Introduction
The failure of surgical treatment in patients with
glioma is mainly due to the invasion of tumor cells into the normal
brain beyond the resection areas. These invasive remaining cells
are inoperable, resistant to radio- and chemotherapy, and
eventually lead to tumor regrowth. Finally, due to the invasion
ability of malignant glioma, patients generally die within 1 year
after the initial diagnosis (1,2).
To infiltrate into adjacent tissues, certain tumor
cells should be able to recognize the barrier matrix, to breach the
matrix and to grow into the ectopic locale. Other tumor cells also
need to control signaling pathways that would contribute to tumor
invasion. It is difficult to directly investigate the specific
phenotype that is associated with invasion ability in vivo.
The dysregulation of the induced motility of tumor cells has been
postulated to be one of the invasion processes that functions in
the transmigration of the barrier matrix. Thus, there is a good
correlation between increased invasiveness in vivo and
increased motility in vitro (1,3,4). The
identification of genes related to increased motility is critical
for understanding the molecular basis of biological behavior in
invasive gliomas.
In a previous study, the authors sought to identify
genes that could serve as determining factors for the mobility of
malignant glioma cells using DD-PCR (5,6). The
results showed that metallothionein 1E was highly expressed in
motile cell lines.
Metallothionein 1E (MT1E) is the name for a family
of low-molecular weight intracellular metalloproteins with a high
affinity for heavy metal ions, such as zinc, cadmium, copper,
mercury, and platinum. A striking feature of these metalloproteins
is that they are involved in heavy metal detoxification,
chemoresistance to anti-cancer drugs and free radical scavenging
for cell protection (7–11). Furthermore, they can modulate the
activities of zinc-dependent regulatory proteins including enzymes
and zinc-finger transcription factors, by the removal and transfer
of zinc (12,13). It is also well known that MT
participates in fundamental cellular processes, such as cell
proliferation and apoptosis (14,15).
However, even though many reports have been issued concerning MT,
its functions associated with the migration and invasion have not
reported in glioma.
Nuclear factor κB (NF-κB) is a transcription factor
that regulates an exceptionally large number of genes, particularly
those involved in immune and inflammatory responses. NF-κB is a
dimer most commonly composed of two protein subunits, p50 and p65,
and normally remains in an inactive form in the cytoplasm bound to
an inhibitory subunit, IκB. Various stimuli activate upstream IκB
kinases, which in turn phosphorylate and degrade IκB. The released
NF-κB then enters the nucleus and activates the transcription of
many different target genes. NF-κB also plays a pivotal role in a
diverse array of cellular activities associated with the regulation
of cell death, growth, and development (16). NF-κB has far reaching importance in
the regulation of cell death as the overexpression of NF-κB renders
cancer cells resistant to chemotherapeutic agents (17–19).
MT1E may act as a potential intracellular modulator of NF-κB
activation by regulating the cellular level and activity of NF-κB
(20,21). Despite the obvious importance of
understanding the MT1E-NF-κB interaction, little is known about the
tentative molecules that coordinate this interaction.
The relationship between tumor cells and host
factors have an effect on invasion, thus a greater understanding of
these factors could be important in the anti-invasive research.
Primary tumor cell invasion of surrounding tissue is the first
stage of a metastasis cascade, and many factors, such as matrix
metalloproteinases (MMPs), are associated with this stage (22). MMPs belong to a family of
zinc-dependent endopeptidases that degrade the extracellular
matrix. Especially, MMPs have well known effects on tumor
proliferation and metastasis. Therefore, we investigated whether MT
actually modulates migration and invasion in human glioma cells by
modulating activity of the NF-κB, MMP-2 and -9.
Materials and methods
Cell lines and cell culture
Human glioma cell lines, U87MG, U251MG, U373MG and
U343MG-A were obtained from the Korean Cell Line Bank, Seoul, Korea
and from the Brain Tumor Research Center, University of California,
San Francisco, CA, USA, respectively. U87MG-10′ was obtained by
repetition scratch to tenth U87MG. All cell lines were routinely
grown in Dulbecco’s modified Eagle’s medium (Gibco-BRL,
Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum
(Gibco-BRL, Gaithersburg, MD, USA) at 37°C in a humidified 95%
air/5% CO2 atmosphere. Samples of astrocytic tumors were
obtained from the patients with the surgery performed at Chonnam
National University Hospital. The specimens were snap-frozen in
liquid nitrogen and stored at −70°C until use.
Migration assay
In order to stop cell proliferation, the media used
to culture cells were replaced with medium containing 5 mM
hydroxyurea. After 24 h of hydroxyurea treatment, the cultures were
scraped with a single-edged razor blade. Cells were washed twice
with PBS and placed in medium containing hydroxyurea. After 48 h of
incubation, the cells were washed twice with PBS, fixed with
absolute alcohol, and stained with 0.1% toluidine blue. Six
microscopic fields were evaluated for each wound injury. The number
of cells migrating across the wound edge and the maximum distance
migrated were determined in each field and averaged for each
injury. These experiments were repeated three times.
RNA isolation from glioma cell
lines
Total RNA was isolated from the malignant glioma
cell lines using TRIzol (Life Technologies, Gaithersburg, MD)
according to the manufacturer’s instructions. After isolating the
RNA, the RNA pellets were frozen and stored at −80°C until use.
Differentially display-polymerase
chain reaction (DD-PCR)
DD-PCR was performed using the
Genefishing™ kit (Seegene Incorp., Seoul, Korea)
according to the manufacturer’s instructions. For first strand
synthesis, 3 μg of the purified total RNA was incubated with 10
μM dT-ACP for 3 min at 80°C, followed by the addition of a
buffer containing 4 μl of 5X RT buffer (Promega, Madison,
WI, USA), 1 μl of dNTP (2 mM each), 2.5 μl of 25 mM
MgCl2, 2 μl of RNase Inhibitor (40 U/μl
Promega), and 1 μl of reverse transcriptase (200 U/μl
Promega). The reaction volume was 20 μl, and the reaction
was allowed to proceed for 90 min at 42°C, and then for 2 min at
90°C. The PCR protocol for second-strand synthesis was one cycle at
94°C for 3 min, followed by 50°C for 3 min, and 72°C for 1 min.
After second-strand DNA synthesis was completed, the PCR products
were subjected to second-stage PCR amplification, which included 40
cycles of 94°C for 40 sec, 65°C for 40 sec, 72°C for 40 sec,
followed by a 5 min final extension step at 72°C. The PCR products
were separated on a 1.2% agarose gel and stained with ethidium
bromide. The differentially expressed bands were extracted from the
gel using a QIAquick Gel extraction kit (Qiagen, Carlsbad, CA,
USA), directly cloned into the pGEMR-T Easy vector (Promega)
without reamplification of the recovered bands, and sequenced.
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was isolated from three cultured glioma
cell lines using TRIzol reagent. A total of 1 μg of RNA was
reverse transcribed to synthesize the cDNA. Reverse transcription
(RT) was followed by polymerase chain reaction (PCR). For the first
strand synthesis, 1 μg of the purified total RNA was
incubated with oligo (dT) (0.5 μg/μl Promega) for 5
min at 70°C, followed by the addition of buffer containing 4
μl of 5X Reaction buffer (Promega), 1 μl of dNTP (10
mM each), 3.5 μl of 25 mM MgCl2, 2 μl of
RNase Inhibitor (40 U/μl Promega), and 1 μl of
reverse transcriptase (200 U/μl Promega). The reaction
volume was 20 μl, and the reaction was carried out for 90
min at 42°C, and then for 2 min at 90°C. β-actin was used as an
internal control. Reactions with varying numbers of polymerase
chain reaction (PCR) cycles were run for each transcript.
Northern blot analysis
Northern blot analysis was performed under
conventional conditions. Total RNA (20 μg) was separated by
electrophoresis on 1.2% agarose formaldehyde gels, then transferred
to a nylon membrane overnight and cross linked with UV irradiation.
The filters were prehybridized at 65°C for 3 h and then hybridized
to a 32P-labeled probe. The probes were prepared from
the clones described above by digestion with restriction enzymes,
and this was followed by gel electrophoresis. The filters were
washed in SSC and then 0.1% SDS for 15 min at 65°C. They were then
exposed to X-ray film at −70°C.
Transfection
Preparation of metallothionein 1E
construct
The complete coding region of the human MT cDNA was
amplified from U87MG cDNA by PCR with synthetic primers. The PCR
primers were designed as follows. MT-sense:
5′-CGGGATCCATGGACCCCAACTGCTCTT-3′, 5′-GCTCTAGAGCTCAG
GCACAGCAGCTGCACT-3′. MT-anti sense: 5′-GCTCTAG
AGCATGGACCCCAACTGCTCTT-3′, 5′-CGGGATCCTCAGGCACAGCAGCTGCACT-3′ The
amplified cDNA was sequenced and directly subcloned into the
pcDNA3.1(+) vector (Invitrogen, San Diego, CA, USA) between the
BamHI and XbaI sites, which is contained with in the
CMV promoter and the neomycin resistance gene. The resulting
vectors were designated as pcDNA3.1-MT-S, pcDNA3.1-MT-AS.
Transfection
U343MG-A and U87MG were maintained
under exponential growth conditions in DMEM supplemented with 10%
fetal bovine serum in the absence of antibiotics
The optimal cell density for transfection is
normally between 50–80% confluency for adherent cells.
pcDNA3.1-MT-S/AS were respectively transfected into human malignant
glioma cell lines U343MG-A and U87MG using the
GeneJuice™ transfection reagent (Novagen, Madison, WI,
USA). Cells in serum-free DMEM were mixed with 15 μg of
plasmid DNA and 45 μl of GeneJuice/serum-free media
according to the manufacturer’s protocol. After incubation at 37°C
(5% CO2) for 5 h, the transfection mixture was replaced
with DMEM supplemented with 10% FBS. After 24 h incubation, the
medium was replaced with DMEM containing 10% FBS and 500
μg/ml G418. U343MG-A transfected with sense MT1E cDNA
plasmid (pcDNA3.1-MT-S) and U87MG transfected with antisense MT1E
cDNA plasmid (pcDNA3.1-MT-AS). Each transfectants were designated
as U343-MT-S and U87-MT-AS.
Separation of cytoplams and nuclear
extract
Cytoplasmic extraction buffer (CEB) consisted of 10
mM HEPES (pH 7.9), 40 mM KCl, 2 mM MgCl2, 10% glycerol
and protease inhibitors. After the cells were scraped into CEB,
they were transferred into a micro-centrifuge tube, lysed by
vortexing a few times and chilled on ice for 15 min. Nuclei were
pelleted by centrifugation at 5,000 rpm for 5 min, and the pellets
were washed twice with PBS. The supernatant was removed, and the
nuclei were lysed in nuclear extraction buffer consisting of 10 mM
HEPES (pH 7.9), 500 mM NaCl, 1% Triton X-100, 10% glycerol and
protease inhibitors. After centrifugation at 12,000 rpm for 5 min,
the supernatants containing the nuclear proteins were stored at
20°C until use.
Preparation of total protein and
conditioned media
For the preparation of total protein, cells were
lysed in a protein extraction buffer [50 mM Tris (pH 8.0), 5 mM
EDTA, 150 mM sodium chloride, 0.5% deoxycholic acid, 0.1% SDS, 1%
NP-40, 1 mM PMSF, and 1 mg/ml protease inhibitor cocktail].
For the preparation of conditioned media, cells were
grown in 60-mm plates until they were subconfluent, and were then
washed three times with ice-cold CMF-PBS. 1 ml of serum-free medium
was added to each plate, and the cells were incubated at 37°C for
48 h. The conditioned media were clarified by centrifugation. The
concentrations of all kinds of protein samples were determined
using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA,
USA).
Western blotting
A total of 20 μg of whole cell lysates were
separated by 15% SDS-PAGE and transferred to a polyvinylidene
difluoride membrane (Pall corporation, USA). The membrane was then
incubated for 2 h at room temperature in TBS-T solution [10 mM
Tris-Cl (pH 8.0), 150 mM NaCl, and 0.05% Tween 20] supplemented
with 5% non-fat dry milk and probed overnight at 4°C with
anti-metallothionein (Santa Cruz Biotechnology, Santa Cruz, CA,
USA), anti-p50, p65 (Cell Signaling Technology, Beverly, MA),
anti-MMP2 and MMP9 (BD Pharmingen, San Diego, CA, USA). The bound
antibodies were visualized with the appropriate horseradish
peroxidaseconjugated secondary antibody (Jackson Immunoresearch
Laboratory, WestGrove, PA, USA) conjugated with horseradish
peroxidase using enhanced chemiluminescence reagents (ECL, Amersham
Biosciences, Bucks, UK).
Cell morphological
characteristics
The transfectants and the parental cells were fixed
with methanol at 4°C for 10 min. Cells were washed twice with PBS,
fixed with cold methanol and stained with 0.1% toluidine blue.
These were examined via light microscopy (Nikon, Garden City, NY)
and digitally photographed to evaluate and record the morphology of
the cell population.
Doubling time of stable
transfectants
The cells were seeded at 3×104 cells in
60 mm culture dish. After serum starvation for 24 h, the cells were
counted each day. The cells were treated with trypsin-EDTA and the
number of viable cells was counted with a hemocytometer. The
doubling-time was calculated from the cell growth curve over 5
days. Equation for doubling time = (t-t°)log2/logN-logN° (t, final
time; unit, hour; t°, initial times; N, final cell number; N°,
initial cell number).
Immunocytochemistry for cytoskeletal
proteins
The cells were cultured on glass coverslips in 35 mm
dishes until reaching subconfluency, washed with phosphate buffer
saline and fixed with 4% p-formaldehyde for 10 min. After washing
in immuno/ DNA buffer (Research Genetics, Huntsville, AL, USA), the
cells were treated with triton X-100 at 0.1% for 5 min and washed
3–5 more times. The cells were incubated with 2% bovine serum
albumin (Sigma-Aldrich, Steinheim, Germany) for 30 min in an effort
to reduce non-specific reactions. Incubation with anti-vimentin (BD
Pharmingen) was performed in a humid chamber for 1 h, Alexa
488-conjugated goat anti-mouse antibody (Molecular Probe, Eugene,
OR, USA) for 40 min. Rhodamineconjugated phalloidin (Molecular
Probe) was used for actin staining. The coverslips were mounted on
slides with Immuno-mount (Shandon, Pittsburg, PA, USA). Confocal
microscopy was performed with a confocal microscope (LaserSharp
2000 version 5.1) equipped with a Plan-Apochromat 63x/1.40 oil
objective. Confocal images were acquired using LSM 5102.3 software.
The wavelength of each antibody is Alexa 488-Emission 519, Alexa
568-Emission 603 nm.
Invasion assay
Matrigel (reconstituted basement membrane; 25
μg) was dried on a polycarbonated filter
(polyvinylpyrrolidone-free; Nucleopore; Whatmann, Madestone, UK).
Cells were harvested by brief exposure to 1 mM/l EDTA, washed with
DMEM containing 0.1% bovine serum albumin, and added to the Boyden
chamber (2x105 cells). Cells were incubated for 24 h at
37°C in a humidified atmosphere of 95% air and 5% CO2.
Cells that transversed the Matrigel layer and attached to the
filter were stained with the Diff Quik kit (Dade Diagnostics,
Aguada, PA, USA) and counted in five randomized fields. The results
are expressed as the mean ± SE of three independent
experiments.
Data analysis
The comparison of the nucleotide sequence homology
of the isolated cDNAs with the registered sequence in Genebank was
carried out using the BLAST algorithm. We measured the statistical
significance of the cell distance and the cell number using the
Mann-Whitney U-test, and the doubling time by repeated measures
ANOVA. P<0.05 was considered to be significant. Statistical
analysis was performed using the SPSS software program (version
12.0 for Windows; SPSS Inc., Chicago, IL).
Results
Comparison of the migration abilities
of three glioma cell lines
The motility of the cell lines was compared using a
simple scratch technique. In order to eliminate any confounding
effects of an experimental agent on cell proliferation, the cell
culture media were replaced with medium containing hydroxyurea.
Treatment with hydroxyurea resulted in the complete inhibition of
cell proliferation. As shown in Fig.
1, the U87MG cells were more motile than the U343MG-A cells,
and the U87MG-10′ cells were the fastest. The difference among the
three groups was statistically significant for the mean cell number
(p<0.021).
Identification of differentially
expressed genes
To obtain a profile of the genes related to
invasion, the author estimated the motility of the three cell lines
that showed different motilities. MT1E showed a higher expression
level in the motile cell lines (Fig.
2A).
Validation of DD-PCR data by RT-PCR
and northern blot analysis
The author verified the significant differences in
gene expression pattern by RT-PCR and northern blot analysis for
the candidate gene. As shown in Fig.
2B and C, there were significant differences in MT expression
between the low and high motility cell lines, and the expression
patterns matched those of both RT-PCR and northern blot
analysis.
Selection of stable transfectants of
sense and antisense MT constructs
To assess the effect of MT on proliferation,
migration and invasion in malignant glioma cells, the pcDNA 3.1(+)
vector containing full length sense and antisense MT1E cDNA was
transfected into malignant glioma cell lines under the control of
the CMV promoter, and neomycin-resistance transfected clones were
obtained. U343MG-A cells expressing low levels of MT1E were
inserted into the sense MT1E cDNA construct. In addition, U87MG
cells expressing high levels of MT1E were inserted into an
antisense MT1E cDNA construct. Western blotting was used to detect
the clone that showed the best characterization after transfection
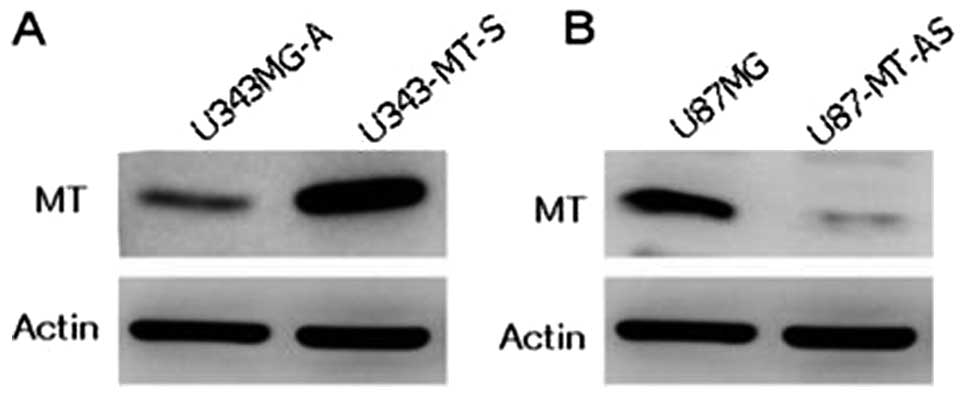
with sense and antisense constructs. The U343-MT-S cell line showed
a strong band when compared with the parental cell line (Fig. 3A). On the other hand, the U87-MT-AS
cell line showed a very faint band when compared with parental cell
line (Fig. 3B).
The effect of MT on migration and
invasion in malignant glioma cells
Each cell line was pretreated with hydroxyurea,
which inhibits cell proliferation, in order to observe changes in
cell motility alone. As shown in Fig.
4A, MT1E sense/ antisense-transfected cell lines showed
positively altered their motilities. U343MG-A is a less motile cell
line. Following sense MT1E transfection, it becomes about 37%
faster than parental cell lines. The maximum distance migrated and
the mean cell number was 261 μm and 78 cells in the U343MG-A
cell line and 358 mm and 148 cells in the U343-MT-S cell line,
respectively. The difference between the two groups was
statistically significant for the mean cell number (p<0.001) and
maximal distance (p<0.001). Likewise, U87MG is a high motility
cell line, and it became less motile after transfection with
antisense MT. The maximum distance migrated and the mean number of
total cells migrated were 844 μm and 495 cells in the U87MG
cell line and 659 μm and 270 cells in the M87-MT-AS cell
line, respectively. The difference between the two groups was also
statistically significant for the mean cell number (p<0.001) and
maximal distance (p<0.001). In addition, MT1E on the invasion in
the sense and antisense transfectants was markedly enhanced in the
Matrigel assay as compared with the parental cell line (p<0.001,
Fig. 4B).
Morphological and cytoskeletal changes
after MT transfection
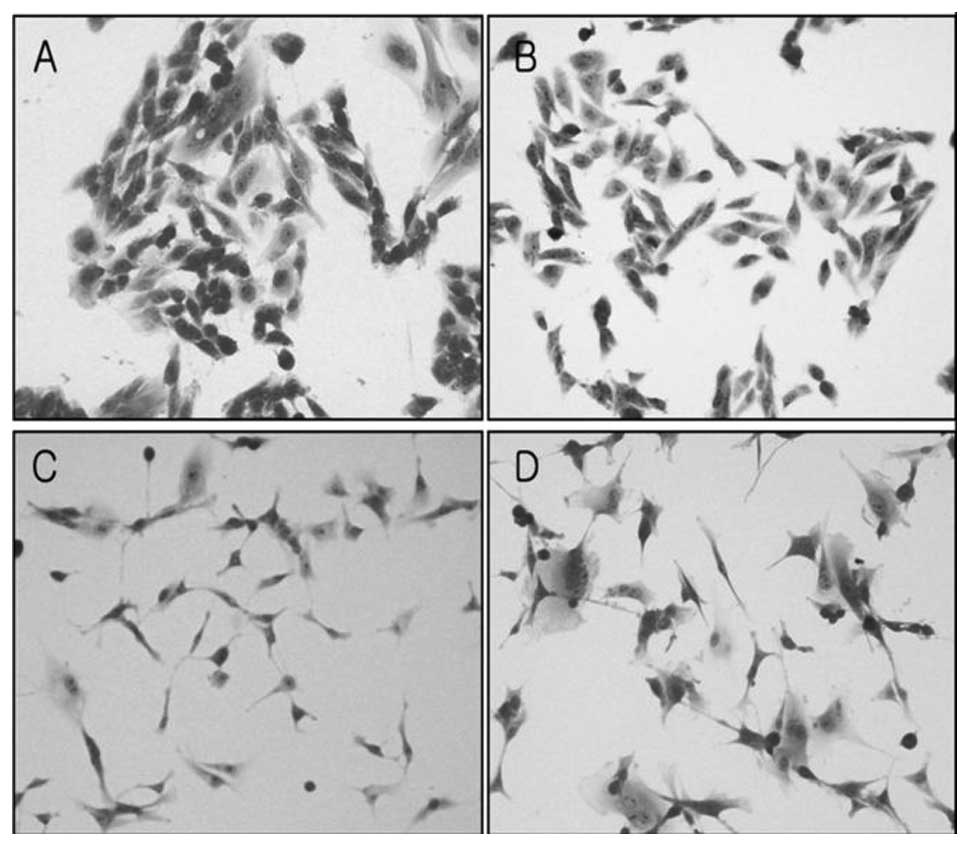
Morphological changes were characterized by light
microscopy. U343MG-A cells generally formed colonies and were round
in shape. On the other hand, U343-MT-S cells grew apart from each
other, and their shape became more spindle-like (Fig. 5A and B). In addition, U87-MT-AS
cells became flat and enlarged in comparison to U87MG cells
(Fig. 5C and D).
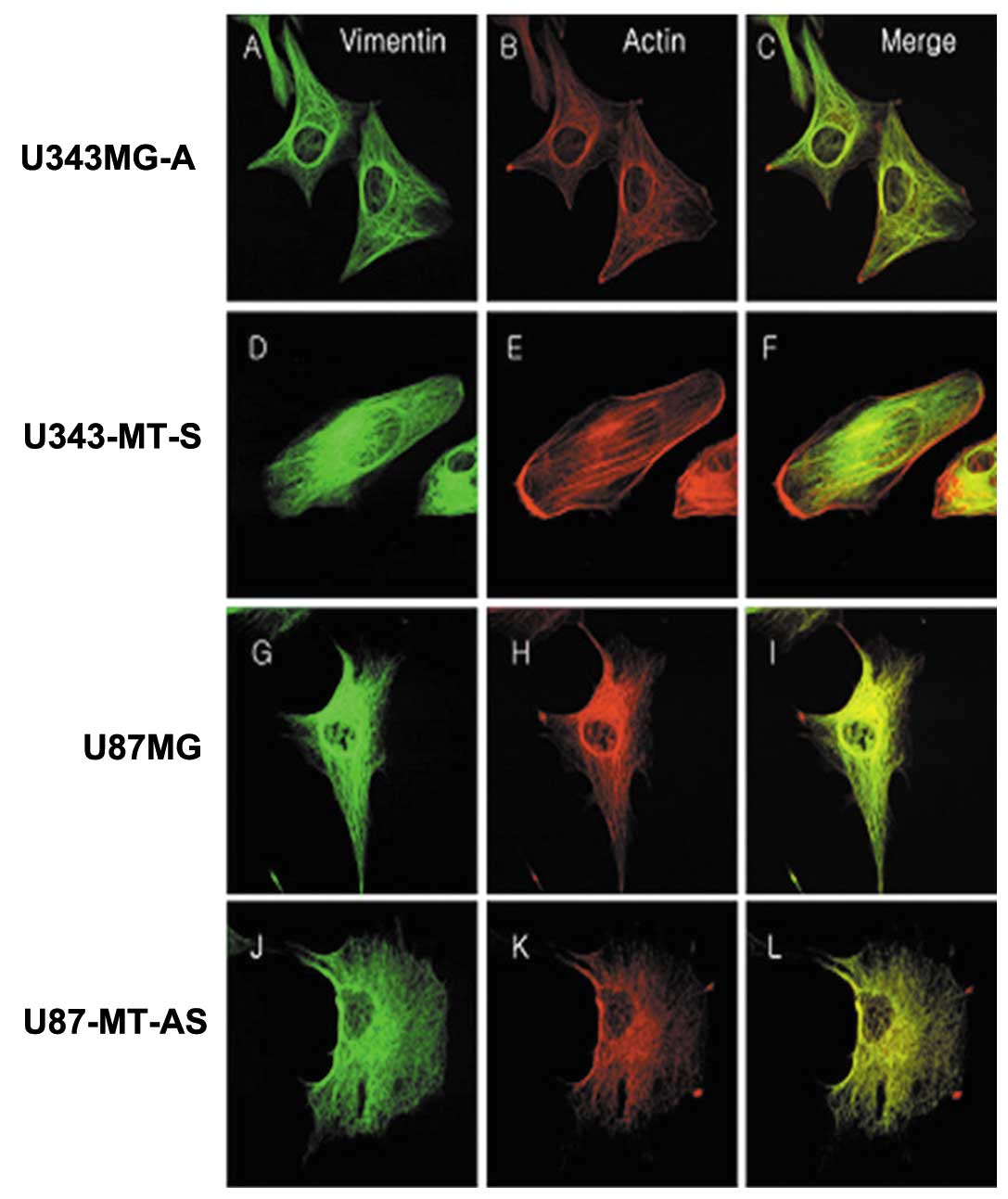
Immunofluorescence staining of actin and vimentin were performed in
order to determine whether the differences in cytoskeletal
alterations were associated with tumor cell motility and invasion.
The expression patterns of actin and vimentin were similar. The
U343-MT-S and U87MG cell lines with high motility formed many
stress fibers and showed extensive lamellipodia and strong positive
staining in comparison with the parental cell lines. The proteins
were distributed on one side around the nucleus and cytoplasm
(Fig. 6F and I). On the other
hand, the U343MG-A and U87-MT-AS cell lines, which were less
motile, showed fewer stress fibers and shorter lamellipodia. The
cytoskeletal proteins were mainly concentrated around the nucleus
and became entangled (Fig. 6C and
L).
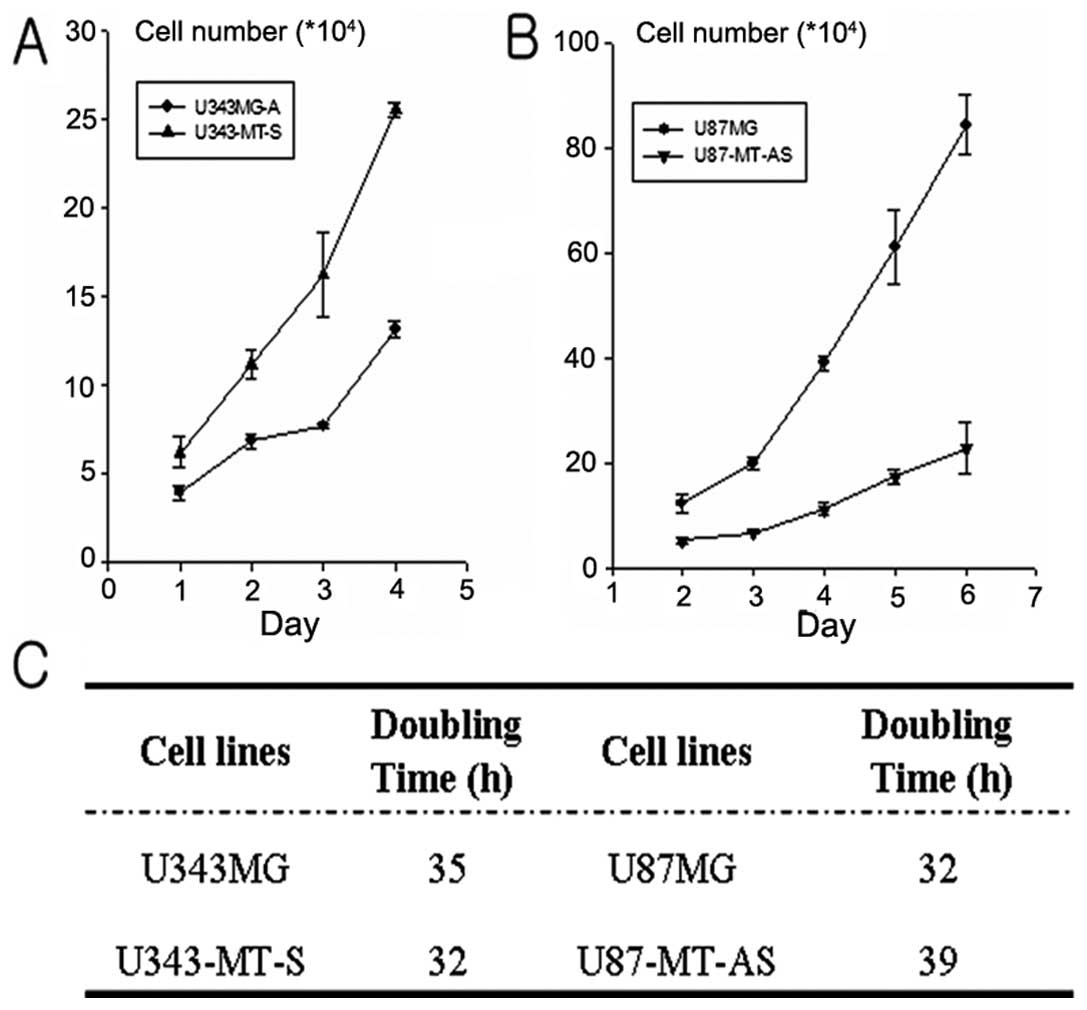
Doubling time test
The growth rate was compared of each cell line with
those of transfected clones. As shown in Fig. 7, the doubling time was 35 and 31 h
in the U343MG-A and U343-MT-S cell lines, respectively. The
doubling time for the U87MG and U87-MT-AS cell lines was 32 and 39
h, respectively. U343-MT-S cell line was reduced by about 4 h and
that of the U87-MT-AS cell line was only about 7 h, compared with
parental cell lines.
Expression of endogenous MT in
malignant glioma cell lines
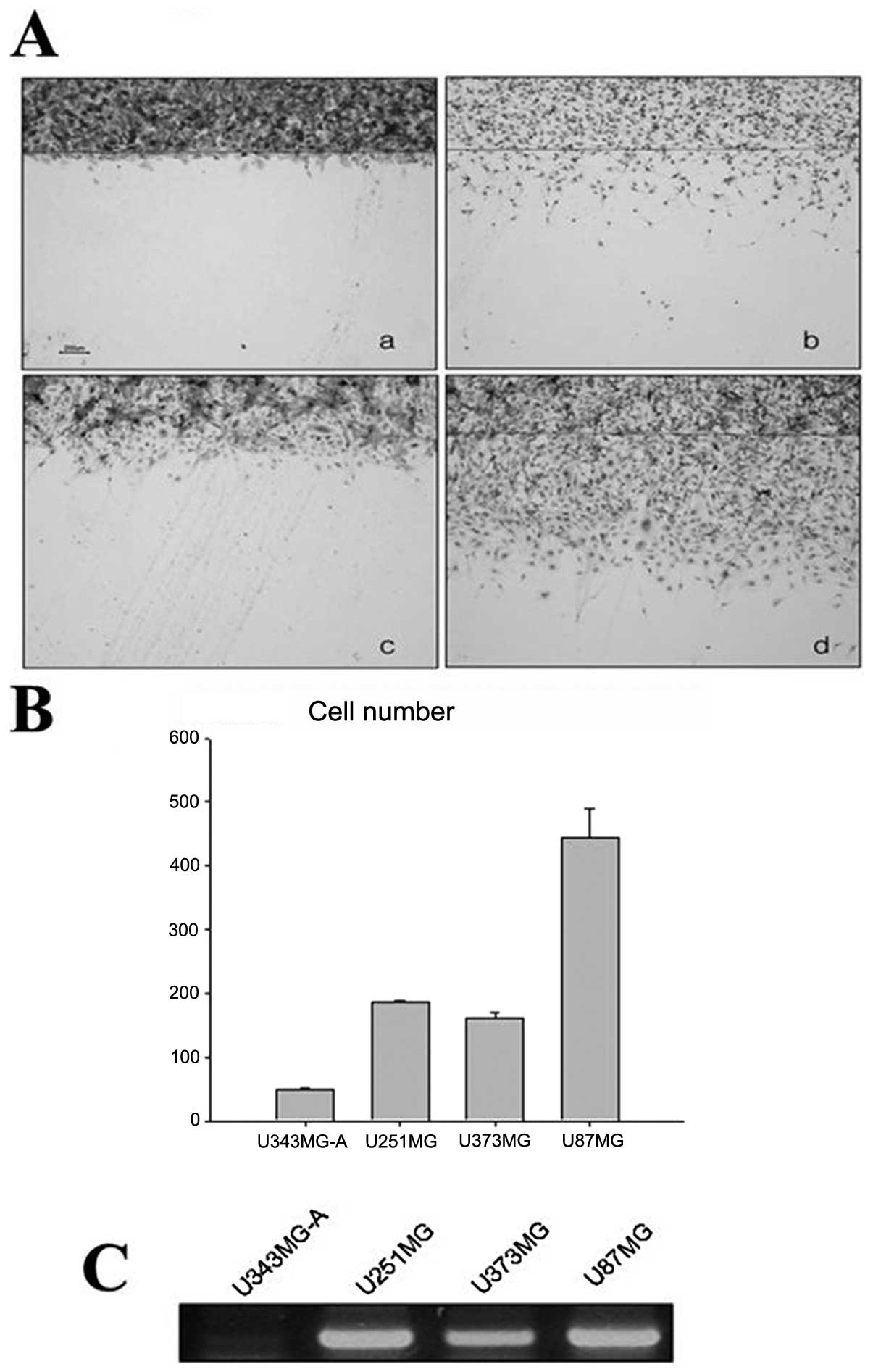
RT-PCR was used to confirm the expression of MT1E in
glioma cell lines with different motilities. The U87MG, U251MG,
U373MG and U343MG-A cell lines showed high motility sequences
(Fig. 8A and B). The levels of
MT1E mRNA were proportional to the motility of the cell lines
(Fig. 8C).
Effects of MT on NF-κB translocation
following the activity of MMP-2 and -9
U87MG cells were stably transfected with antisense
MT1E cDNA, and the best two clones were selected (U87-MT-AS). In
order to examine the relationship between MT1E and NF-κB activity,
the p50 and p65 subunits were detected in the nuclear and cytoplasm
fraction by western blot analysis. As shown in Fig. 9A-b, U87-MT-AS showed decreased
translocation of the p50 subunit of NF-κB from the cytoplasm to the
nucleus when compared with the parental cells. However, the
translocation of NF-κB p65 was not particularly changed in
comparison with the parental cells. In addition, total IκB was
increased in U87-MT-AS (Fig.
9A-a). On the contrary, total IκB was only slightly increased
in U343-MT-S. Thus, we checked phospho-IκB expression level in
U343-MT-S. As shown in Fig. 9B,
the expression of phospho-IκB was increased with total IκB. The
expression of MMP-2 in U87MG and U87-MT-AS were detected by western
blotting and zymography. But the expression of MMP-9 was only
confirmed by western blotting because it could not be detected by
zymography. There was no difference in MMP-2 expression between
U87MG and U87-MT-AS. However, the active and latent forms of MMP-9
were remarkably decreased in the conditioned media of U87-MT-AS
(Fig. 9A-a), otherwise U343-MT-S
had increased active forms of MMP-9 in conditioned media.
Discussion
In a previous in vitro study, we identified
motility-related genes that would be representative of in
vivo invasiveness in glioma cell lines with different
motilities using the Genefishing technology (6). Among the identified genes, MT1E was
overexpressed in highly motile malignant glioma cell lines,
indicating that this gene may be a positive regulator of tumor cell
motility. In this study, the author investigated whether MT1E is
actually associated with migration and invasion in human glioma
cell lines. Firstly, two glioma cell lines were selected with
similar origin, but showed a marked difference of their motility
(Fig. 4). Then a sense MT1E cDNA
construct was transferred into the glioma cell line with less
motility, and an antisense MT1E cDNA construct was transferred into
glioma cell line with high motility. The effect of MT1E on the
migratory and invasive abilities of human glioma cell lines was
evaluated by an in vitro assay that involved a simple
scratch technique and the Boyden chamber system. The results showed
that the proliferation rate in the U343-MT-S cell line, an
MT1E-overexpressing cell line, was two times faster, whereas the
opposite effect was observed in the U87-MT-AS cell line, which
expressed lower levels of MT1E. In addition, some malignant glioma
cell lines with different motilities expressed MT in proportion to
their mobility. Generally, cell movement is accompanied by various
changes, such as morphology, cytoskeleton, and proliferation rate.
To determine whether the differences associated with cytoskeletal
alterations were also associated with tumor cell motility,
immunofluorescence staining for actin and vimentin was performed
and the morphological changes in the shape and size of the cells
following transfection were examined. The U343-MT-S cells grew
apart from each other and showed well-defined stress fibers. On the
other hand, when this cell line was transfected with antisense
MT1E, the cells became flatter and larger, and the cytoskeletal
proteins became entangled. In the process of several experiments,
the antisense transfectants was more sensitive to trypsin than the
parental cell line (data not shown), while the sense transfectants
were resistance to it. This may indicate that these cells are ready
to migrate.
MT is related to tumor grade, malignancy and
prognosis in certain tumors. MT is correlated with the histological
grade and proliferative potential of astrocytic tumors (23,24).
In addition, MT is overexpressed in invasive and in situ
breast cancer cells, and it may be a biomarker of tumor
differentiation and aggressiveness (25,26).
In order to verify whether MT1E affects the proliferation of glioma
cell lines, doubling time test in U343-MT-S and U87-MT-AS, was
performed compared with each parental cell line. Actually,
U343-MT-S increased cell proliferation rate, while U87-MT-AS was
the opposite. On the basis of the above results, MT-1E may be
directly or indirectly involved in cell motility and invasion.
Structural studies of MT have shown that this
unusual protein with 61 amino acids (mammalian MT) can bind with
both essential metals (zinc and copper) and toxic metals (cadmium
and mercury) in two distinct cluster structures in the molecule
(27). Due to this characteristic,
MTs act as controllers of zinc and copper, by participating in cell
proliferation processes (28). MT
could donate zinc/copper to various metallo-enzymes and
transcription factors (29,30).
The protective role of MT in oxidative stress and metal toxicity
suggests that MT may also have a functional role in tumor cell
survival and growth. According to some reports, MT could facilitate
tumor cell growth by two potential mechanisms. One possible
mechanism is that MT may act as a zinc donor to various
transcription factors, including tumor suppressor gene products,
such as p53. In vitro studies have shown that thionein can
modulate the transcriptional activation of Sp1, a zinc finger
transcription factor. Another possible mechanism is that MT may
protect the cells from radiation and chemotherapeutic agents by
virtue of its free radical scavenging property (31–33).
We placed major emphasis on first hypothesis, and
then desired to look for any factors which have Zn associated site
in their structure and associated in tumor cell invasion. It may be
related with MT1E in the change of the motility of malignant glioma
cell lines.
MMPs are a part of a family of Zn-dependent enzymes
and have a well known activity in tumor invasion and metastasis
(39). The change of the
expression level of MMP-2 and -9 was examined in both total cell
lysates and conditioned media in U87MG and U87-MT-AS. The activity
of MMP-9 was decreased in U87-MT-AS, while the activity of MMP-2
was similar, in comparison to the parental cell line. In 1996, Haga
et al reported that metallothionein increased the activity
of MMP, which may change in conformation by chelation with
metallothionein (40).
NF-κB is also a zinc-dependent transcription factor,
plays a pivotal role in a diverse array of cellular activities and
gene activations (30). There are
strong pathophysiologic functional similarities between MT and
NF-κB. Both are anti-apoptotic entities, especially in cancer cells
and both have regulatory roles in inflammation (34,35).
Moreover, many stimulators of their expression overlap, including
tumor necrosis factor-α, lipopolysaccharide, and interleukin-1
(36). Thus, it is plausible that
there is a close molecular relationship between the two molecules.
A previous study showed that MT regulates the cellular level and
activity of NF-κB as a potential intracellular modulators of NF-κB
activation. In addition, MT overexpression was found to up-regulate
DNA binding of the NF-κB (21,31,37),
and it plays an important role in MMP-9 gene expression (38).
In this study, when MT1E expression was decreased in
glioma cell line with high motility, the levels of NF-κB p50 in the
nuclear fraction significantly decreased. Most reports on the
association between MT and NF-κB have suggested that it involves
the p65 subunit of NF-κB. However, the present study showed that MT
affected the p50 subunit of NF-κB rather than the p65 subunit. A
1998 report suggested that MT specifically interacts with the p50
subunit of the p50/RelA heterodimer and that MT may be required for
the stabilization of its DNA complex formation, thus allowing for
the potent transactivation of target genes (39). According to the last report, MT2A
promotes breast cancer cell invasion by upregulating MMP-9 via AP-1
and NF-κB (41).
On the basis of our results and reports, MT1E can
modulate the motility and invasion in a human glioma cell line. It
may involve two mechanisms. MT1E may induce the change of
morphology via the modification of cytoskeletal proteins in human
glioma cell lines, or MT1E may modulate the activity of MMP-9 by
NF-κB in malignant glioma cell lines. This series of events
enhances migration and invasion in malignant glioma cell lines.
More experiments should be performed to clarify these
mechanisms.
Acknowledgements
This research was supported by Leading
Foreign Research Institute Recruitment Program through the National
Research Foundation of Korea (NRF) funded by the Ministry of
Education, Science and Technology (MEST) (2011-0030034).
References
|
1
|
Kassis J, Lauffenburger DA, Turner T and
Wells A: Tumor invasion as dysregulated cell motility. Semin Cancer
Biol. 11:105–117. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mariani L, McDonough WS, Hoelzinger DB,
Beaudry C, Kaczmarek E, Coons SW, Giese A, Moghaddam M, Seiler RW
and Berens ME: Identification and validation of P311 as a
glioblastoma invasion gene using laser capture microdissection.
Cancer Res. 61:4190–4196. 2001.PubMed/NCBI
|
|
3
|
Coffey DS: Prostate cancer metastasis:
talking the walk. Nat Med. 2:1305–1306. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Levine MD, Liotta LA and Stracke ML:
Stimulation and regulation of tumor cell motility in invasion and
metastasis. EXS. 74:157–179. 1995.PubMed/NCBI
|
|
5
|
Kim YJ, Kwak CI, Gu YY, Hwang IT and Chun
JY: Annealing control primer system for identification of
differentially expressed genes on agarose gels. Biotechniques.
36:424–430. 2004.PubMed/NCBI
|
|
6
|
Ryu HH, Jung S, Sun HS, Jung TY, Jin SG,
Jin YH, Kim IY, Jeong YI and Kang SS: Screening for
motility-associated genes in malignant astrocytoma cell lines. J
Neurooncol. 82:125–131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cai L and Cherian MG: Zinc-metallothionein
protects from DNA damage induced by radiation better than
glutathione and copper- or cadmium-metallothioneins. Toxicol Lett.
136:193–198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ebadi M, Leuschen MP, el Refaey H, Hamada
FM and Rojas P: The antioxidant properties of zinc and
metallothionein. Neurochem Int. 29:159–166. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kelley SL, Basu A, Teicher BA, Hacker MP,
Hamer DH and Lazo JS: Overexpression of metallothionein confers
resistance to anticancer drugs. Science. 241:1813–1815. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roesijadi G: Metal transfer as a mechanism
for metallothionein-mediated metal detoxification. Cell Mol Biol
(Noisy-le-grand). 46:393–405. 2000.PubMed/NCBI
|
|
11
|
Satoh M, Cherian MG, Imura N and Shimizu
H: Modulation of resistance to anticancer drugs by inhibition of
metallothionein synthesis. Cancer Res. 54:5255–5257.
1994.PubMed/NCBI
|
|
12
|
Maret W, Jacob C, Vallee BL and Fischer
EH: Inhibitory sites in enzymes: zinc removal and reactivation by
thionein. Proc Natl Acad Sci USA. 96:1936–1940. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meplan C, Richard MJ and Hainaut P:
Metalloregulation of the tumor suppressor protein p53: zinc
mediates the renaturation of p53 after exposure to metal chelators
in vitro and in intact cells. Oncogene. 19:5227–5236. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jayasurya A, Bay BH, Yap WM and Tan NG:
Correlation of metallothionein expression with apoptosis in
nasopharyngeal carcinoma. Br J Cancer. 82:1198–1203.
2000.PubMed/NCBI
|
|
15
|
Nagel WW and Vallee BL: Cell cycle
regulation of metallothionein in human colonic cancer cells. Proc
Natl Acad Sci USA. 92:579–583. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ghosh S, May MJ and Kopp EB: NF-kappa B
and Rel proteins: evolutionarily conserved mediators of immune
responses. Annu Rev Immunol. 16:225–260. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Andela VB, Gordon AH, Zotalis G, Rosier
RN, Goater JJ, Lewis GD, Schwarz EM, Puzas JE and O’Keefe RJ:
NFkappaB: a pivotal transcription factor in prostate cancer
metastasis to bone. Clin Orthop Relat Res. S75–S85. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baldwin AS: Control of oncogenesis and
cancer therapy resistance by the transcription factor NF-kappaB. J
Clin Invest. 107:241–246. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Karin M, Cao Y, Greten FR and Li ZW:
NF-kappaB in cancer: from innocent bystander to major culprit. Nat
Rev Cancer. 2:301–310. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Butcher HL, Kennette WA, Collins O, Zalups
RK and Koropatnick J: Metallothionein mediates the level and
activity of nuclear factor kappa B in murine fibroblasts. J
Pharmacol Exp Ther. 310:589–598. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim CH, Kim JH, Lee J and Ahn YS:
Zinc-induced NF-kappaB inhibition can be modulated by changes in
the intracellular metallothionein level. Toxicol Appl Pharmacol.
190:189–196. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakajima M and Chop AM: Tumor invasion and
extracellular matrix degradative enzymes: regulation of activity by
organ factors. Semin Cancer Biol. 2:115–127. 1991.PubMed/NCBI
|
|
23
|
Hiura T, Khalid H, Yamashita H, Tokunaga
Y, Yasunaga A and Shibata S: Immunohistochemical analysis of
metallothionein in astrocytic tumors in relation to tumor grade,
proliferative potential, and survival. Cancer. 83:2361–2369. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maier H, Jones C, Jasani B, Ofner D,
Zelger B, Schmid KW and Budka H: Metallothionein overexpression in
human brain tumours. Acta Neuropathol. 94:599–604. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gallicchio LM, Flaws JA, Fowler BA and
Ioffe OB: Metallothionein expression in invasive and in situ breast
carcinomas. Cancer Detect Prev. 29:332–337. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin R, Bay BH, Chow VT and Tan PH:
Metallothionein 1F mRNA expression correlates with histological
grade in breast carcinoma. Breast Cancer Res Treat. 66:265–272.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Winge DR: Limited proteolysis of
metallothioneins. Methods Enzymol. 205:438–447. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Florianczyk B: Copper and metallothioneins
in cancer cells. Ann Univ Mariae Curie Sklodowska Med. 58:390–393.
2003.PubMed/NCBI
|
|
29
|
Nartey NO, Banerjee D and Cherian MG:
Immunohistochemical localization of metallothionein in cell nucleus
and cytoplasm of fetal human liver and kidney and its changes
during development. Pathology. 19:233–238. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sato M and Bremner I: Oxygen free radicals
and metallothionein. Free Radic Biol Med. 14:325–337. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cherian MG, Jayasurya A and Bay BH:
Metallothioneins in human tumors and potential roles in
carcinogenesis. Mutat Res. 533:201–209. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shimoda R, Achanzar WE, Qu W, Nagamine T,
Takagi H, Mori M and Waalkes MP: Metallothionein is a potential
negative regulator of apoptosis. Toxicol Sci. 73:294–300. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeng J, Vallee BL and Kagi JH: Zinc
transfer from transcription factor IIIA fingers to thionein
clusters. Proc Natl Acad Sci USA. 88:9984–9988. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamamoto Y and Gaynor RB: Therapeutic
potential of inhibition of the NF-kappaB pathway in the treatment
of inflammation and cancer. J Clin Invest. 107:135–142. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zangger K, Oz G, Haslinger E, Kunert O and
Armitage IM: Nitric oxide selectively releases metals from the
amino-terminal domain of metallothioneins: potential role at
inflammatory sites. FASEB J. 15:1303–1305. 2001.PubMed/NCBI
|
|
36
|
Coyle P, Philcox JC, Carey LC and Rofe AM:
Metallothionein: the multipurpose protein. Cell Mol Life Sci.
59:627–647. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kanekiyo M, Itoh N, Kawasaki A, Tanaka J,
Nakanishi T and Tanaka K: Zinc-induced activation of the human
cytomegalovirus major immediate-early promoter is mediated by
metallothionein and nuclear factor-kappaB. Toxicol Appl Pharmacol.
173:146–153. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Miyamoto H, Altuwaijri S, Cai Y, Messing
EM and Chang C: Inhibition of the Akt, cyclooxygenase-2, and matrix
metalloproteinase-9 pathways in combination with androgen
deprivation therapy: potential therapeutic approaches for prostate
cancer. Mol Carcinog. 44:1–10. 2005. View Article : Google Scholar
|
|
39
|
Abdel-Mageed AB and Agrawal KC: Activation
of nuclear factor kappaB: potential role in
metallothionein-mediated mitogenic response. Cancer Res.
58:2335–2338. 1998.PubMed/NCBI
|
|
40
|
Haga A, Nagase H, Kito H and Sato T:
Enhanced invasiveness of tumour cells after host exposure to heavy
metals. Eur J Cancer. 32A:2342–2347. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim HG, Kim JY, Han EH, Hwang YP, Choi JH,
Park BH and Jeong HG: Metallothionein-2A overexpression increases
the expression of matrix metalloproteinase-9 and invasion of breast
caner cells. FEBS Lett. 585:421–428. 2011. View Article : Google Scholar : PubMed/NCBI
|