | 26.9±13.5 | | 49.7±28.3 | | 48.7±25.8 | | 2 | 18 | |
| O | 14.8±19 | | 20.6±18.4 | | 71.8±18.3 | | 14 | 0 | |
| OA | 10.8±11.2 | | 67.8±28.5 | | 61.1±19 | | 1 | 8 | |
Immunohistochemical detection of EGFR
isoforms and EGFRvIII mutant
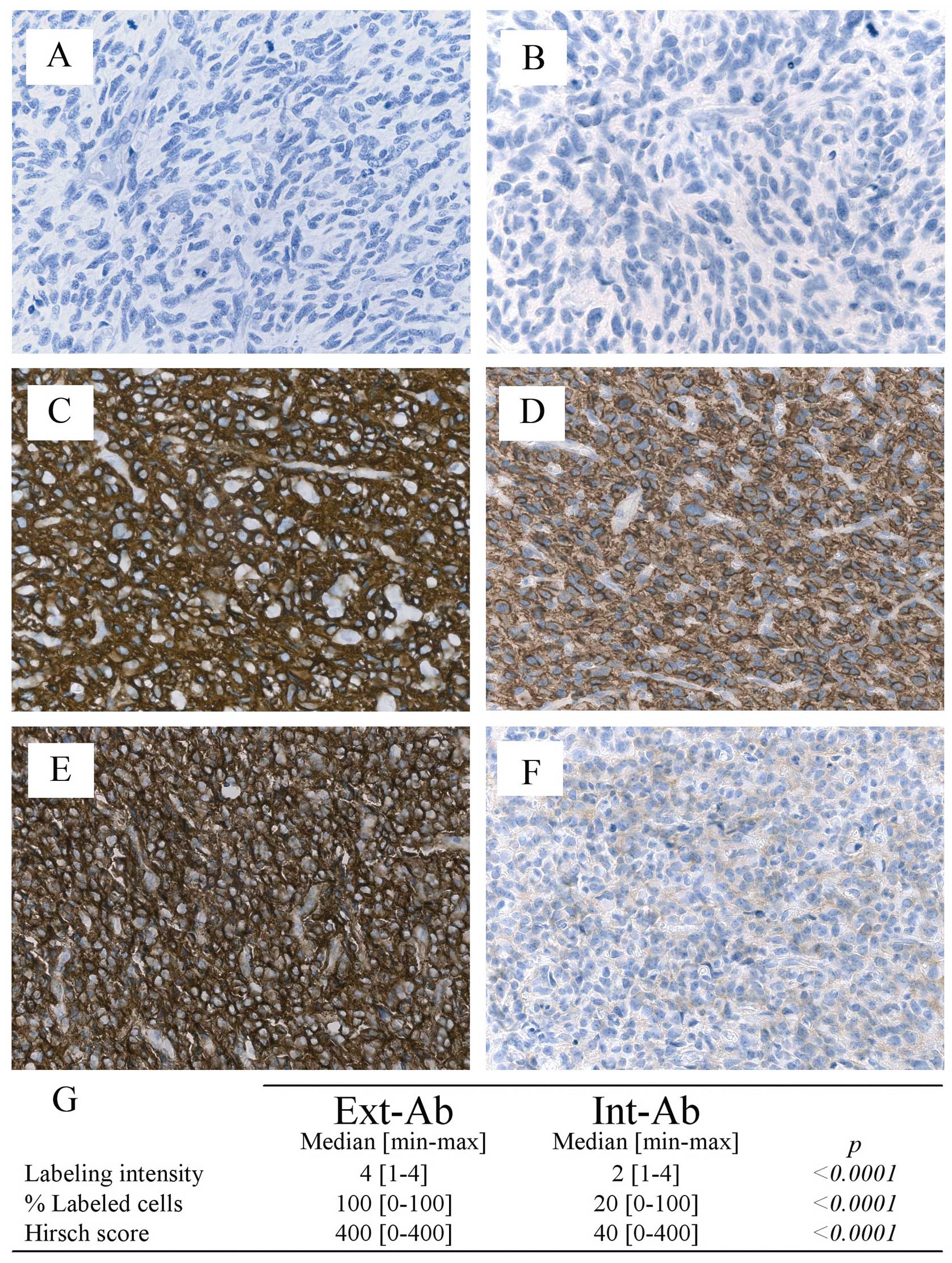
The percentage of gliomas stained by Ext-Ab was 98%
(44/45), whereas only 78% (35/45) of the gliomas were stained by
Int-Ab. The Ext-Ab and Int-Ab antibodies generated very different
staining patterns in the gliomas (Fig.
2A-F). Glioma staining by Int-Ab was significantly lower than
that of Ext-Ab in terms of intensity, percentage of positive cells
and Hirsch score (p<0.0001; Fig.
2G).
Neither antibody detected any significant
differences in EGFR staining with respect to patient sex or age
(data not shown). Staining intensities, percentages of labeled
cells and Hirsch scores obtained with Ext-Ab did not significantly
differ according to histological types. In contrast, Int-Ab scores
were significantly higher in glioblastomas, astrocytomas or
oligodendrogliomas compared to oligoastrocytomas (Table IV). In non-tumor tissue, we found
no or very weak EGFR expression whatever the antibody used.
 | Table IVAssociation between Ext-Ab and Int-Ab
labeling and pathological parameters. |
Table IV
Association between Ext-Ab and Int-Ab
labeling and pathological parameters.
| | Ext-Ab | Int-Ab |
|---|
| |
|
|
|---|
| No. | Labeling
intensity | p-value | Percentage of
labeled cells | p-value | Hirsch score | p-value | Labeling
intensity | p-value | Percentage of
labeled cells | p-value | Hirsch score | p-value |
|---|
| All | 45 | 4 [1–4] | | 100 [0–100] | | 400 [0–400] | | 2 [1–4] | | 20 [0–100] | | 40 [0–400] | |
| Histological
type |
| A | 3 | 4 [3–4] | 0.73 | 70 [20–90] | 0.33 | 360 [60–400] | 0.69 | 2 [2–3] | 0.006 | 30 [5–40] | 0.01 | 60 [10–120] | 0.01 |
| GBM | 19 | 4 [2–4] | | 100 [10–100] | | 400 [20–400] | | 3 [1–4] | | 30 [0–100] | | 90 [0–400] | |
| O | 14 | 4 [1–4] | | 100 [0–100] | | 380 [0–400] | | 2 [1–3] | | 45 [0–70] | | 90 [0–210] | |
| OA | 9 | 4 [2–4] | | 100 [20–100] | | 300 [80–400] | | 1 [1–2] | | 0 [0–10] | | 0 [0–20] | |
Quantitation of EGFR variants 1, 2, 3, 4
and EGFRvIII mRNAs
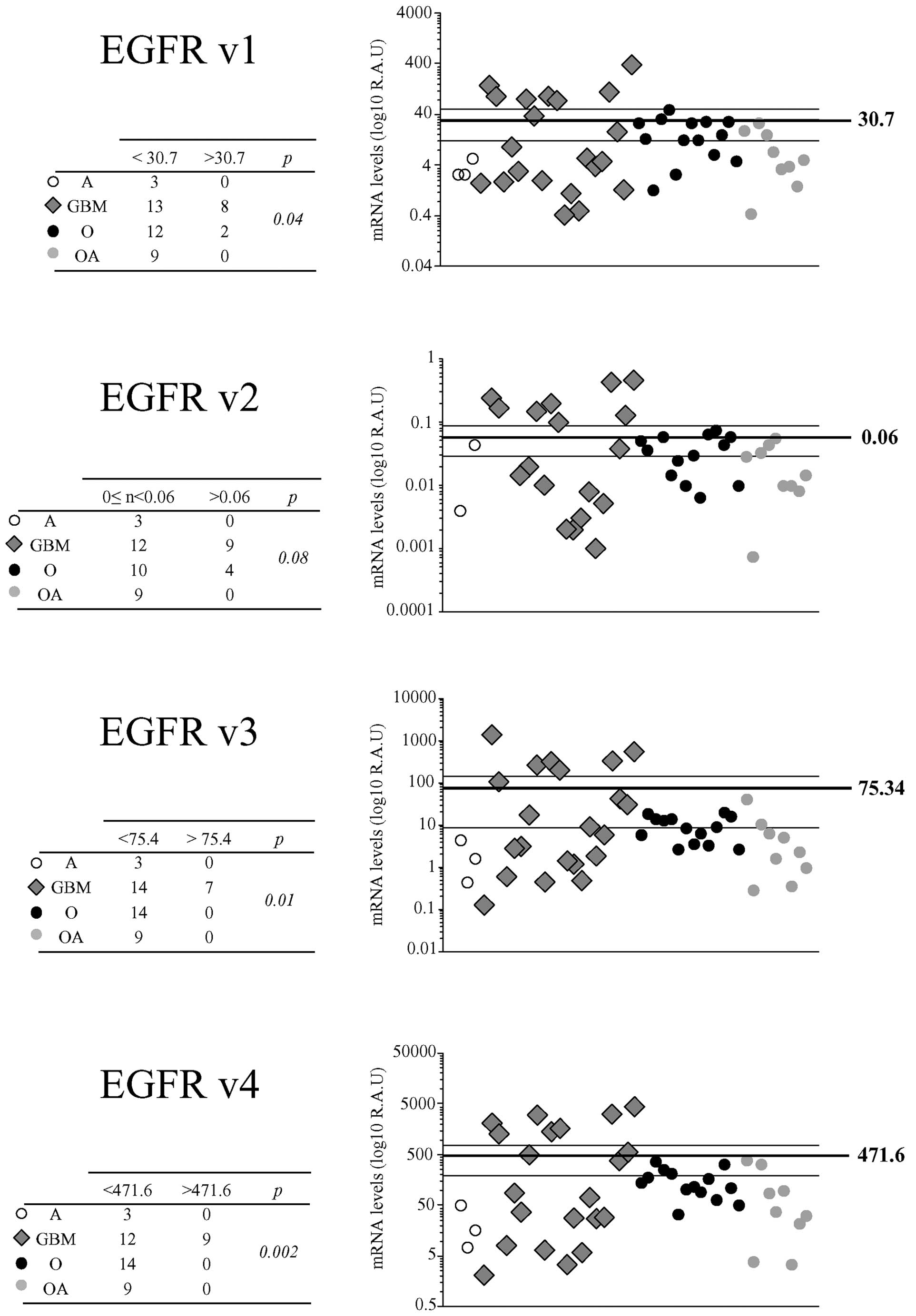
EGFR mRNA levels varied widely among the tumor
samples. Median R.A.U. values were 7.3 (0.4–390.2) for EGFRv1+vIII
mRNA, 0.02 (0–0.5) for EGFRv2 mRNA, 6.2 (0.1–1396.8) for EGFRv3
mRNA and 94.6 (2.1–4445.2) for EGFRv4 mRNA. Straight correlations
were found for all comparisons between EGFRv1+vIII, -v2, -v3 and
-v4 mRNA levels (p<0.0001 for all, Fig. 3). EGFRvIII mutant expression was
qualitatively detected in 26% (12/47) of the gliomas. In non-tumor
tissue mRNA variant were very weakly expressed with 0.7 R.A.U. for
EGFRv1+vIII and EGFRv3, 6.6 R.A.U. for EGFRv4 and no EGFRvIII and
EGFRv2 expression.
Association of EGFR variant and EGFRvIII
mRNA expression with other parameters
EGFRv1+vIII, -v2, -v3, and -v4 mRNA levels were not
influenced by patient sex and age (data not shown). EGFRv1+vIII,
-v3 and -v4 mRNA levels were higher in glioblastomas than in other
tumor types when mean values were taken as a cut-off (p=0.04, 0.01
and 0.002, respectively) (Fig. 4).
EGFRvIII mRNA was significantly associated with histological type,
it was found in one astrocytoma, ten glioblastomas and one
oligoastrocytoma, but in none oligodendroglioma (p=0.01, data not
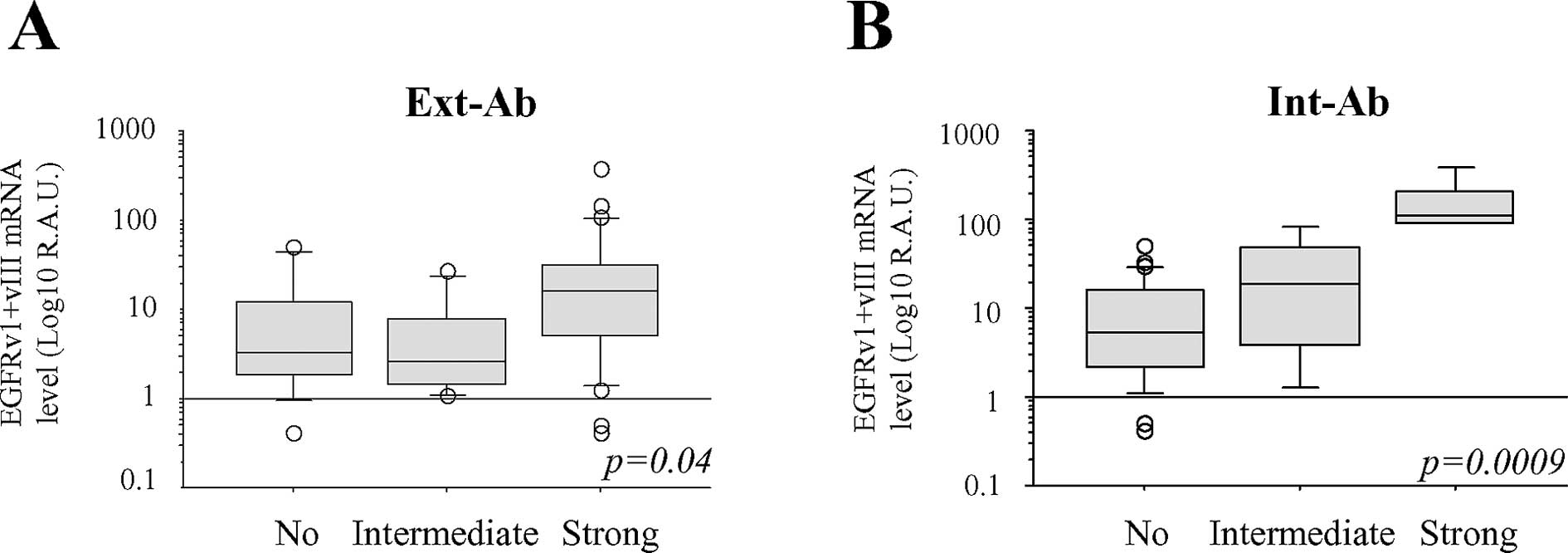
shown). We also observed that EGFRv1+vIII mRNA levels more
straightly correlated with Int-Ab staining than with Ext-Ab
staining (Fig. 5).
EGFR gene amplification in gliomas
We detected EGFR gene amplification in 8 out
of the 20 glioblastomas, but not in the other glioma types
(p=0.007, Table V). Glioblastomas
with EGFR gene amplification expressed significantly
stronger EGFRv1, -v2, -v3 and -v4 mRNA levels than gliomas with no
EGFR amplification (Table
V). The presence of mutant EGFRvIII mRNA was significantly
associated with EGFR amplification (p<0.0001).
 | Table VEGFR amplification. |
Table V
EGFR amplification.
| EGFR
amplification |
|---|
|
|
|---|
| No. | Yes | p-value |
|---|
| Histological
type |
| A | 3 | 0 | 0.007 |
| GBM | 12 | 8 | |
| O | 13 | 0 | |
| OA | 9 | 0 | |
| EGFRv1+vIII
mRNA |
| < mean | 34 | 2 | 0.0001 |
| > mean | 3 | 6 | |
| EGFRv2 mRNA |
| < mean | 32 | 2 | 0.001 |
| > mean | 5 | 6 | |
| EGFRv3 mRNA |
| < mean | 36 | 1 | <0.0001 |
| > mean | 3 | 5 | |
| EGFRv4 mRNA |
| < mean | 36 | 1 | <0.0001 |
| > mean | 1 | 7 | |
| EGFRvIII mRNA |
| Present | 4 | 7 | <0.0001 |
| Absent | 33 | 1 | |
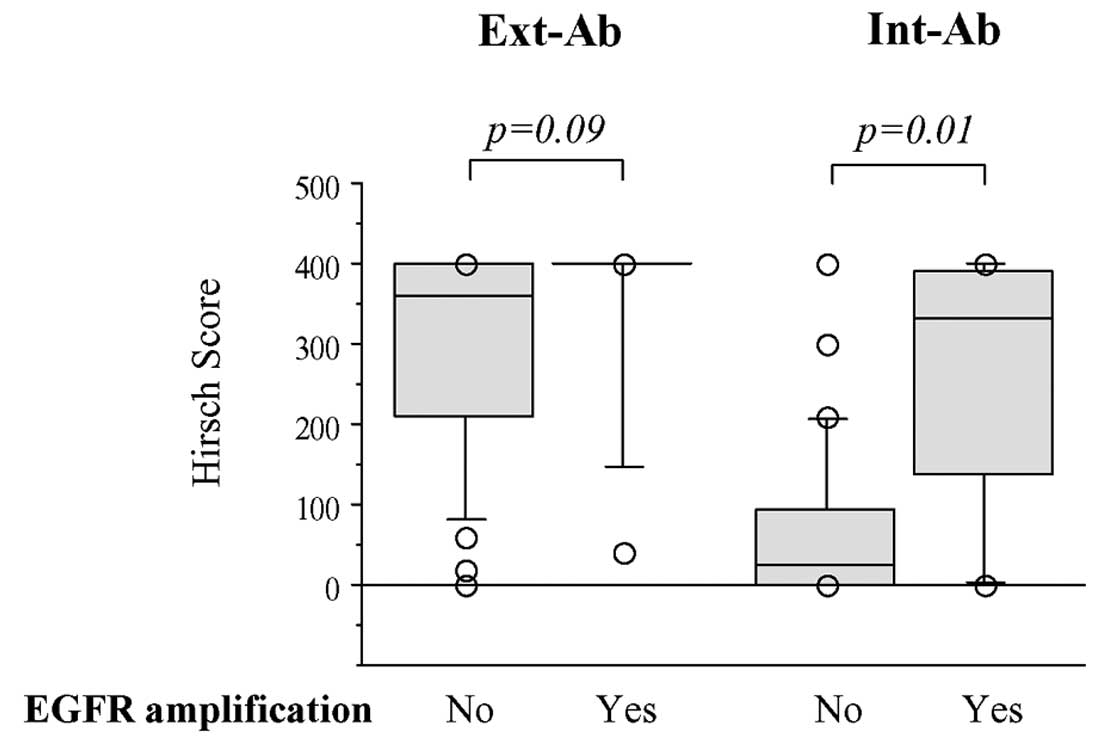
Weak Ext-Ab staining was more closely associated
with the absence of EGFR gene amplification than with its
presence (p=0.09), whereas strong Int-Ab staining was significantly
associated with gene amplification (p=0.01; Fig. 6).
Prognostic values
PFS and OS were significantly shorter for patients
diagnosed with glioblastoma and astrocytoma than for those with
oligodendroglioma and oligoastrocytoma (p<0.0001 for both)
(Table VI, Fig. 7A). OS and PFS were better for
patients with tumors showing a 1p36–19q13 loss and an absence of
EGFR amplification (Table
VI).
 | Table VIPFS and OS according to histological
and molecular parameters. |
Table VI
PFS and OS according to histological
and molecular parameters.
| PFS | OS |
|---|
|
|
|
|---|
| Median
(months) | p-value | Median
(months) | p-value |
|---|
| Histological
types |
| A | 16.4 | <0.0001 | 34.9 | <0.0001 |
| GBM | 5.4 | | 10.9 | |
| O | 45.1 | | 93.3 | |
| OA | 41 | | nr | |
| EGFR
amplification |
| Yes | 4.7 | <0.0001 | 8.8 | 0.0003 |
| No | 21.6 | | 93.3 | |
| 1p36-19q13
loss |
| Yes | 21.9 | 0.04 | 93.3 | 0.09 |
| No | 16.4 | | 24 | |
| Ext-Ab |
|
No/intermediate | 22.7 | 0.25 | 52.8 | 0.7 |
| Strong | 16.4 | | 93.3 | |
| Int-Ab |
|
No/intermediate | 21.4 | 0.006 | 93.3 | 0.07 |
| Strong | 4.7 | | 21.6 | |
| EGFRv1 mRNA |
| Weak | 21 | 0.02a | 93.3 | 0.1 |
| Strong | 5.4 | | 21.6 | |
| EGFRv2 mRNA |
| Weak | 21.4 | 0.01a | 52.8 | 0.007 |
| Strong | 9 | | 15.1 | |
| EGFRv3 mRNA |
| Weak | 21 | 0.0007 | 93.3 | 0.004 |
| Strong | 4.7 | | 10.9 | |
| EGFRv4 mRNA |
| Weak | 21.4 | <0.0001 | 93.3 | 0.0006 |
| Strong | 4.7 | | 10.9 | |
| EGFRvIII mRNA |
| No | 21.4 | 0.0007 | 52.8 | 0.03 |
| Yes | 4.7 | | 10.9 | |
No/intermediate or strong tumor labeling with EGFR
Ext-Ab did not influence OS and PFS times whereas no/intermediate
labeling with EGFR Int-Ab was associated with longer OS and PFS
(Table VI). In addition, PFS and
OS were longer when gliomas expressed weak EGFRv1+vIII, -v2, v3, or
-v4 mRNA levels and showed no mutant EGFRvIII mRNA expression
(Table VI, Fig. 7B-F).
In glioblastomas (data not shown), PFS was
significantly better for patients with no EGFR amplification
(5.4 vs. 8.4 months, p=0.01), no expression of EGFRvIII mutant mRNA
(3.7 vs. 8.4 months, p=0.04), or weak EGFRv2 (3.3 vs. 5.6 months,
p=0.04) or -v4 mRNA levels (8.4 vs. 4.7 months, p=0.05). OS did not
change according to these parameters.
Discussion
Based on the present results, we report that diffuse
gliomas expressed truncated EGFR protein isoforms, based on: i)
immunohistochemical data and ii) EGFRv1, -v2, -v3, -v4 variants and
EGFRvIII mutant mRNA detection.
Immunohistochemical results varied according to the
antibody used and favored the hypothesis of an expression of the
truncated isoforms. The stronger EGFR staining obtained with Ext-Ab
reflected their expression since, in addition to functional EGFR
and the EGFRvIII mutant targeted by both antibodies, Ext-Ab also
recognized the truncated EGFR isoforms b, c, and d whereas Int-Ab
did not. The detection of truncated isoforms, depending on the
antibody used, could explain some of the discrepancies found in
literature regarding EGFR expression in gliomas (9–15).
In our series, glioblastomas and oligodendrogliomas were strongly
labeled by both Ext-Ab and Int-Ab, whereas oligoastrocytomas were
moderately or strongly labeled by Ext-Ab but weakly by Int-Ab.
Thus, in combination with other markers such as Olig2, p53 or 1p19q
deletion, the study of EGFR expression might be useful to further
characterize the diffuse gliomas (32–36).
The transcriptomic analysis showed that
alternatively spliced EGFRv2, -v3 and -v4 transcript variants,
encoding EGFR isoforms b, c and d, respectively, were expressed in
addition to the EGFRv1 and EGFRvIII mutant mRNAs. In accordance
with immunohistochemistry, detection of EGFRv1+vIII mRNA associated
more closely with Int-Ab staining than with Ext-Ab staining.
However, we also found an association between Int-Ab staining and
EGFRv2, -v3 and -v4 transcript expressions, although Int-Ab did not
detect the isoform they encode. This association is probably the
consequence of the strong link existing between the expressions of
each EGFR transcript (Fig. 3).
Nevertheless, alternatively spliced EGFR transcript variants and
EGFRvIII mRNA were produced at different levels according to the
histological type of glioma. The glioblastomas had a peculiar
profile. They expressed the highest levels of EGFRv3 and -v4 mRNA
transcripts and, in addition, EGFRvIII mRNA expression was related
to this tumor type.
EGFR gene amplification was detected in eight
glioblastomas. As previously reported (28), weak Ext-Ab staining was associated
with the lack of EGFR amplification. In contrast, Int-Ab
staining intensity directly correlated with EGFR gene
amplification, as already shown (37). This suggests that EGFR gene
amplification is tightly associated with high expression of EGFR
receptor isoforms that contain the intracellular tyrosine kinase
domain, i.e., EGFR isoform a and the EGFRvIII mutant. Regarding the
relationships with the prognosis in our series, the histological
type (astrocytoma and glioblastoma), a strong staining with Int-Ab
and the presence of EGFR amplification and of mutant vIII
were associated with shorter PFS and OS times. High levels of
EGFRv2, -v3 and -v4 transcript expression were also related to a
shortened OS and PFS.
Our data tend to indicate that the role of EGFR
pathway in glioblastoma oncogenesis is more complex than expected.
In our series, in addition to the known molecular alterations i.e.,
EGFR gene amplification and expression of vIII mutant, we
observed a strong staining with Int-Ab and high levels of EGFR mRNA
variants. Actually, the functional roles of the truncated EGFR
isoforms are poorly known, particularly in vivo. In vitro,
it has been described that soluble isoforms may regulate EGFR
signaling in normal and tumor cells (26,38).
Paradoxically, these isoforms have been shown to decrease cellular
proliferation (22,23). The known mechanisms responsible for
growth inhibition include competitive binding of ligands to soluble
isoforms and formation of inactive heterodimers which inhibit the
formation of holoreceptor dimers and/or intracellular kinase
activity (24–26).
The presence of truncated EGFR isoforms in adult
infiltrating gliomas must be considered in therapeutic management.
The interactions between truncated EGFR isoforms and EGFR-targeted
therapeutics are not well understood. The presence of
non-functional receptors could contribute to the failure of
therapeutics which target the EGFR extracellular domain (39). Conversely, it has been reported
that the presence of truncated EGFR isoforms may be predictive of
the therapeutic response to gefitinib in endometrious
adenocarcinomas (40). Lafky and
coworkers speculated that the soluble vascular endothelial growth
factor receptor (sVEGFR) represents a paradigm for understanding
the function and potential application of EGFR isoforms as novel
therapeutic molecules (41).
Soluble VEGFR isoforms have been presented as effective therapeutic
molecules (42) and a similar
application for certain EGFR truncated isoforms may be
possible.
To our knowledge, this is the first report that
gliomas express EGFR transcripts other than EGFRv1 mRNA, which
encodes the full-length and functional EGFR isoform a. The role of
EGFR isoforms in glioma pathogenesis remains to be clarified, but
their expression makes them potential targets of future diagnostic
and therapeutic strategies.
Acknowledgments
This work was supported by grants from the ‘Comité
Départemental de la Ligue contre le Cancer de la Corrèze’. We thank
Mrs. Marion Porcheron for technical assistance, the ‘Tumorothèque
du Limousin’ for providing all tissue samples and the ‘Plateforme
d’Oncologie Moléculaire’ of the Limoges Dupuytren University
Hospital.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Krex D, Klink B, Hartmann C, von Deimling
A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger
G, Weller M and Schackert G: Long-term survival with glioblastoma
multiforme. Brain. 130:2596–2606. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Andersson U, Guo D, Malmer B, Bergenheim
AT, Brannstrom T, Hedman H and Henriksson R: Epidermal growth
factor receptor family (EGFR, ErbB2-4) in gliomas and meningiomas.
Acta Neuropathol. 108:135–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bredel M, Pollack IF, Hamilton RL and
James CD: Epidermal growth factor receptor expression and gene
amplification in high-grade non-brainstem gliomas of childhood.
Clin Cancer Res. 5:1786–1792. 1999.PubMed/NCBI
|
|
5
|
McLendon RE, Turner K, Perkinson K and
Rich J: Second messenger systems in human gliomas. Arch Pathol Lab
Med. 131:1585–1590. 2007.PubMed/NCBI
|
|
6
|
Potti A, Forseen SE, Koka VK, Pervez H,
Koch M, Fraiman G, Mehdi SA and Levitt R: Determination of
HER-2/neu overexpression and clinical predictors of survival in a
cohort of 347 patients with primary malignant brain tumors. Cancer
Invest. 22:537–544. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schlegel J, Stumm G, Brandle K, Merdes A,
Mechtersheimer G, Hynes NE and Kiessling M: Amplification and
differential expression of members of the erbB-gene family in human
glioblastoma. J Neurooncol. 22:201–207. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tabori U, Rienstein S, Dromi Y,
Leider-Trejo L, Constantini S, Burstein Y, Dvir R, Amariglio N,
Toren A, Rechavi G, Izraeli S and Aviram A: Epidermal growth factor
receptor gene amplification and expression in disseminated
pediatric low-grade gliomas. J Neurosurg. 103:357–361.
2005.PubMed/NCBI
|
|
9
|
Gil-Benso R, Lopez-Gines C, Benito R,
Lopez-Guerrero JA, Callaghan RC, Pellin A, Roldan P and
Cerda-Nicolas M: Concurrent EGFR amplification and TP-53 mutation
in glioblastomas. Clin Neuropathol. 26:224–231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakamura JL: The epidermal growth factor
receptor in malignant gliomas: pathogenesis and therapeutic
implications. Expert Opin Ther Targets. 11:463–472. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Okada Y, Ohno C, Ueki K, Ogino M, Kawamoto
S and Kim P: Comparison of numerical change of epidermal growth
factor receptor gene among pre- and postradiation glioma, and
gliosis, and its clinical use. Brain Tumor Pathol. 24:15–18. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schober R, Bilzer T, Waha A, et al: The
epidermal growth factor receptor in glioblastoma: genomic
amplification, protein expression, and patient survival data in a
therapeutic trial. Clin Neuropathol. 14:169–174. 1995.
|
|
13
|
Shinojima N, Tada K, Shiraishi S, Kamiryo
T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, Oka
K, Ishimaru Y and Ushio Y: Prognostic value of epidermal growth
factor receptor in patients with glioblastoma multiforme. Cancer
Res. 63:6962–6970. 2003.PubMed/NCBI
|
|
14
|
Torp SH, Helseth E, Dalen A and Unsgaard
G: Epidermal growth factor receptor expression in human gliomas.
Cancer Immunol Immunother. 33:61–64. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Torp SH, Helseth E, Ryan L, Stolan S,
Dalen A and Unsgaard G: Amplification of the epidermal growth
factor receptor gene in human gliomas. Anticancer Res.
11:2095–2098. 1991.PubMed/NCBI
|
|
16
|
Gan HK, Kaye AH and Luwor RB: The EGFRvIII
variant in glioblastoma multiforme. J Clin Neurosci. 16:748–754.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lal A, Glazer CA, Martinson HM, Friedman
HS, Archer GE, Sampson JH and Riggins GJ: Mutant epidermal growth
factor receptor up-regulates molecular effectors of tumor invasion.
Cancer Res. 62:3335–3339. 2002.PubMed/NCBI
|
|
18
|
Sampson JH, Heimberger AB, Archer GE,
Aldape KD, Friedman AH, Friedman HS, Gilbert MR, Herndon JE II,
McLendon RE, Mitchell DA, Reardon DA, Sawaya R, Schmittling RJ, Shi
W, Vredenburgh JJ and Bigner DD: Immunologic escape after prolonged
progression-free survival with epidermal growth factor receptor
variant III peptide vaccination in patients with newly diagnosed
glioblastoma. J Clin Oncol. 28:4722–4729. 2010. View Article : Google Scholar
|
|
19
|
Maihle NJ, Baron AT, Barrette BA, Boardman
CH, Christensen TA, Cora EM, Faupel-Badger JM, Greenwood T, Juneja
SC, Lafky JM, Lee H, Reiter JL and Podratz KC: EGF/ErbB receptor
family in ovarian cancer. Cancer Treat Res. 107:247–258.
2002.PubMed/NCBI
|
|
20
|
Reiter JL and Maihle NJ: Characterization
and expression of novel 60-kDa and 110-kDa EGFR isoforms in human
placenta. Ann NY Acad Sci. 995:39–47. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reiter JL, Threadgill DW, Eley GD, Strunk
KE, Danielsen AJ, Sinclair CS, Pearsall RS, Green PJ, Yee D,
Lampland AL, Balasubramaniam S, Crossley TD, Magnuson TR, James CD
and Maihle NJ: Comparative genomic sequence analysis and isolation
of human and mouse alternative EGFR transcripts encoding truncated
receptor isoforms. Genomics. 71:1–20. 2001. View Article : Google Scholar
|
|
22
|
Flickinger TW, Maihle NJ and Kung HJ: An
alternatively processed mRNA from the avian c-erbB gene encodes a
soluble, truncated form of the receptor that can block
ligand-dependent transformation. Mol Cell Biol. 12:883–893.
1992.PubMed/NCBI
|
|
23
|
Weber W, Gill GN and Spiess J: Production
of an epidermal growth factor receptor-related protein. Science.
224:294–297. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cadena DL and Gill GN: Expression and
purification of the epidermal growth factor receptor extracellular
domain utilizing a polycistronic expression system. Protein Expr
Purif. 4:177–186. 1993. View Article : Google Scholar
|
|
25
|
Greenfield C, Hiles I, Waterfield MD,
Federwisch M, Wollmer A, Blundell TL and McDonald N: Epidermal
growth factor binding induces a conformational change in the
external domain of its receptor. EMBO J. 8:4115–4123.
1989.PubMed/NCBI
|
|
26
|
Basu A, Raghunath M, Bishayee S and Das M:
Inhibition of tyrosine kinase activity of the epidermal growth
factor (EGF) receptor by a truncated receptor form that binds to
EGF: role for interreceptor interaction in kinase regulation. Mol
Cell Biol. 9:671–677. 1989.PubMed/NCBI
|
|
27
|
Hirsch FR, Varella-Garcia M, Bunn PA Jr,
Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C and Franklin WA:
Epidermal growth factor receptor in non-small-cell lung carcinomas:
correlation between gene copy number and protein expression and
impact on prognosis. J Clin Oncol. 21:3798–3807. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guillaudeau A, Durand K, Pommepuy I,
Robert S, Chaunavel A, Lacorre S, De Armas R, Bourtoumieux S, El
Demery M, Moreau JJ and Labrousse F: Determination of EGFR status
in gliomas: usefulness of immunohistochemistry and fluorescent in
situ hybridization. Appl Immunohistochem Mol Morphol. 17:220–226.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Korshunov A, Sycheva R and Golanov A:
Molecular stratification of diagnostically challenging high-grade
gliomas composed of small cells: the utility of fluorescence in
situ hybridization. Clin Cancer Res. 10:7820–7826. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marks RA, Zhang S, Montironi R, McCarthy
RP, MacLennan GT, Lopez-Beltran A, Jiang Z, Zhou H, Zheng S,
Davidson DD, Baldridge LA and Cheng L: Epidermal growth factor
receptor (EGFR) expression in prostatic adenocarcinoma after
hormonal therapy: a fluorescence in situ hybridization and
immunohistochemical analysis. Prostate. 68:919–923. 2008.
View Article : Google Scholar
|
|
32
|
Durand K, Guillaudeau A, Pommepuy I,
Mesturoux L, Chaunavel A, Gadeaud E, Porcheron M, Moreau JJ and
Labrousse F: Alpha-internexin expression in gliomas: relationship
with histological type and 1p, 19q, 10p and 10q status. J Clin
Pathol. 64:793–801. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Durand KS, Guillaudeau A, Weinbreck N, De
Armas R, Robert S, Chaunavel A, Pommepuy I, Bourthoumieu S, Caire
F, Sturtz FG and Labrousse FJ: 1p19q LOH patterns and expression of
p53 and Olig2 in gliomas: relation with histological types and
prognosis. Mod Pathol. 23:619–628. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hirose T, Ishizawa K and Shimada S:
Utility of in situ demonstration of 1p loss and p53 overexpression
in pathologic diagnosis of oligodendroglial tumors. Neuropathology.
30:586–596. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ligon KL, Alberta JA, Kho AT, Weiss J,
Kwaan MR, Nutt CL, Louis DN, Stiles CD and Rowitch DH: The
oligodendroglial lineage marker OLIG2 is universally expressed in
diffuse gliomas. J Neuropathol Exp Neurol. 63:499–509.
2004.PubMed/NCBI
|
|
36
|
Okada M, Yano H, Hirose Y, Nakayama N, Ohe
N, Shinoda J and Iwama T: Olig2 is useful in the differential
diagnosis of oligodendrogliomas and extraventricular neurocytomas.
Brain Tumor Pathol. 28:157–161. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Coulibaly B, Nanni I, Quilichini B,
Gaudart J, Metellus P, Fina F, Boucard C, Chinot O, Ouafik L and
Figarella-Branger D: Epidermal growth factor receptor in
glioblastomas: correlation between gene copy number and protein
expression. Hum Pathol. 41:815–823. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ilekis JV, Stark BC and Scoccia B:
Possible role of variant RNA transcripts in the regulation of
epidermal growth factor receptor expression in human placenta. Mol
Reprod Dev. 41:149–156. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Voelzke WR, Petty WJ and Lesser GJ:
Targeting the epidermal growth factor receptor in high-grade
astrocytomas. Curr Treat Options Oncol. 9:23–31. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Albitar L, Pickett G, Morgan M, Wilken JA,
Maihle NJ and Leslie KK: EGFR isoforms and gene regulation in human
endometrial cancer cells. Mol Cancer. 9:1662010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lafky JM, Wilken JA, Baron AT and Maihle
NJ: Clinical implications of the ErbB/epidermal growth factor (EGF)
receptor family and its ligands in ovarian cancer. Biochim Biophys
Acta. 1785:232–265. 2008.PubMed/NCBI
|
|
42
|
Shibata MA, Ambati J, Shibata E,
Albuquerque RJ, Morimoto J, Ito Y and Otsuki Y: The endogenous
soluble VEGF receptor-2 isoform suppresses lymph node metastasis in
a mouse immunocompetent mammary cancer model. BMC Med. 8:692010.
View Article : Google Scholar : PubMed/NCBI
|