Introduction
Ovarian cancer is the leading cause of death from
gynaecological malignancies (1,2). It
accounts for 5% of all cancer deaths among women with an estimated
21,880 new cases and 13,850 deaths from ovarian cancer in the
United States in 2010 (2). The
poor prognosis and high mortality rate associated with the disease
have not significantly improved over the last 30 years despite
advances in treatment (3). Current
therapies are effective for patients with early stage disease (FIGO
stage I/II) where 5-year survival rates range from 73% to 93%.
Their usefulness, however, is limited for patients with advanced
stage disease where the 5-year survival is only about 30% (2). Because so many ovarian cancer
patients are diagnosed at a later stage, it is important to find
methods by which to improve treatments for more advanced
disease.
The selective recognition of tumor antigens by the
immune system provides a powerful means to screen for
tumor-associated antigens (TAAs). The identification of tumor
antigens has yielded an array of target molecules for diagnosis,
monitoring, and immunotherapy of human cancer (4). The serologic analysis of recombinant
cDNA expression libraries (SEREX) was designed to combine serologic
analysis with antigenic cloning techniques to identify human tumor
antigens eliciting high-titered IgG antibodies (5) and has provided a powerful approach to
identify immunogenic tumor antigens. To date, over 2,500 tumor
antigens have been identified from a variety of malignancies using
SEREX, which include gastric cancer (6), colon cancer (7), melanoma (8), breast cancer (9), renal cell carcinoma (10), lung cancer (11) and leukemia (12). These antigens can be classified
into several categories, including mutational antigens (5,10),
differentiation antigens (10,14),
over-expressed antigens (15) and
cancer/testis (CT) antigens (16,17).
CT antigens are the products of transcripts present only in
developing germ cells and human cancers of diverse origins that
elicit spontaneous cellular and humoral immune responses in some
cancer patients (18,19). Because of their tissue-restricted
expression and immunogenicity, CT antigens are potential targets
for vaccine-based immunotherapy (20).
Previous SEREX analysis of ovarian cancer by Stone
et al (21) resulted in the
detection of 25 distinct antigens. The majority of these antigens
were recognized only by autologous serum, however 6 antigens were
found to be immunogenic in at least 2 of the 25 patient sera.
Additional studies on ovarian cancer have been performed by Luo
et al (22) and Lokshin
et al (23), who identified
12 and 20 ovarian cancer associated antigens, respectively. OVA-66
antigen identified Jin et al (24) was assessed for immunogenicity by
ELISA using 48 control sera and 113 cancer sera from patients with
various malignancies including ovarian cancer. OVA-66 reacted with
6 out of 27 sera from ovarian cancer patient (22.2%). The homeobox
genes HOX-A7 and HOX-B7 (25,26)
reacted with serum samples from 16/24 (66%) and 13/39 (33%) ovarian
cancer patients, respectively, while normal individuals showed
little or no reactivity toward these antigens. Expression of these
gene products is not tissue-restricted at the mRNA level, and it is
therefore unlikely that these antigens represent viable vaccine
targets. These SEREX-defined ovarian cancer related antigens were
known as TAAs but were not found to be significant CT antigens.
In the present study, the SEREX methodology was
applied to further define the spectrum of immunogenic proteins in
serous ovarian cancer patients. A specific focus was given to the
KP-OVA-52 gene to determine its potential as a possible CT
antigen.
Materials and methods
Human tissues, sera and cell lines
Human tumor tissues and sera were obtained from the
Department of Gynecologic Oncology, Roswell Park Cancer Institute
and Department of Pathology, Pusan National University Hospital
after diagnosis and staging. The tissues were frozen in liquid
nitrogen and stored at −80°C until use. Human ovarian cancer cell
line SK-OV-3; human colon cancer cell lines SNU-C1 and SNU-C2A;
human lung cancer cell lines SK-LC-5 and SK-LC-14; human breast
cancer cell line MCF7; and human small cell lung cancer cell lines
NCI-H82, NCI-H146, and NCI-H189 were obtained from the Korean Type
Culture Collection and the American Type Culture Collection. All
these cell lines were maintained in RPMI-1640 (Gibco-BRL Life
Technologies Inc., Grand Island, NY, USA) medium supplemented with
10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin
and 100 μg/ml streptomycin.
Total-RNA extraction from tissues and
cell lines
Total-RNA was isolated from human tissue samples and
human tumor cell lines using the standard TRIzol reagent (Life
Technologies, Gaithersburg, MD, USA) and RNA isolation kit (RNeasy
maxi kit, Qiagen) following manufacturer’s instructions. The amount
of isolated RNAs was measured by spectrophotometer (Ultrospec 2000,
Pharmacia Biotech) at 260 nm. Normal tissue total-RNAs were
purchased from Clontech Laboratories Inc., (Palo Alto, CA, USA) and
Ambion Inc. (Austin, TX, USA).
Total-RNA from various cell lines were obtained from
the cell repository of the Ludwig Institute for Cancer Research,
New York Branch at the Memorial Sloan-Kettering Cancer Center and
analyzed with reverse transcriptase-polymerase chain reaction
(RT-PCR).
Preparation of cDNA library and sera
Poly(A)+ RNA from normal testis was
purchased from Clontech Laboratories Inc. and Poly(A)+
RNAs from SK-OV-3 and SNU 840 ovarian cancer cell lines were
prepared by using the Fast Track mRNA purification kit (Invitrogen,
Life Technologies, Carlsbad, CA, USA). mRNA 5 μg was used to
construct a cDNA library in the ZAP Express vector (Stratagene, La
Jolla, CA, USA), following the manufacturer’s instructions. The
library contained approximately one million recombinants and was
used for immunoscreening without prior amplification. Five
preparations of sera from ovarian cancer patients were used
independently. The patients ages ranged from 45–64 years and all
patients had advanced (stages III/IV) disease, with serous
histology. Each serum sample was diluted 1:200 for SEREX analysis.
To remove serum antibodies reactive with E.
coli/bacteriophage-related antigens, sera were absorbed against
E. coli/bacteriophage lysates as described by Lee et
al (11).
Immunoscreening
Immunoscreening of the cDNA library was performed as
described (11,17). Briefly, E. coli XL1 blue MRF
cells were transfected with the recombinant phages, plated at a
density of approximately 5,000 pfu/150-mm plate (NZCYM-IPTG agar),
incubated for 8 h at 37°C, and transferred to nitrocellulose
filters (PROTRAN BA 85, 0.45 μm, Schleicher & Schuell).
Then the filters were incubated with a 1:200 dilution of the
patient sera, which had been preabsorbed with E. coli-phage
lysate. The serum-reactive clones were detected with AP-conjugated
secondary antibody and visualized by incubation with
5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium
(BCIP/NBT). After screening, the isolated positive clones were
removed from the plate and conserved in suspension medium (SM)
buffer with 25 μl of chloroform. Positive phages were mixed
with a helper phage to co-infect XL-1 Blue MRF, and they were
rescued into pBluescript phagemid forms by in vivo excision.
The excised phagemids were transformed into the host bacteria
(XLOLR) to multiply for plasmid extraction and stock. The size of
the inserted cDNA was determined primarily by double restriction
enzyme digestion with EcoRI and XhoI. The cDNA was sequenced
commercially (Macrogen, Seoul, Korea).
RT-PCR
The cDNA preparations used as templates for RT-PCR
reactions were prepared using 1 μg of total-RNA in
conjunction with the Superscript first strand synthesis kit
(Invitrogen, Life Technologies). PCR primers were as follows:
KP-OVA-52 5′/3′:AGGAGAAGCGGCAGAGTGGC/AGGTCCTTCCGTC TGGAGCG.
KP-OVA-35 5′/3′:TGAGGAGGAGCAAGAGCC GA/TGGTGCTGTGTGAAGCACAC.
KP-OVA-38 5′/3′:TGAT GACGCCACGTGGGCCG/GCTCTGGTGGCTTGGGTGGC. The
cDNA templates were normalized on the base amplification of GAPDH.
For PCR, 20 μl reaction mixture consisting of 2 μl
cDNA, 0.2 mM dNTP, 1.5 mM MgCl2, 0.25 μM gene
specific forward and reverse primers, and 3 units Taq DNA
polymerase (Solgent, Daejun, Korea) was preheated to 94°C for 5
min, followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec and
72°C for 1 min followed by a final elongation step of 72°C for 5
min. Amplified PCR products were analyzed on 1.5% agarose gels
stained with ethidium bromide.
Real-time quantitative RT-PCR
Oligonucleotide primers for the real-time
quantitative PCR were synthesized commercially. PCR reaction
mixtures (20 μl), consisting of 2 μl cDNA (or 2.0
μl of genomic DNA amplification controls), 0.2 mM dNTP, 1.5
mM MgCl2, 0.25 μM gene specific forward and
reverse primers, and 2.5 units Platinum Taq DNA polymerase
(Invitrogen, Life Technologies), were heated to 55°C for 5 min,
94°C for 10 min, followed by 40 thermal cycles of 94°C for 30 sec,
65°C for 30 sec and 54°C for 30 sec and a final cycle of 72°C for
30 sec. Thermal cycling was performed using a Perkin Elmer GeneAmp
PCR System 9700. Resultant PCR products were analyzed in 2%
agarose/Tris-acetate-EDTA gels. Primer sequences were as follows:
KP-OVA-52 TaqMan primer: 5′-ACCAGCACTCAGAA CAACAGC-3′. KP-OVA-52
TaqMan primer: 3′-TCCTCCGAT GCCAGAAGAGTC-5′. KP-OVA-52 Taq Man
probe: 5′-CTGC GTCAGTGAGGTCCTTCCGTCT-3′. The parameter Ct was
defined as the threshold cycle number at which the fluorescence
generated by cleavage of the probe exceeded the baseline. The
target message was quantified by measuring the Ct value. GAPDH
transcripts were quantified as endogenous RNA control using TaqMan
human GAPDH control regents (Applied Biosystems).
Serum antibody detection assay
(SADA)
Phages corresponding to 48 antigens that were
identified in SEREX screen were screened with individual sera by
the SADA method as described by Lee et al (17). Briefly, 50 μl of phages (500
pfu) were placed in duplicate wells of a 96-well microtitre plate.
The phages were then transferred to a rectangle shaped agar plate
covered previously with E. coli XL1-Blue MRF, top agarose
and 10 mM IPTG by replica pin. The plates were incubated overnight
at 37°C and were incubated with nitrocellulose transfer membranes
for an additional 4 h. Membranes were used immediately for
immunoscreening with each human serum.
Generation of recombinant KP-OVA-52
fusion proteins
The open reading frame (ORF) cDNA inserts of the
hyphotetical protein, KP-OVA-52, from gene bank (MN001042367), were
subcloned into the pET23a expression plasmids containing a
poly-histidine tag (Novagene). The expected protein size was about
29.5 kDa. The induction of recombinant protein synthesis by
isopropyl β-D-thiogalactoside was performed at a low culture
temperature (20°C). Protein synthesis was monitored by
SDS/Poly-acrylamide gel electrophoresis and Coomassie Blue
staining.
Western blot analysis
About 100 ng of purified X6 His-proteins were
separated on 10% SDS-PAGE and transferred onto nitrocellulose
membranes (Hybond-ECL, GE Healthcare). After blocking with TBST
(TBS, 0.1% Tween-20) containing 5% skim milk for 1 h at room
temperature, the membrane was incubated in sera diluted to 1:200
overnight at room temperature. The membranes were washed and
incubated with horseradish peroxidase-conjugated sheep anti-human
IgG antibody (GE healthcare) diluted to 1:3,000 for 1 h at room
temperature. After washing with TBST and mounting with the
chemiluminescence reagent plus (Perkin Elmer), the membrane was
exposed to Kodak medical X-ray film.
Treatment of cells with ADC
The various cell lines were grown in RPMI-1640 (Life
Technologies) and DMEM supplemented with 10% FBS in a 5%
CO2 humidified atmosphere. In experiments designed to
determine the effect of demethylation on the expression of
KP-OVA-52 gene, actively replicating cells were treated with 1
μM of ADC (Sigma, St. Louis, MO, USA) for 5 days. At the end
of treatment, total-RNAs was purified and KP-OVA-52 gene expression
was analyzed by RT-PCR.
Results
Identification of ovarian cancer antigens
by SEREX
Immunoscreening of two ovarian cancer cell lines and
testis cDNA expression libraries with the selected serum led to the
isolation of 151 seroreactive cDNA clones. These clones include 75
independent antigens from testis and ovarian cancer cell lines, and
were designated KP-OVA-1 through KP-OVA-75 (Tables I and II). When the cDNA sequences encoding the
75 ovarian cancer antigens were compared to those deposited in
cancer immunome database (27).
Forty-nine of the 75 antigens (65%) had been previously identified
by SEREX analysis with any cDNA/serum combination whereas 26 (35%)
have not been previously reported (Tables I and II). These 75 antigens comprise of 66
known proteins together with 9 uncharacterized gene products, which
does not present in the sequences designated in the databases as
expressed sequence tags (ESTs), KIAA series clones, FLJ series
clones, MGC series clones, DKFZ series clones, and anonymous open
reading frames (ORFs). The immunomic pattern of the tumor antigens
that were previously identified in SEREX by other groups in the
cancer immunome database was analyzed. One antigen
(KP-OVA-40/MAGE10A) out of 49 previously identified antigens
reacted with sera from ovarian cancer patients. The remaining 48
antigens are known to be associated with other tumor types
including breast cancer, glioma, leukemia, melanoma, sarcoma,
Hodgkin’s disease, breast, lung, hepatocellular, renal cell and
prostate cancer. However, these antigens have not been known to be
associated with ovarian cancers.
 | Table IAntigens identified through the
screening of testis with individual serum. |
Table I
Antigens identified through the
screening of testis with individual serum.
| | | | Seroreactivity
|
|---|
| KP-OVA-number | Genes (Unigene
cluster)a | Serum source
(no.) | Previously
identified by SEREXb | Normal (20) | Ovarian cancer
patients (20) |
|---|
| 1 |
ZNF234(Hs.235992) | 14547 | Y | 0/20 | 1/20 |
| 2 |
BRAP(Hs.530940) | 14547 | Y | 3/20 | 5/20 |
| 3 |
MYNN(Hs.507025) | 14547 | N | 0/20 | 1/20 |
| 4 |
GARS(Hs.404321) | 14547 | N | 0/20 | 1/20 |
| 5 |
TXNRD1(Hs.690011) | 14547 | Y | 0/20 | 1/20 |
| 6 |
PRPF38B(Hs.342307) | 14547 | Y | 0/20 | 1/20 |
| 7 |
ZFP95(Hs.110839) | 14547 | Y | 0/20 | 1/20 |
| 8 |
TEKT3(Hs.414648) | 14547 | N | 0/20 | 1/20 |
| 9 |
RBPJ(Hs.479396) | 14547 | Y | 0/20 | 1/20 |
| 10 | GNL2(Hs.75528) | 14547 | Y | 2/20 | 1/20 |
| 11 |
EID3(Hs.659857) | 14547 | Y | 0/20 | 1/20 |
| 12 |
RPL26(Hs.644794) | 14547 | Y | 0/20 | 1/20 |
| 13 | RPGR(Hs.61438) | 14547 | N | 0/20 | 1/20 |
| 14 |
PAFAH1B1(Hs.77318) | 14547 | Y | 2/20 | 7/20 |
| 15 | PRM1(Hs.2909) | 16100 | Y | 0/20 | 1/20 |
| 16 |
RANBP5(Hs.712598) | 16100 | N | 0/20 | 1/20 |
| 17 |
FNBP1(Hs.189409) | 16100 | Y | 0/20 | 1/20 |
| 18 |
MRPL45(Hs.462913) | 16100 | Y | 1/20 | 5/20 |
| 19 |
PIK3R3(Hs.655387) | 16100 | Y | 2/20 | 6/20 |
| 20 |
TEX2(Hs.175414) | 16100 | N | 1/20 | 6/20 |
| 21 |
TMBIM1(Hs.591605) | 16100 | Y | 0/20 | 1/20 |
| 22 |
RPL3(Hs.119598) | 16100 | Y | 0/20 | 1/20 |
| 23 |
GTF2H1(Hs.577202) | 16100 | N | 0/20 | 1/20 |
| 24 |
GART(Hs.473648) | 16100 | N | 1/20 | 3/20 |
| 25 |
TBL2(Hs.647044) | 16100 | N | 0/20 | 4/20 |
| 26 |
AHNAK(Hs.502756) | 16100 | N | 0/20 | 1/20 |
| 27 |
RPS10(Hs.645317) | 16086 | Y | 0/20 | 1/20 |
| 28 | FLJ21228
fis(Hs.677287) | 16086 | Y | 0/20 | 1/20 |
| 29 |
STXBP3(Hs.530436) | 16086 | N | 0/20 | 1/20 |
| 30 |
PARP2(Hs.409412) | 16086 | N | 0/20 | 1/20 |
| 31 |
GKAP1(Hs.522255) | 16086 | N | 0/20 | 1/20 |
| 32 |
CD164(Hs.520313) | 16086 | N | 0/20 | 1/20 |
| 33 |
CSNK1D(Hs.631725) | 16086 | Y | 0/20 | 1/20 |
| 34 |
PDHX(Hs.502315) | 16086 | Y | 0/20 | 1/20 |
| 35 | TNP1(Hs.3017) | 16086 | Y | 0/20 | 6/20 |
| 36 |
CCDC104(Hs.264208) | 16086 | N | 0/20 | 2/20 |
| 37 |
TPT1(Hs.374596) | 16086 | Y | 2/20 | 6/20 |
| 38 |
C10orf62(Hs.662302) | 16160 | N | 0/20 | 1/20 |
| 39 |
KIF27(Hs.697514) | 16160 | N | 0/20 | 1/20 |
| 40 |
MAGEA10(Hs.18048) | 16160 | N | 0/20 | 1/20 |
| 41 |
BPTF(Hs.444200) | 16160 | N | 1/20 | 6/20 |
| 42 |
SETDB2(Hs.631789) | 16160 | Y | 0/20 | 1/20 |
| 43 |
C16orf80(Hs.532755) | 16160 | N | 0/20 | 1/20 |
| 44 |
JAKMIP2(Hs.184323) | 16160 | Y | 0/20 | 1/20 |
| 45 |
CREM(Hs.200250) | 16160 | N | 0/20 | 1/20 |
| 46 |
FAM128B(Hs.469925) | 16160 | Y | 0/20 | 1/20 |
| 47 |
ZBTB44(Hs.178499) | 16160 | N | 0/20 | 1/20 |
| 48 |
LRRC6(Hs.591865) | 16160 | N | 0/20 | 1/20 |
| 49 |
HSF2(Hs.158195) | 16160 | Y | 0/20 | 1/20 |
| 50 | USP1(Hs.35086) | 16160 | Y | 0/20 | 1/20 |
| 51 |
EIF4G3(Hs.467084) | 16160 | Y | 0/20 | 1/20 |
| 52 |
C15orf60(Hs.730877) | 16160 | Y | 0/20 | 1/20 |
 | Table IIAntigens identified through the
screening of ovarian cancer cell lines with individual serum. |
Table II
Antigens identified through the
screening of ovarian cancer cell lines with individual serum.
| | | | Seroreactivity
|
|---|
| KP-OVA-number | Genes (Unigene
cluster)a | Source of library
and serum | Previously
identified by SEREXb | Normal (20) | Ovarian cancer
patients (20) |
|---|
| 53 | KRT17(Hs.2785) |
SNU840/no.14495 | Y | 0/20 | 1/20 |
| 54 |
HMGA1(Hs.518805) |
SNU840/no.14495 | N | 0/20 | 1/20 |
| 55 |
FAM63B(Hs.591122) |
SNU840/no.14495 | Y | 3/20 | 1/20 |
| 56 |
NHP2L1(Hs.182255) |
SNU840/no.14495 | N | 0/20 | 1/20 |
| 57 | TK1(Hs.515122) |
SNU840/no.14495 | N | 0/20 | 1/20 |
| 58 |
DDB1(Hs.290758) |
SNU840/no.14495 | N | 0/20 | 1/20 |
| 59 |
NME1(Hs.463456) |
SNU840/no.14547 | Y | 0/20 | 1/20 |
| 60 | OGFR(Hs.67896) |
SNU840/no.14547 | Y | 0/20 | 1/20 |
| 61 |
RPL14(Hs.730621) |
SNU840/no.14547 | Y | 0/20 | 1/20 |
| 62 | EIF3S5(Hs.516023
) |
SNU840/no.14547 | N | 0/20 | 1/20 |
| 63 |
Hist1h1c(Hs.7644) |
SNU840/no.14547 | Y | 0/20 | 1/20 |
| 64 |
GUK1(Hs.376933) |
SNU840/no.14547 | N | 1/20 | 1/20 |
| 65 |
HOMER3(Hs.720208) |
SNU840/no.14547 | N | 0/20 | 1/20 |
| 66 |
GSDMD(Hs.118983) |
SNU840/no.14547 | Y | 0/20 | 1/20 |
| 67 |
KIF7(Hs.513134) |
SK-OV-3/no.16160 | N | 0/20 | 1/20 |
| 68 |
C16orf42(Hs.134846) |
SK-OV-3/no.16160 | Y | 0/20 | 3/20 |
| 69 |
TINP1(Hs.482526) |
SK-OV-3/no.16160 | N | 0/20 | 1/20 |
| 70 |
SSNA1(Hs.530314) |
SK-OV-3/no.16160 | Y | 1/20 | 5/20 |
| 71 |
FTH1(Hs.524910) |
SK-OV-3/no.16160 | N | 0/20 | 1/20 |
| 72 |
TSPAN4(Hs.654836) |
SK-OV-3/no.16160 | Y | 0/20 | 1/20 |
| 73 |
S100A4(Hs.654444) |
SK-OV-3/no.16160 | Y | 0/20 | 3/20 |
| 74 | PRP6(Hs.31334) |
SK-OV-3/no.16160 | Y | 0/20 | 1/20 |
| 75 |
MRPS26(Hs.18946) |
SK-OV-3/no.16160 | Y | 0/20 | 1/20 |
Putative CT-like antigens and RT-PCR in
normal tissues and cancer cell lines
A preliminary in silico mRNA expression
profile and characterization of gene products identified in this
study was undertaken based on the tissue distribution of expressed
sequence tags (ESTs) and serial analysis of gene expression (SAGE)
tags in the Cancer Genome Anatomy Database (CGAP: http://cgap.nci.nih.gov/) as well as the information
contained in the GeneCards database (http://bioinfo.weizmann.ac.il/cards-bin/). Four
antigens, KP-OVA-35, -38, -40 and -52 were identified as CT-like
antigens that could potentially serve as targets for ovarian cancer
immunotherapy. Of four CT-like antigens, KP-OVA-40/MAGE10A was
previously reported as a CT antigen (28). To examine expression of the novel
CT-like KP-OVA-35, -38 and -52, conventional RT-PCR was performed
using mRNA from normal tissues. These three antigens were expressed
strongly only in normal testis (Fig.
1A). The mRNA expression profiles of the KP-OVA-35 and
KP-OVA-38 were analyzed in various cancer cell lines and malignant
cancer tissues by RT-PCR. Transcripts encoding KP-OVA-35 and -38
were not detected in cancer cell lines and tumor tissues (data not
shown). Therefore, KP-OVA-35 and -38 antigens are a virtual CT
antigen, as its expressions in both normal tissues and cancer cell
lines could not be verified by RT-PCR analysis. To examine the
distribution of KP-OVA-52 gene expression in detail, conventional
and real time RT-PCR were performed from various cancer cell lines
including ovarian cancer cell lines. KP-OVA-52 was expressed in 3
out of 9 ovary cancer cell lines (Fig.
1B) and not expressed in other cancer cell lines including
breast, colon, hepatoma, lung, renal, sarcoma, and thyroid
anaplastic cell line. Also, quantitative real-time RT-PCR
demonstrated KP-OVA-52 in one of the 16 ovarian cancer tissues
(Fig. 2).
 | Figure 1Conventional RT-PCR analysis of
antigens in normal tissues and ovarian cancer lines. (A) From the
left: 1, spleen; 2, thymus; 3, prostate; 4, ovary; 5, small
intestine; 6, colon; 7, leukocyte; 8, heart; 9, brain; 10,
placenta; 11, lung; 12, pancreas; 13, liver; 14, skeletal muscle;
15, kidney and 16, testis. The cDNA templates were normalized using
GAPDH as shown in the bottom panel. (B) From the left: 1, A-10; 2,
OV-2774; 3, OV-CAR-3; 4, SK-OV-1; 5, SK-OV-3; 6, SK-OV-4; 7,
SK-OV-6; 8, SNU-8 and 9, SNU-840. |
ADC activates KP-OVA-52 expression in
cancer cell lines
Despite the expression of KP-OVA-52 in ovarian
cancer cell lines and ovarian tumors, it is expressed with low
frequency and is not expressed in most cancer cells. We determined
whether silencing of the KP-OVA-52 gene expression in cancer cell
lines was mediated by DNA hypermethylation. For this purpose, we
utilized a methylation-specific PCR program (www.urogene.org/methprimer/index1.html) to determine
the methylation status of the CpG island in the promoter region of
KP-OVA-52. Ten cancer cell lines were treated with 1 μM of
ADC for 5 days, and the mRNA expression levels of KP-OVA-52 were
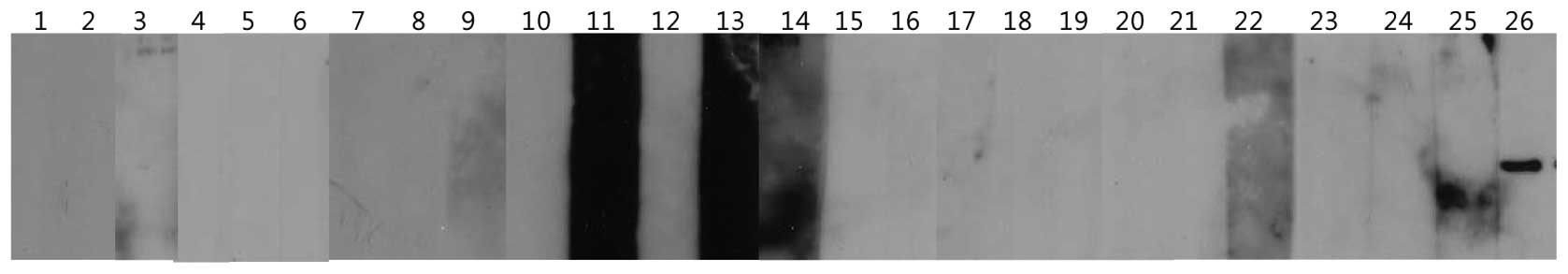
analyzed. As demonstrated in Fig.
3, KP-OVA-52 non expressing six cell lines treated ADC
demonstrated KP-OVA-52 mRNA expression, while KP-OVA-52 mRNA was
not restored expression in three cell lines (NCI-H23, MCF-7 and
NCI-H146). In the KP-OVA-52-expressing cell line, SK-OVA-3,
KP-OVA-52 mRNA expression with ADC treatment was slightly increased
(Fig. 3). These results suggest
that the expression of the KP-OVA-52 is activated by the
demethylating agent.
Seroreactivity of isolated antigens by
SADA and KP-OVA-52 by western blot analysis
To determine whether immune recognition of these
expressed cDNA clones was cancer related, allogeneic sera samples
obtained from 20 normal blood donors and 20 patients with ovarian
cancer were tested for their reactivity by SADA. SADA is based on
the SEREX method and is used to determine seroreactivity against
antigens. Each antigen was spotted twice and antigens were only
considered positive if the duplicates were positive. A spot was
scored as positive if it was clearly darker than the spots
corresponding to the negative phage. Of the 75 antigens screened,
13 reacted with a subset of sera from both normal and cancer
patients, 14 reacted with two or more allogeneic serum samples from
ovarian cancer patients, and the remaining 58 reacted only with the
screening sera.
The 17 clones that reacted with sera from both
normal and cancer patients or cancer patients alone are listed in
Table III. Sera from ovarian
cancer patients compared to those from healthy donors showed a high
percent reactivity (ranging from 15% to 35%) with 13 antigens
(KP-OVA-2, 14, 18, 19, 20, 24, 25, 35, 37, 41, 68, 70 and 73). Of
these antigens, four antigens, KP-OVA-25 (TBL2), KP-OVA-35 (TNP1),
KP-OVA-68 (C16orf42) and KP-OVA-73 (S100A4) reacted with 20%, 30%,
15%, and 15% ovarian cancer sera, respectively, and not with sera
from normal individuals. Of the four genes, the S100A4 (KP-OVA-73)
has been shown to function as an autocrine/paracrine factor that
plays an important role in the aggressive behavior of ovarian
carcinoma (29). Although
KP-OVA-52 reacted with 1 of 20 ovarian cancer sera (5%) and not
with sera from normal individuals (Table I), it is considered a new CT
antigen. Moreover, KP-OVA-52 protein expression was also tested in
20 ovarian cancer sera and six normal sera with western blot
analysis. KP-OVA-52 protein expression was only detected in one
screening serum and was not detected in normal sera (Fig. 4).
 | Table IIIAntigens with high
seroreactivity. |
Table III
Antigens with high
seroreactivity.
| | Seroreactivity (%)
|
|---|
| KP-OVA-number | Genes (Unigene
cluster) | Normal | Ovarian cancer
patients |
|---|
| 2 |
BRAP(Hs.530940) | 15 | 25 |
| 14 |
PAFAH1B1(Hs.77318) | 10 | 35 |
| 18 |
MRPL45(Hs.462913) | 5 | 25 |
| 19 |
PIK3R3(Hs.655387) | 10 | 30 |
| 20 |
TEX2(Hs.175414) | 5 | 30 |
| 24 |
GART(Hs.473648) | 5 | 15 |
| 25 |
TBL2(Hs.647044) | 0 | 20 |
| 35 | TNP1(Hs.3017) | 0 | 30 |
| 36 |
CCDC104(Hs.264208) | 0 | 10 |
| 37 |
TPT1(Hs.374596) | 0 | 30 |
| 41 |
BPTF(Hs.444200) | 5 | 30 |
| 68 |
C16orf42(Hs.134846) | 0 | 15 |
| 70 |
SSNA1(Hs.530314) | 5 | 25 |
| 73 |
S100A4(Hs.654444) | 0 | 15 |
Discussion
The characterization of tumor-associated antigens,
recognized by cellular or humoral effectors of the immune system,
represents a new perspective for diagonosis and cancer
immunotherapy (20). In this
study, SEREX methodology was applied to further definine the
spectrum of immunogenic proteins in serous ovarian cancer patients.
Immunoscreening of cDNA expression libraries from testis and two
ovarian cancer cell lines led to the isolation of 75 independent
antigens, designated KP-OVA-1 through KP-OVA-75. The 75 antigens
identified in this SEREX analysis of ovarian cancer represent a
broad spectrum of cellular components, with 66 being the products
of known genes and 9 representing uncharacterized gene products. A
striking feature of these antigens was the diversity of gene
products recognized by the immune system of ovarian cancer
patients, i.e., transcription factors, mRNA splicing factors,
translation factors, DNA binding proteins, metabolic enzymes,
molecular chaperones, signaling molecules, cytoskeleon proteins and
membrane-associated proteins. However, the majority of these
SEREX-defined antigens have no known association with cancer or
with autoimmunity. CT antigens are immunogenic proteins expressed
in normal testis and different type of tumors. Therefore, CT
antigens are promising candidates for cancer immunotherapy and the
identification of novel CT antigens is a prerequisite for the
development of cancer vaccines (20). On the basis of digital expression
analysis, four tissue-restricted antigens were identified KP-OVA-35
(TNP1), KP-OVA-38 (C10orf62), KP-OVA-40 (MAGEA10), and KP-OVA-52
(C15orf60).
MAGEA10 is a highly immunogenic member of the MAGEA
gene family located in the q28 region of chromosome X and belongs
to the CT antigen family. The MAGE-A10 gene has two splice variants
which encode the same protein (30) and is expressed by different
histological types of tumors, such as non-small cell lung cancer,
urothelial carcinoma, head and neck squamous cell carcinoma,
hepatocellular carcinoma and melanoma (31,32,33).
Transition protein-1(TNP1, KP-OVA-35) is a
spermatid-specific product of the haploid genome which replaces
histone and is itself replaced in the mature sperm by the
protamines (34). TNP1 encodes a
small peptide protein (55 amino acids) that constitutes one exon.
The TNP1 gene regulates conversion of nucleosomal chromatin to the
compact, non-nucleosomal form found in the sperm nucleus. TNP1 is
associated with the appearance of a small set of basic chromosomal
transition proteins in the elongating spermatids of mammals. Two
uncharacterized proteins, C10orf62 and C15orf60, encode 223 and 266
amino acids, respectively, and the in vivo function of these
genes is not yet known.
Of these four antigens, our studies focused on
KP-OVA-35, -38 and -52. Conventional RT-PCR demonstrated strong
mRNA expression in normal testis alone from the three antigens of
interest (Fig. 1). However,
transcripts encoding KP-OVA-35 and -38 were not detected in cancer
cell lines and tumor tissues (data not shown). KP-OVA-52 was
expressed in 3 out of 8 ovarian cancer cell lines and with low
frequency in ovarian cancer tissues (1/16), but the gene was not
expressed in various other cancer cell lines. These results suggest
that KP-OVA-52 is likely to be ovarian cancer specific.
For many CT antigens, especially those encoded by
the X chromosome (CT-X antigens), expression is regulated by
epigenetic mechanisms such as hypermethylation of CpG islands
within the promoter region (35).
Recently, it has been reported that treatment of ovarian cancer
cell lines with ADC increased the expression of the CT antigens
tested, including MAGE-A1, MAGE-A3, MAGE-A4, MAGE-A6, MAGE-A10,
MAGE-A12, NY-ESO-1, TAG-1, TAG-2a, TAG-2b, and TAG-2c. Thus,
treatment with ADC may provide a new therapeutic strategy for
modulating CT antigen expression in combination with
immunotherapeutic approaches (36).
Using a methylation-specific PCR program (www.urogene.org/methprimer/index1.html)
CpG island in the promoter region of KP-OVA-52 was identified.
Thus, we investigated whether silencing of the KP-OVA-52 gene
expression was mediated by hypermethylation. We found evidence that
the expression of KP-OVA-52 is induced by hypomethylation after
treatment with the demethylating agent ADC in cancer cell lines not
expressing KP-OVA-52 (Fig. 3). Our
results clearly indicate a significant correlation between the
expression of KP-OVA-52 and hypomethylation.
With regard to immunogenicity, we analyzed anti-IgG
antibodies in sera from normal and ovarian cancer patients against
the SEREX defined antigens using SADA. Fifty-eight of the 75
antigens were reactive with the screening sera; 13 of these were
reactive with sera from both normal donors and cancer patients, and
4 other antigens (KP-OVA-25/TBL2, KP-OVA-35/TNP1,
KP-OVA-68/C16orf42 and KP-OVA-73/S100A4) reacted exclusively with
sera from cancer patients (ranging from 15% to 30%) (Table III). Despite no expression of the
TNP1 gene (KP-OVA-35) in ovarian tumors and several cancer cell
lines, TNP1 demonstrated a high percent reactivity (30%). S100A4,
which plays an important role in the aggressiveness of ovarian
carcinoma cells, was detected in 3 out of 20 allogenic sera.
Although results of these two antigens were interesting, a broader
analysis will need to be performed in order to determine whether
these antigens act as immunogens in ovarian cancer patients.
Interesting, KP-OVA-52, considered a new CT antigen, reacted with 1
of 20 ovarian cancer sera by SADA and with 1 of 20 ovarian cancer
sera by western blot analysis.
In summary, the current SEREX analysis of ovarian
cancer led to the isolation of 4 tissue-restricted gene products,
including one known CT antigen (KP-OVA-40/MAGEA10). Two of these
differentially expressed antigens (KP-OVA-38/C10orf62 and
KP-OVA-52/C15orf60) are novel gene products, and the remaining
tissue-restricted antigen (KP-OVA-35/TNP1) has not been previously
studied in relation to cancer. Their tissue restricted expression
profile and immunogenicity indicate that these four antigens should
be further analyzed with regard to their immunotherapeutic
potential. Of particular interest is KP-OVA-52, which represents a
recently defined CT antigen expressed exclusively in normal testis
as well as ovarian cancer. Unlike several other CT antigens,
KP-OVA-52 is expressed with low frequency in ovarian cancer and is
not expressed in most other cancer cells. Our results demonstrate
that the silencing of KP-OVA-52 expression is restored by the
demethylating agent ADC and the expression of KP-OVA-52 is
suppressed by hypermethylation. Thus, we suggest that the KP-OVA-52
is a CT antigen and may be useful as a potential target for cancer
immunotherapy.
Acknowledgements
The study was supported by a grant
from Industrialization Support Program for Bio-technology of
Agriculture and Forestry (20110301-061-532-001-05-00), Ministry for
Food, Agriculture, Forestry and Fisheries, Republic of Korea and a
grant to the Ovarian Cancer Working Group of the Cancer Research
Institute, USA.
References
|
1.
|
Ovarian cancer in Australia: An overview,
2010. Australian Institute of Health and Welfare and National
Breast and Ovarian Cancer Centre; Canberra: 2010
|
|
2.
|
Altekruse SF, Kosary CL, Krapcho M, Neyman
N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H,
Mariotto A, Eisner MP, Lewis DR, Cronin K, Chen HS, Feuer EJ,
Stinchcomb DG and Edwards BK: SEER Cancer Statistics Review
1975–2007. National Cancer Institute; Bethesda, MD: 2011
|
|
3.
|
Hennessy BT, Coleman RL and Markman M:
Ovarian cancer. Lancet. 374:1371–1382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Jaras K and Anderson K: Autoantibodies in
cancer: prognostic biomarkers and immune activation. Expert Rev
Proteomics. 8:577–589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Sahin U, Tureci O, Schmitt H, Cochlovius
B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I and
Pfreundschuh M: Human neoplasms elicit multiple specific immune
responses in the autologous host. Proc Natl Acad Sci USA.
92:11810–11813. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Obata Y, Takahashi T, Sakamoto J, Tamaki
H, Tominaga S, Hamajima N, Chen YT and Old LJ: SEREX analysis of
gastric cancer antigens. Cancer Chemother Pharmacol. 46(Suppl):
S37–S42. 2000. View Article : Google Scholar
|
|
7.
|
Song MH, Ha JC, Lee SM, Park YM and Lee
SY: Identification of BCP-20 (FBXO39) as a cancer/testis antigen
from colon cancer patients by SEREX. Biochem Biophys Res Commun.
408:195–201. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Chen YT, Gure AO, Tsang S, Stockert E,
Jager E, Knuth A and Old LJ: Identification of multiple
cancer/testis antigens by allogeneic antibody screening of a
melanoma cell line library. Proc Natl Acad Sci USA. 95:6919–6923.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Jager D, Stockert E, Scanlan MJ, Gure AO,
Jager E, Knuth A, Old LJ and Chen YT: Cancer-testis antigens and
ING1 tumor suppressor gene product are breast cancer antigens:
characterization of tissue-specific ING1 transcripts and a
homologue gene. Cancer Res. 59:6197–6204. 1999.PubMed/NCBI
|
|
10.
|
Scanlan MJ, Gordan JD, Williamson B,
Stockert E, Bander NH, Jongeneel V, Gure AO, Jager D, Jager E,
Knuth A, Chen YT and Old LJ: Antigens recognized by autologous
antibody in patients with renal-cell carcinoma. Int J Cancer.
83:456–464. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Lee SY, Williamson B, Caballero OL, Chen
YT, Scanlan MJ, Ritter G, Jongeneel CV, Simpson AJ and Old LJ:
Identification of the gonad-specific anion transporter SLCO6A1 as a
cancer/testis (CT) antigen expressed in human lung cancer. Cancer
Immun. 4:132004.PubMed/NCBI
|
|
12.
|
Chen G, Zhang W, Cao X, Li F, Liu X and
Yao L: Serological identification of immunogenic antigens in acute
monocytic leukemia. Leuk Res. 29:503–509. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Chen YT, Scanlan MJ, Sahin U, Tureci O,
Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M and Old
LJ: A testicular antigen aberrantly expressed in human cancers
detected by autologous antibody screening. Proc Natl Acad Sci USA.
94:1914–1918. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Scanlan MJ, Gout I, Gordon CM, Williamson
B, Stockert E, Gure AO, Jager D, Chen YT, Mackay A, O’Hare MJ and
Old LJ: Humoral immunity to human breast cancer: antigen definition
and quantitative analysis of mRNA expression. Cancer Immun.
1:42001.PubMed/NCBI
|
|
15.
|
Jager D, Stockert E, Gure AO, Scanlan MJ,
Karbach J, Jager E, Knuth A, Old LJ and Chen YT: Identification of
a tissue-specific putative transcription factor in breast tissue by
serological screening of a breast cancer library. Cancer Res.
61:2055–2061. 2001.PubMed/NCBI
|
|
16.
|
Scanlan MJ, Gure AO, Jungbluth AA, Old LJ
and Chen YT: Cancer/testis antigens: an expanding family of targets
for cancer immunotherapy. Immunol Rev. 188:22–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Lee SY, Obata Y, Yoshida M, Stockert E,
Williamson B, Jungbluth AA, Chen YT, Old LJ and Scanlan MJ:
Immunomic analysis of human sarcoma. Proc Natl Acad Sci USA.
100:2651–2656. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Simpson AJ, Caballero OL, Jungbluth A,
Chen YT and Old LJ: Cancer/testis antigens, gametogenesis and
cancer. Nat Rev Cancer. 5:615–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Scanlan MJ, Simpson AJ and Old LJ: The
cancer/testis genes: review, standardization, and commentary.
Cancer Immun. 4:12004.PubMed/NCBI
|
|
20.
|
Caballero OL and Chen YT: Cancer/testis
(CT) antigens: potential targets for immunotherapy. Cancer Sci.
100:2014–2021. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Stone B, Schummer M, Paley PJ, Thompson L,
Stewart J, Ford M, Crawford M, Urban N, O’Briant K and Nelson BH:
Serologic analysis of ovarian tumor antigens reveals a bias toward
antigens encoded on 17q. Int J Cancer. 104:73–84. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Luo LY, Herrera I, Soosaipillai A and
Diamandis EP: Identification of heat shock protein 90 and other
proteins as tumour antigens by serological screening of an ovarian
carcinoma expression library. Br J Cancer. 87:339–343. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Lokshin AE, Winans M, Landsittel D,
Marrangoni AM, Velikokhatnaya L, Modugno F, Nolen BM and Gorelik E:
Circulating IL-8 and anti-IL-8 autoantibody in patients with
ovarian cancer. Gynecol Oncol. 102:244–251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Jin S, Wang Y, Zhang Y, Zhang HZ, Wang SJ,
Tang JQ, Chen HJ and Ge HL: Humoral immune responses against
tumor-associated antigen OVA66 originally defined by serological
analysis of recombinant cDNA expression libraries and its
potentiality in cellular immunity. Cancer Sci. 99:1670–1678. 2008.
View Article : Google Scholar
|
|
25.
|
Naora H, Montz FJ, Chai CY and Roden RB:
Aberrant expression of homeobox gene HOXA7 is associated with
mullerian-like differentiation of epithelial ovarian tumors and the
generation of a specific autologous antibody response. Proc Natl
Acad Sci USA. 98:15209–15214. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Naora H, Yang YQ, Montz FJ, Seidman JD,
Kurman RJ and Roden RB: A serologically identified tumor antigen
encoded by a homeobox gene promotes growth of ovarian epithelial
cells. Proc Natl Acad Sci USA. 98:4060–4065. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
The Academy of Cancer Immunology and the
Ludwig Institute for Cancer Research: Cancer Immunome Database.
Avaible online: http://ludwig-sun5.unil.ch/CancerImmunomeDB,
1997.
|
|
28.
|
Valmori D, Dutoit V, Rubio-Godoy V,
Chambaz C, Lienard D, Guillaume P, Romero P, Cerottini JC and
Rimoldi D: Frequent cytolytic T-cell responses to peptide
MAGE-A10(254–262) in melanoma. Cancer Res. 61:509–512.
2001.PubMed/NCBI
|
|
29.
|
Kikuchi N, Horiuchi A, Osada R, Imai T,
Wang C, Chen X and Konishi I: Nuclear expression of S100A4 is
associated with aggressive behavior of epithelial ovarian
carcinoma: an important autocrine/paracrine factor in tumor
progression. Cancer Sci. 97:1061–1069. 2006. View Article : Google Scholar
|
|
30.
|
Lin J, Lin L, Thomas DG, Greenson JK,
Giordano TJ, Robinson GS, Barve RA, Weishaar FA, Taylor JM,
Orringer MB and Beer DG: Melanoma-associated antigens in esophageal
adenocarcinoma: identification of novel MAGE-A10 splice variants.
Clin Cancer Res. 10:5708–5716. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Groeper C, Gambazzi F, Zajac P, Bubendorf
L, Adamina M, Rosenthal R, Zerkowski HR, Heberer M and Spagnoli GC:
Cancer/testis antigen expression and specific cytotoxic T
lymphocyte responses in non small cell lung cancer. Int J Cancer.
120:337–343. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Sharma P, Shen Y, Wen S, Bajorin DF,
Reuter VE, Old LJ and Jungbluth AA: Cancer-testis antigens:
expression and correlation with survival in human urothelial
carcinoma. Clin Cancer Res. 12:5442–5447. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Figueiredo DL, Mamede RC, Proto-Siqueira
R, Neder L, Silva WA Jr and Zago MA: Expression of cancer testis
antigens in head and neck squamous cell carcinomas. Head Neck.
28:614–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Shirley CR, Hayashi S, Mounsey S,
Yanagimachi R and Meistrich ML: Abnormalities and reduced
reproductive potential of sperm from Tnp1- and Tnp2-null double
mutant mice. Biol Reprod. 71:1220–1229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Park JH, Song MH, Lee CH, Lee MK, Park YM,
Old L and Lee SY: Expression of the human cancer/testis antigen
NY-SAR-35 is activated by CpG island hypomethylation. Biotechnol
Lett. 33:1085–1091. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Adair SJ and Hogan KT: Treatment of
ovarian cancer cell lines with 5-aza-2′-deoxycytidine upregulates
the expression of cancer-testis antigens and class I major
histocompatibility complex-encoded molecules. Cancer Immunol
Immunother. 58:589–601. 2009.
|