Introduction
The persistent infection of the head and neck
epithelium by high-risk human papillomaviruses (HR-HPVs), mainly
HPV type 16, predominantly in the region of the oropharynx, is
associated with an increased risk of head and neck squamous cell
carcinoma (HNSCC) (1). HR-HPVs
define a distinct clinical subpopulation that displays improved
disease-free and overall survival (reviewed in ref. 1 and references therein). These
observations may be explained by the molecular mechanisms that
underlie HPV-driven cell transformation. HR-HPV induces
carcinogenesis by interfering with the p53 and pRb signaling
pathways (1). The HPV E6
oncoprotein is known to induce p53 ubiquitination and subsequent
degradation. The TP53 gene is not mutated in the majority of
HPV-related HNSCCs, and data from cell culture assays suggest that
non-degraded p53 persisting in HPV-positive cells may be sufficient
to induce the expression of p53 target genes and cell death upon
treatment with ionizing radiation (2).
Genomic and transcriptomic analyses have shown that
HR-HPV-positive HNSCC display specific chromosomal gains and
losses, as well as changes in gene expression relative to their
HPV-negative counterparts. HR-HPV-related tumors have fewer genomic
aberrations, but display specific imbalances, such as a loss of the
16q region (3,4). Of note, this aberration correlates
with improved local control (4).
We have previously shown that the expression of the
NEDD8-activating enzyme 1/amyloid β precursor protein-binding
protein 1 (NAE1/APP-BP1) gene, which is located on
chromosome 16q22, is expressed at lower levels in HPV-positive
oropharyngeal tumors (3). Together
with the UBA3 protein, NAE1/APP-BP1 forms the NEDD8-activating
enzyme, which has E1 ubiquitinligase activity, and initiates the
NEDD8 conjugation pathway. In this complex, NAE1/APP-BP1 plays the
role of a regulatory subunit for NEDD8 E1 enzyme activity (5,6).
Post-translational NEDD8 conjugation has mainly been described for
members of the cullin family of proteins (7). Previously, a function of the NEDD8
conjugation pathway in the regulation of the p53 transcriptional
activity has been unraveled: NAE1/APP-BP1 is required for the NEDD8
conjugation of p53 via MDM2, which results in the inhibition of p53
transcriptional activity without triggering its degradation
(8).
In this study, we investigated the possibility that
the expression of NAE1/APP-BP1 affects p53-dependent
transcriptional activity and sensitivity to ionizing radiation in
head and neck cancer cells. We used the naturally HPV16-infected
oropharyngeal cell line, UPCI:SCC90, that expresses wild-type p53
(9), and the HPV-negative SQ20B
cell line that expresses mutant p53 as the control. UPCI:SCC90
cells are more radiosensitive than SQ20B cells (10). We show that UPCI:SCC90 cells
express lower levels of NAE1/APP-BP1 mRNA, and that the
overexpression of NAE1/APP-BP1 in these cells induces
radioresistance. Conversely, SQ20B cells become radiosensitive when
wild-type TP53 is expressed. The inhibition of
NAE1/APP-BP1 potentiates the effect of wild-type p53. These
results point to the importance of NEDDylation in the
radiosensitivity of tumor cells that express wild-type p53.
Materials and methods
Cell culture and transfections
The HNSCC SQ20B cell line was purchased from the
American Type Culture Collection (ATCC; Rockville, MD, USA). The
radioresistant SQ20B cells have been reported to express
non-funtional p53 protein (10).
The hHNSCC UPCI:SCC90 cell line (9) was a kind gift from Professor Susan
Gollin (University of Pittsburgh, Pittsburgh, PA, USA). UPC1:SCC90
cells originate from a HPV16-positive oropharyngeal tumor, and bear
wild-type TP53 (9).
UPCI:SCC90 cells have been shown to be more radiosensitive than
SQ20B cells (11). The cells were
grown in Dulbecco’s modified Eagle’s medium (DMEM; PAN Biotech
GmbH, Dominique Dutscher, Brumath, France) according to standard
conditions.
All transfection experiments were carried out using
Lipofectamine™ 2000 (Invitrogen, Fisher Scientific, Illkirch,
France) according to the manufacturer’s instructions. UPCI:SCC90
cells (5×105) were transfected with 4 μg of the
NAE1/APP-BP1 expression vector (12) (kind gift from Dr Dimitris
Xirodimas). Mock transfections with 4 μg of the empty pcDNA3 vector
(Invitrogen) or the empty pCI-neo vector (Promega,
Charbonnières-les-bains, France) were used as the negative
controls. p53 was overexpressed by transfection of 5×105
SQ20B cells with 4 μg of pC53-C1N3 plasmid.
siRNA experiments
The NAE1/APP-BP1 downregulation experiments
were performed using the ON-TARGET plus SMARTpool
(L-006401-00-0005; Dharmacon, Fisher Scientific, Illkirch, France).
This pool is composed by the combination of 4 single siRNAs:
siRNA-1, upgrade siRNA NAE1/APP-BP1 J-006401-05; siRNA-2,
upgrade siRNA NAE1/APP-BP1 J-006401-06; siRNA-3, upgrade
siRNA NAE1/APP-BP1 J-006401-07; and siRNA-4, upgrade siRNA
NAE1/APP-BP1 J-006401-08. The ON-TARGET plus non-targeting
pool (D-001810-10-05) was used as the negative control. siRNAs (40
pmol) were transfected into 4×104 SQ20B cells using
Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s
instructions.
RNA extraction and quantitative real-time
RT-PCR (qRT-PCR)
RNA was extracted from 5×105 cells and
cDNA synthesis was performed as previously described (3). Cycle threshold (Ct) levels were
normalized to the average Ct values of 3 internal controls
(housekeeping genes): ubiquitin B (UBB), 18S rRNA and hypoxanthine
phospho-ribosyltransferase 1 (HPRT1). Primers specific for the
genes of interest were used: HR-HPV16 E6/E7 mRNA forward,
5′-GTTACTGCGACGTGAGGTA TATG-3′ and reverse,
5′-CATTTATCACATACAGCATATGG ATTC-3′ (13); NAE1/APP-BP1 forward,
5′-CTGGAAAGCT GCTCAAGG-3′ and reverse, 5′-TCTTCTCCGCTGACCT GATT-3′;
CDKN2Ap16 forward, 5′-GCTGCCCAACGCACCGA ATA-3′
and reverse, 5′-ACCACCAGCGTGTCCAGGAA-3′;
WAF1/CIP1p21 forward, 5′-ATGAAATTCACCCCCTTTCC-3′
and reverse, 5′-CCCTAGCTGTGCTCACTTC-3′; PUMA forward,
5′-ACGACCTCAACGCACAGTACGA-3′ (14)
and reverse, 5′-GTAAGGGCAGGAGTCCCATGATGA-3′ (14); MDM2 forward,
5′-GGTGGGAGTGATCAAAAGGA-3′ and reverse, 5′-GTGGCGTTTTCTTTGTCGTT-3′;
HPRT1 forward, 5′-TGCTCGAGATGTGATGAAGG-3′ and reverse, 5′-GT
CCCCTGTTGACTGGTCATT-3′; UBB forward, 5′-GCTTTG
TTGGGTGAGCTTGT-3′ and reverse, 5′-CGAAGATCTGCA TTTTGACCT-3′; 18S
rRNA forward, 5′-TGTGGTGTTGA GGAAAGCAG-3′ and reverse,
5′-TCCAGACCATTGGCT AGGAC-3′. Gene expression levels observed in the
non-irradiated mock-transfected cells were artificially set to 100,
and other expression values were normalized with respect to this
level. Differences were considered statistically significant when
p<0.05 (ANOVA).
Irradiation procedures, clonogenic
survival assay and surviving fraction at 2 Gy (SF2)
calculation
Cells in the exponential growth phase were detached,
counted and plated in 6-well plates in 2 ml of growth medium 24 to
48 h prior to irradiation, in order to allow them to attach and
undergo transfection. X-ray irradiation was performed with 6 MV
photons at the Department for Radiation Therapy of the Paul Strauss
Cancer Centre (Strasbourg, France). Clonogenic assays were
performed as previously described (15). In brief, cells were collected 24 h
after irradiation, diluted, seeded and allowed to grow for up to 3
to 4 weeks after irradiation. Clones were stained with 0.05%
crystal violet (Sigma-Aldrich, Lyon, France) in a 5% ethanol
solution, and positive colonies (>64 cells) were counted. The
SF2 was calculated by dividing the number of positive colonies by
the number of cells that were seeded, multiplied by the plating
efficiency. The plating efficiency was calculated by dividing the
number of positive colonies that grew in the absence of irradiation
by the number of cells that were seeded. Differences were
considered statistically significant when p<0.05 (ANOVA).
Western blot analyses
Cells (4×104 to 5×106) were
harvested in lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris-Cl pH
8.0, 0.5% DOC, 0.1% SDS) and proteins were analyzed by SDS-PAGE
according to standard methods. Proteins were detected with the
following antibody dilutions: polyclonal mouse AO1 anti-APP-BP1
(1:1000; Abnova, Tebu-Bio, Le-Perray-en-Yvelines, France),
monoclonal mouse DO1 anti-p53 (1:1000; Santa Cruz Biotechnologies,
CliniSciences, Montrouge, France), polyclonal rabbit anti-p53
FL-393 (1:200; Santa Cruz Biotechnologies), anti-NEDD8 rabbit
polyclonal (1:2000; Alexis Biochemicals, Enzo Life Sciences,
Villeurbanne, France), mouse monoclonal CM1 anti-β actin (1:1000;
Abnova), anti-β tubulin (1:5000; Sigma-Aldrich), enhanced
chemiluminescence (ECL) sheep anti-mouse IgG, HRP-conjugated
(1:10000; GE Healthcare, Saclay, France), and anti-rabbit IgG,
HRP-linked antibodies (1:20000; Cell Signaling, Ozyme,
Saint-Quentin-en-Yveline, France). Proteins were revealed with the
ECL western blot analysis system (GE Healthcare) according to the
manufacturer’s instructions. Quantification of the signals was
performed using ImageJ software (National Institutes of Health,
Bethesda, MD, USA) (16,17).
p53 transcriptional activity assay
With the use of the Lipofectamine™ 1000 transfection
system, 5×105 UPCI:SCC90 cells were co-transfected with
0.75 μg of PG13-Luc plasmid (kind gift from Dr Arnold Levine) or
MG15-Luc plasmid, 0.75 μg of RSV-LacZ vector [used as a
transfection efficiency control (18)], and increasing amounts (0, 1.0,
1.5, 2.5 μg) of either the NAE1/APP-BP1 expression vector or
of the empty vector (pcDNA3). The total amount of transfected
plasmid was made up to 4 μg with pUC19 (New England Biolabs). Cells
were harvested 36 h after transfection in lysis buffer (25 mM
Tris-phosphate, pH 7.8; 2 mM EDTA; 1 mM DTT; 10% glycerol; 1%
Triton X-100), and luciferase activity was evaluated by mixing 50
μl of protein extract with luciferase assay buffer (265 μM ATP; 235
μM luciferin; 135 μM coenzyme A; 20 mM tricine; 1.07 mM
MgCl2; 2.70 mM MgSO4; 0.10 mM EDTA; 33.3 mM
DTT) using a MikroWin 2000 microplate reader (Mikrotek Laboratories
GmbH, Overath, Germany). The luminescence levels were normalized to
the β-galactosidase activity from the RSV-LacZ control plasmid that
was measured with an o-nitrophenyl-β-galactoside (ONPG)
colorimetric reaction. In brief, β-galatcosidase activity was
evaluated by mixing 25 µl of cell extract with 150 µl of ONPG
staining solution (0.666 mg/ml ONPG; 2% β-mercaptoethanol; 60 mM
Na2HPO4; 40 mM NaH2PO4;
10 mM KCl; 1 mM MgSO4), and o-nitrophenol was
monitored on a Berthold 900 spectrophotometer using Gen5 Data
analysis software. Fluorescence observed in cells transfected with
0 μg of NAE1/APP-BP1 expression vector was set at a value of
100, and other measures were normalized with respect to that point.
Differences were considered significant when p<0.05
(Student-Newman-Keuls test for pairwise comparisons).
Immunoprecipitations
Cells (1.5×106) were harvested in
denaturing lysis buffer (1% SDS, 5 mM EDTA, 10 mM DTT, 15 U/ml
DNase, PMSF). A total of 5 μl of each cell lysate was diluted in 45
μl of non-denaturing buffer (20 mM Tris-HCl pH 8.0, 137 mM NaCl,
10% glycerol, 1% Triton X-100, 2 mM EDTA, PMSF). Diluted cells
lysates were pre-incubated with 0.4 μg of anti-p53 antibody (DO1
anti-p53; Santa Cruz Biotechnologies) or with 2 μl of anti-NEDD8
antibody (Alexis Biochemicals). A total of 10 μl of Protein A/G
PLUS-Agarose Immunoprecipitation Reagent (sc-2003; Santa Cruz
Biotechnologies) was further mixed with extracted protein, pelleted
and resuspended in loading buffer and subjected to western blot
analysis with an anti-NEDD8 antibody and a polyclonal rabbit
anti-p53 FL-393 antibody (Santa Cruz Biotechnologies). Normal mouse
IgG-AC (10 μl) (sc-2343; Santa Cruz Biotechnologies) was incubated
with protein lysate as the negative control.
Apoptosis detection by Annexin V
staining
UPCI:SCC90 cells were transfected and irradiated as
described above. Annexin V staining was monitored by flow cytometry
on an Agilent Bioanalyzer 2100 (Agilent Technologies Deutschland,
Waldbronn, Germany), according to the manufacturer’s instructions.
In brief, cells were incubated first with Annexin V-Biotin (Annexin
V-Biotin apoptosis detection kit, Oncogene Research Product, Merck
Chemicals, Nottingham, UK), and later with streptavidin-Cy5
(Amersham, GE Healthcare). In order to discriminate early apoptotic
cells from dead cells, samples were counterstained with Calcein-AM
(Molecular Probes, Invitrogen, Cergy Pontoise, France). Cell
samples were loaded on Cell Fluorescence LabChip® kits,
and flow cytrometry data were analyzed with Agilent Cell
Fluorescence software. Data were analyzed with one-way ANOVA,
together with a Student-Newman-Keuls test of all pairwise possible
comparisons, and differences were considered significant when
p<0.05.
Results
NAE1/APP-BP1 gene and protein expression
are lower in the UPCI:SCC90 cells than in SQ20B cells
In order to evaluate the involvement of
NAE1/APP-BP1 gene expression levels in HR-HPV-positive
oropharyngeal squamous cell carcinoma radiosensitivity, we selected
2 different cell lines: the HPV-negative SQ20B cell line that
originates from a laryngeal tumor, harboring a TP53 mutant
allele and that is radioresistant (10); and the HR-HPV16-positive UPCI:SCC90
cell line that was established from an oropharyngeal squamous cell
carcinoma, that bears a wild-type TP53 locus and is
radiosensitive as compared to the SQ20B cell line (11). Using a qRT-PCR approach with 3
independent total RNA extracts from the 2 cell lines, we confirmed
that the mRNA that encodes the HPV16 E6 and E7 oncoproteins is
expressed in UPCI:SCC90 cells, whereas it was not detectable in RNA
extracts from SQ20B cells (Fig.
1A). As expected, the expression of the
CDKN2Ap16 gene, which is used as a surrogate
marker of HPV infection, was found to be dramatically increased in
the UPCI:SCC90 cells as compared to the SQ20B cells (Fig. 1A). These results confirm that HPV16
is biologically active in UPCI:SCC90 cells. We measured the
expression of WAF1/CIP1p21, whose expression is
regulated by p53. In agreement with their respective TP53
statuses, the expression of WAF1/CIP1p21 was
found to be higher in the UPCI:SCC90 cells as compared to the SQ20B
cells (Fig. 1A). Finally, the
expression of the NAE1/APP-BP1 gene was found to be lower in
the UPCI:SCC90 cells than in the SQ20B cells. We further evaluated
the expression levels of the NEA1/APP-BP1 protein in these 2 cells
lines by western blot analysis. Blots were probed with an
anti-NEA1/APP-BP1 antibody, and quantification of the signals
obtained from total protein extracts confirmed that the
NEA1/APP-BP1 protein displays higher expression levels in SQ20B
cells than in UPCI:SSC90 cells (Fig.
1B). Thus, these 2 cell lines display NAE1/APP-BP1 expression
patterns that are similar to those reported in HR-HPV-positive and
HPV-negative HNSCC. They were used to evaluate the influence of the
NAE1/APP-BP1 gene on their response to ionizing
radiation.
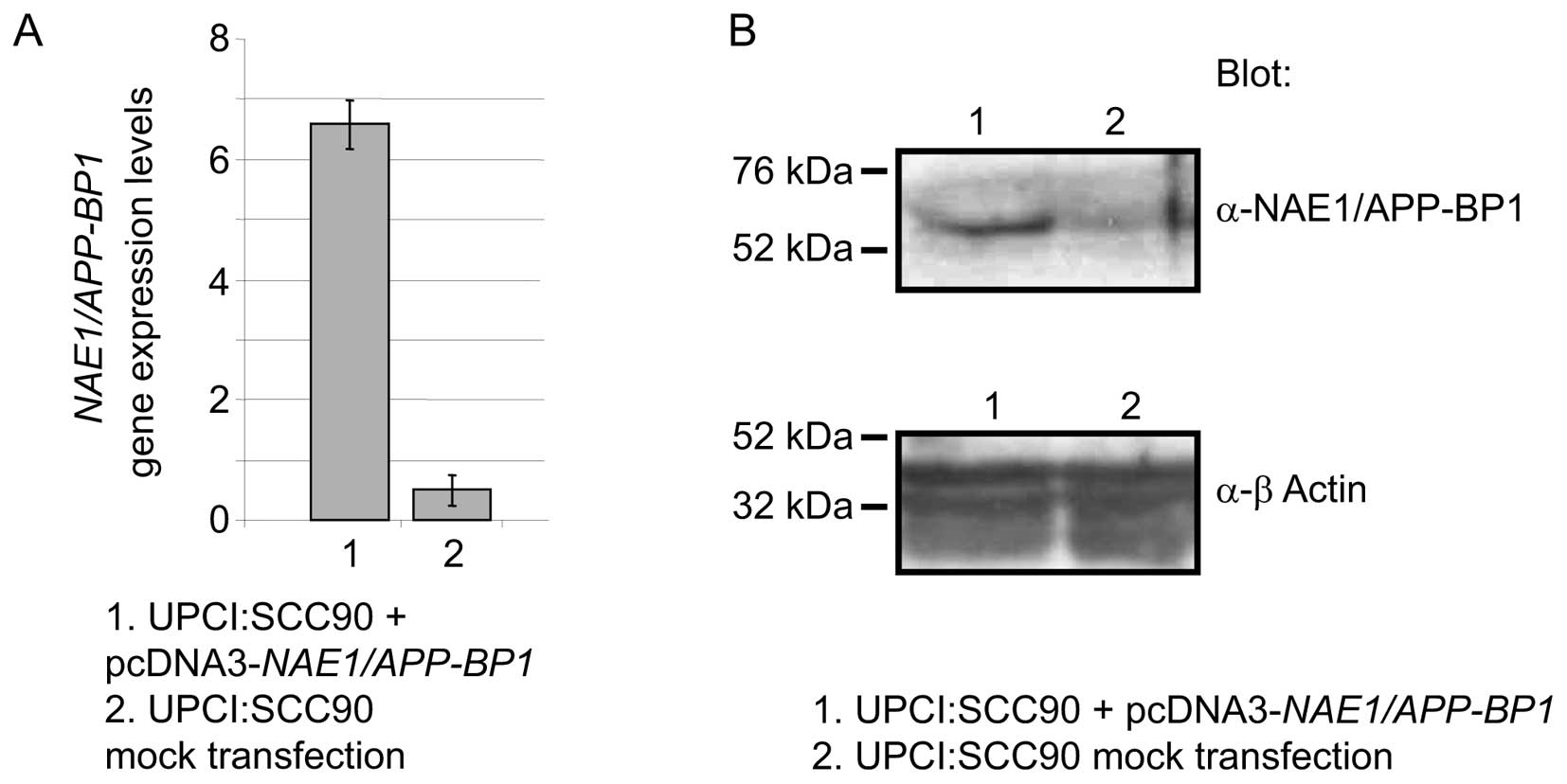
Forced expression of NAE1/APP-BP1
enhances resistance to ionizing radiation
We examined the effect of modulating
NAE1/APP-BP1 expression levels in UPCI:SCC90 cells on
clonogenic survival. NAE1/APP-BP1 was overexpressed by the
transfection of an expression vector. Transfection with the
NAE1/APP-BP1 construct resulted in an increase in
NAE1/APP-BP1 mRNA levels compared to the mock-transfected
cells, as measured by qRT-PCR in 3 independent extracts of cells
harvested 24 h after transfection (Fig. 2A). A western blot analysis of
protein extracts from the same cells, using an anti-NAE1/APP-BP1
antibody, showed that NAE1/APP-BP1 protein levels also increased
(Fig. 2B). In a clonogenic
survival assay, the cells were irradiated with a single 2-Gy dose
of X-rays, and the number of cells that were able to grow colonies
was measured. Non-irradiated cells were used as the control to
calculate the fraction of cells that survived the treatment. The
SF2 of mock-transfected cells and that of cells that overexpressed
NAE1/APP-BP1, calculated from 6 independent measures, was
0.36 and 0.89, respectively (Table
I; p<0.001), showing that NAE1/APP-BP1 overexpression
augments resistance to ionizing radiation in UPCI:SCC90 cells.
 | Table IEffects of NAE1/APP-BP1 and
TP53 expression on the fraction of UPCI:SCC90 and SQ20B
cells that survived after irradiation with 2 Gy X-Rays (SF2). |
Table I
Effects of NAE1/APP-BP1 and
TP53 expression on the fraction of UPCI:SCC90 and SQ20B
cells that survived after irradiation with 2 Gy X-Rays (SF2).
| Cell
transfection | SF2 Gy | p-value ANOVA |
|---|
| UPCI:SCC90
cells | | |
| Mock
transfection | 0.36±0.16 | <0.001 |
|
pcDN3-NAE1/APP-BP1 | 0.89±0.07 | |
| SQ20B cells | | |
| Single
transfection | | |
| Mock
transfection | 0.46±0.01 | |
| Control
siRNA | 0.64±0.10 | NS |
|
NAE1/APP-BP1 siRNA | 0.53±0.12 | NS |
|
pC53-C1N3 | 0.35±0.02 | 0.030 |
| Double
transfection | | |
|
pC53-C1N3 + control
siRNA | 0.46±0.14 | |
|
pC53-C1N3 +
NAE1/APP-BP1 siRNA | 0.26±0.04 | 0.045 |
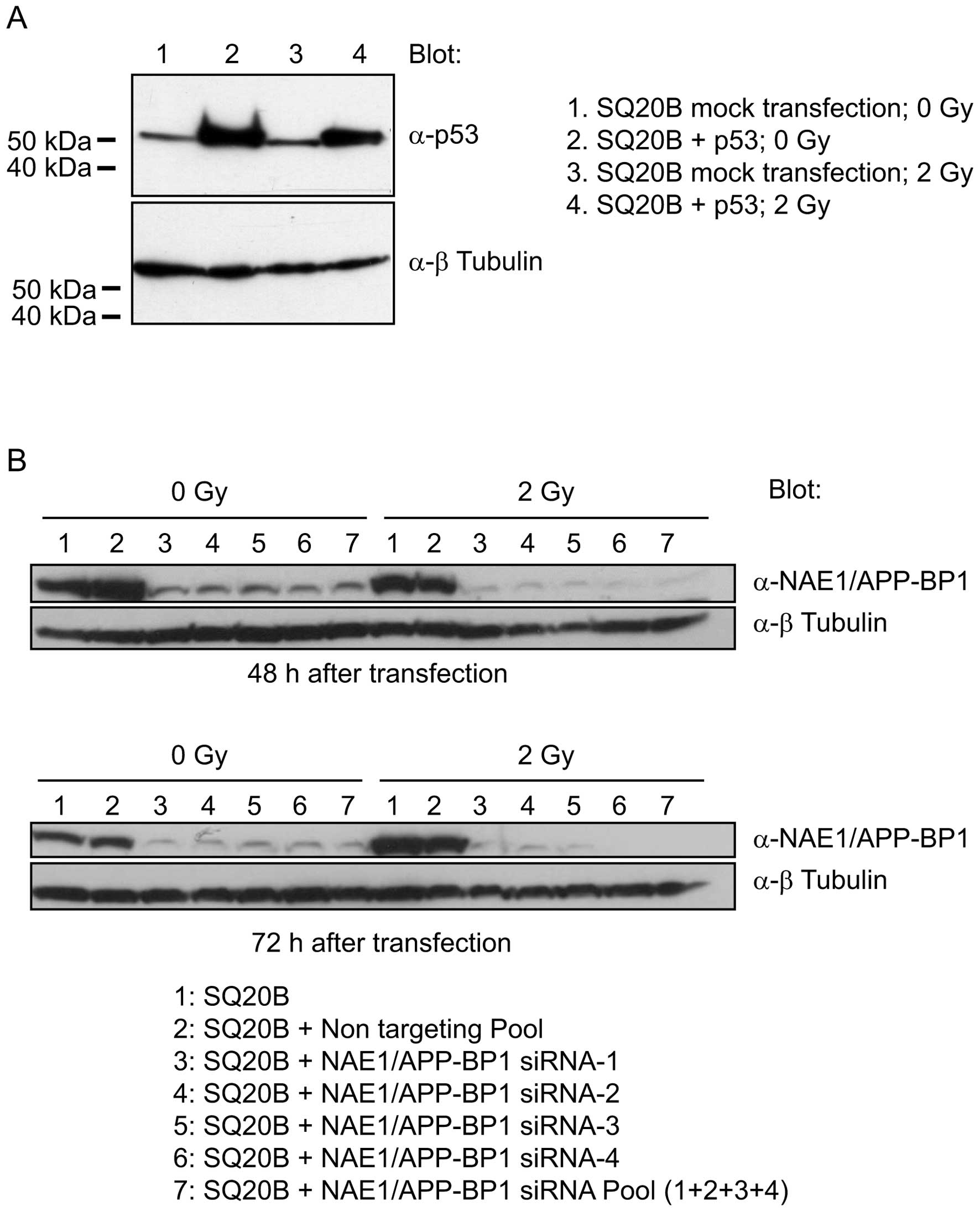
Downregulation of NAE1/APP-BP1 increases
radiosentivity in a p53-dependent manner
We reasoned that if NAE1/APP-BP1 is involved
in cell survival after irradiation, then the response of SQ20B
cells to ionizing radiation may be modulated by both restoring
functional TP53 and downregulating NAE1/APP-BP1
expression. Consequently, SQ20B cells were co-transfected with a
wild-type TP53 expression vector and siRNAs that
specifically target NAE1/APP-BP1. A non-related siRNA was
used as the negative control. SQ20B cells bear a point mutation in
the TP53 gene that results in the production of a
non-functional p53 protein, which can be detected by western blot
analysis (Fig. 3A, lanes 1 and 3).
The transfection of SQ20B cells with an expression vector for
wild-type TP53 further increased p53 levels. Non-irradiated
and irradiated transfected cells displayed increased amounts of p53
as compared to the mock-transfected cells (Fig. 3A, lanes 2 and 4). The SF2 of
mock-transfected SQ20B cells and that of SQ20B cells that
overexpressed p53, was 0.46 and 0.35, respectively (Table I; p=0.030), indicating that
exogenous p53 function is functional in SQ20B cells. The expression
of the NAE1/APP-BP1 protein was downregulated up to 48 and 72 h
after transfecting SQ20B cells with an anti-NAE1/APP-BP1 siRNA pool
(Fig. 3B, lane 7). This pool is
composed of 4 individual siRNAs, which all were found to
efficiently downregulate the expression of the NAE1/APP-BP1
gene (Fig. 3A, lanes 3–6).
Transfection with a non-related siRNA, used as the negative
control, had no effect on NAE1/APP-BP1 protein levels (Fig. 3B, lane 2). Neither the
anti-NAE1/APP-BP1 siRNA, nor the control siRNA increased
cell sensitivity to ionizing radiation. The SF2 in these conditions
was 0.53 and 0.64, respectively (Table
I). Strikingly, the irradiation of SQ20B cells that were
co-transfected with wild-type TP53 and
anti-NAE1/APP-BP1 siRNA resulted in a drop in the SF2 to
0.26, whereas cells that were co-transfected with TP53 and
the control siRNA displayed a SF2 of 0.46 (Table I; p=0.045). Taken together, these
observations suggest that the inhibition of NAE1/APP-BP1 in
SQ20B cells restores cell sensitivity to ionizing radiation in a
p53-dependent manner.
Overexpression of NAE1/APP-BP1 in
UPCI:SCC90 cells inhibits p53 transcriptional activity and impairs
apoptosis
In order to examine whether the modulation of
NAE1/APP-BP1 expression levels in UPCI:SCC90 cells has an
effect on p53 transcriptional activity, we used the pG13-Luc
reporter, in which the gene that encodes the luciferase protein is
under the control of p53-dependent cis-regulatory elements.
UPCI:SCC90 cells were co-transfected with equal amounts of pG13-Luc
and increasing amounts of either the NAE1/APP-BP1 expression
vector, or the empty control vector. Cells were harvested 36 h
after transfection and luciferase activity was measured with 3
independent protein extracts. There was no change in p53
transcriptional activity in the cells that were mock-transfected
with various amounts of control vector (Fig. 4A, left histograms). By contrast,
p53 transcriptional activity was markedly and significantly
repressed in UPCI:SCC90 cells transfected with increasing amounts
of the NAE1/APP-BP1 vector, in a dose-dependent manner
(Fig. 4A; 1.5 and 2.5 μg of
NAE1/APP-BP1 vector; p<0.05). In the control experiment,
we used the MG15-Luc reporter plasmid, which contains mutated
p53-binding elements upstream of the luciferase gene. The
fluorescence that we observed was markedly reduced, and no
differences were observed between the cells transfected with either
the empty plasmid or the NAE1/APP-BP1 expression construct
(Fig. 4A, right histograms). The
UPCI:SCC90 mock-transfected cells displayed rapid and significant
(p<0.05) induction of p53 target genes, such as
WAF1/CIP1p21, PUMA and MDM2, 30 min
after a 2-Gy irradiation (Fig.
4B). This induction was not observed in UPCI:SCC90 cells that
overexpressed NAE1-APP-BP1 (Fig. 4B). In order to evaluate whether
this modulation of the p53 dependent transcriptional activity
depends on a NAE1/APP-BP1-dependent increase in NEDDylation, we
performed a western blot analysis to detect NEDDylated p53 in
proteins extracted under denaturating conditions. NEDD8-conjugated
proteins were immunoprecipitated with a p53 antibody, and the blots
were probed with an anti-NEDD8 antibody (Fig. 4C-a). Conversely, NEDD8 was
immunoprecipitated from the protein extracts, and the blots were
then probed with an anti-p53 antibody (Fig. 4C-b). NEDDylated proteins were
present in similar amounts in the extracts from the
mock-transfected cells and from the cells transfected with
NAE1/APP-BP1 (Fig. 4C-c). In both
cases, we observed a band whose size corresponded to NEDDylated p53
(approximately 63 kDa; Fig. 4C-a and
-b; asterisk). This signal was stronger in the UPCI:SCC90 cells
that overexpressed NAE1/APP-BP1 than in the mock-transfected
UPCI:SCC90 cells (Fig. 4C-a and
-b). No detectable signal corresponding to a NEDDylated p53
protein was observed when transfected UPCI:SCC90 cell lysates were
immunoprecipitated with an unrelated IgG (Fig. 4C-a). These results indicate that
the overexpression of NAE1/APP-BP1 in UPCI:SCC90 cells
results in an increased NEDDylation of p53. These observations are
consistent with the decreased transcriptional activity of p53
(Fig. 4A and B).
We evaluated whether the modulation of
radiosensitivity by NAE1/APP-BP1 expression levels and p53
activity is associated with changes in apoptosis in the control and
transfected UPCI:SCC90 cells, prior to and 6 h following a 4-Gy
irradiation. A flow cytometry approach was used in which living and
dead cells were discriminated by staining with a calcein probe, and
apoptotic cells by Annexin V. Calcein-positive and -negative cells
were cross-gated to cells that displayed high levels of Annexin V
(>102; Fig. 5A), and
the proportion of each cell category was counted. The graph
represents the statistical analysis of 3 independent experiments
(Fig. 5B). The proportion of
calcein-negative Annexin V-positive cells was found to be decreased
when NAE1/APP-BP1 was overexpressed in irradiated UPCI:SCC90
cells as compared to irradiated mock-transfected cells (p<0.05).
Taken together, these observations suggest that NAE1/APP-BP1
regulates cell sensitivity to ionizing radiation by modulating
apoptosis.
Discussion
A hallmark of patients bearing HR-HPV-positive
HNSCCs is improved disease-free and overall survival as compared to
their HPV-negative counterparts (1), which may be due to increased tumor
sensitivity to chemo- and radiation-therapy (19). We have previously described the
HPV-positive tumor-specific loss of genetic material in the 16q
region (3), which has been
reported to be linked to improved local control (4). We have shown that the gene that
encodes NAE1/APP-BP1, and which is located on chromosome 16q22, is
expressed at lower levels in HPV-positive lesions (3). In the present study, we addressed the
role of NAE1/APP-BP1 in the regulation of the cellular
response to ionizing radiation in HPV-positive and -negative cell
lines. NAE1/APP-BP1 is a negative regulator of p53 transcriptional
activity. The majority of HR-HPV-related oropharyngeal carcinomas
express wild-type TP53 (1)
and have a decreased expression of NAE1/APP-BP1 (3). Thus, we reasoned that the loss of
NAE1/APP-BP1 may be responsible for the increased p53 activity and
subsequent tumor cell death upon genotoxic stress.
Similar to the data that has previously been
presented for HPV-positive oropharyngeal tumors, in the present
study, we show that the expression of NAE1/APP-BP1 is
downregulated in radiosensitive UPCI:SCC90 cells (11) as compared to SQ20B radioresistant
cells (10). The radiosensitive
phenotype of UPCI:SCC90 cells was rescued by the overexpression of
NAE1/APP-BP1, and conversely, the radioresistance of SQ20B
cells was inhibited by the decreasing NAE1/APP-BP1
expression. This effect appears to be p53-dependent.
The NEDDylation of p53 in other cancer cell lines
has been reported. FBXO11, a component of the SCF E3
ubiquitin-ligase complex, has NEDD8-ligase activity. It physically
interacts with p53 in the human lung carcinoma H1299 cell line,
triggers p53 NEDDylation and inhibits its transcriptional activity
(20). NAE1/APP-BP1
expression levels correlate with cell death in Drosophila
melanogaster. Clones of homozygous dAPP-BP1 mutant
cells, generated in heterozygous surrounding wild-type tissue, have
an unusual small size due to a reduction in the number of cells.
This phenotype is due to a dramatic increase of apoptosis in mutant
cells (21).
The NEDD8 conjugation pathway has been implicated in
the regulation of the activity of the SCF ubiquitin-ligase complex
(7,22). Of note, a link between the activity
of the SCF complex and sensitivity to ionizing radiation has
recently been established (23).
The sensitive to apoptosis gene (SAG) protein is a component of the
SCF E3 ubiquitine ligase that is overexpressed in certain human
cancers. The silencing of SAG in cancer cell lines impairs cell
growth and colony formation, and results in increased sensitivity
to radiation. Of note, the pro-apoptotic protein, NOXA, has been
found to be more abundant in cells in which SAG expression
is downregulated, and apoptosis is increased, suggesting that SAG
is required for the degradation of NOXA. In our study, we show that
the NEDD8 conjugation pathway has an influence on p53-dependent
radiation-induced apoptosis. However, we cannot rule out the
possibility that the effects observed are also partially linked to
effects of the SCF complex on the turn-over of anti- or
pro-anti-apoptotic factors and cell cycle regulators (22). In addition, it has previously been
shown that TAp73 transcriptional activity is inhibited by
NEDDylation in the ts41 or wild-type CHO cells (24). Therefore, we cannot exclude that
the inhibition of TAp73 in transfected UPCI:SCC90 cells partially
accounts for the effect of NAE1/APP-BP1 on luciferase and p53
target gene expression.
The radiosensitivity of cancer cells varies during
the cell cycle (25,26). However, we did not observe changes
in the G1 and G2 phase distribution of UPCI:SCC90 cells that
overex-press NAE1/APP-BP1 (data not shown), suggesting that
the effects were not due to changes in the cell cycle.
As a consequence of our findings, the correlation
between NAE/APP-BP1 expression levels and local control and
disease-free survival of irradiated patients is currently being
investigated. More generally, several studies have unambiguously
demonstrated that HR-HPV is a major prognostic factor with respect
to both disease-free and overall survival, and this prognosis is
independent of treatment modality (1). The question as to whether
HPV-positive patients should receive dose de-escalation (decreased
irradiation dose delivered to the tumor area, or decreased area
that receives the irradiation dose) in order to improve the
therapeutic index, while sparing them long-term toxicity, is
currently a matter of debate (29). Alternatively, HPV-positive tumors
could be managed by modulating existing therapies, in order to
potentiate the efficacy of ionizing radiation.
Our data highlight the relevance of the NEDDylation
pathway to the cellular response to radiotherapy, and indicate this
pathway is a potential drug target to potentiate existing
therapies. Recently, a novel class of pharmacological inhibitors of
the NAE1/APP-BP1/UBA3 complex has been tested in preclinical and
clinical tumor models (27,28).
In the light of our findings, it would be of interest to determine
whether a functional copy of p53 is required for the efficacy of
this novel treatment. In addition, these kind of therapeutic
approaches could improve the efficacy of the treatment delivered to
wild type TP53 HPV-negative tumors.
HR-HPV detection (by immunohistochemistry) has been
incorporated into an algorithm that could be used to predict
overall survival (29). A
stratification of patients based on that type of algorithm would be
a first step to randomized trials. In HPV-related oropharyngeal
cancer, the loss of genetic material in the 16q region is related
to improved local control (3,4). It
would be of great interest to further dissect this locus, in order
to identify molecular markers that predict tumor response to
therapy.
Besides increasing our knowledge of the molecular
mechanisms of cell sensitivity to ionizing radiation, the
identification of relevant biomarkers, combined with already
established clinical features, may provide physicians with tools to
select patients for tailored and more appropriate treatment
strategies.
Acknowledgements
The UPCI:SCC90 cells were kindly
provided by Professor Susan Gollin. The pcDNA3-NAE1/APP-BP1
vector was a kind gift from Dr Dimitris Xirodimas. The PG13-luc
plasmid was a kind gift from Dr Arnold Levine. We thank Dr Erwan
Pencreach for critically reading the manuscript. We are most
grateful to Mrs. Christine Wasylyk for her invaluable help with the
immunoprecipitation experiments and luciferase assays. S.G. is
grateful to the Institut National du Cancer for its support (grant:
‘Soutien à la formation à la recherche translationnelle en
cancérologie d’étudiants en médecine et de jeunes médecins’).
Financial support for this study was provided by the Université de
Strasbourg and by the Comité Départemental du Bas-Rhin de la Ligue
Contre le Cancer. We are most grateful to the ‘Carte d’Identité des
Tumeurs’ program of the Ligue Nationale contre le Cancer for their
long-standing support.
References
|
1
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar
|
|
2
|
Butz K, Whitaker N, Denk C, Ullmann A,
Geisen C and Hoppe-Seyler F: Induction of the p53-target gene
GADD45 in HPV-positive cancer cells. Oncogene. 18:2381–2386. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jung AC, Briolat J, Millon R, et al:
Biological and clinical relevance of transcriptionally active human
papillomavirus (HPV) infection in oropharynx squamous cell
carcinoma. Int J Cancer. 126:1882–1894. 2010.PubMed/NCBI
|
|
4
|
Klussmann JP, Mooren JJ, Lehnen M, et al:
Genetic signatures of HPV-related and unrelated oropharyngeal
carcinoma and their prognostic implications. Clin Cancer Res.
15:1779–1786. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gong L and Yeh ET: Identification of the
activating and conjugating enzymes of the NEDD8 conjugation
pathway. J Biol Chem. 274:12036–12042. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Osaka F, Kawasaki H, Aida N, et al: A new
NEDD8-ligating system for cullin-4A. Genes Dev. 12:2263–2268. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Petroski MD and Deshaies RJ: Function and
regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol.
6:9–20. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xirodimas DP, Saville MK, Bourdon JC, Hay
RT and Lane DP: Mdm2-mediated NEDD8 conjugation of p53 inhibits its
transcriptional activity. Cell. 118:83–97. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ragin CC, Reshmi SC and Gollin SM: Mapping
and analysis of HPV16 integration sites in a head and neck cancer
cell line. Int J Cancer. 110:701–709. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maalouf M, Alphonse G, Colliaux A, et al:
Different mechanisms of cell death in radiosensitive and
radioresistant p53 mutated head and neck squamous cell carcinoma
cell lines exposed to carbon ions and x-rays. Int J Radiat Oncol
Biol Phys. 74:200–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gupta AK, Lee JH, Wilke WW, et al:
Radiation response in two HPV-infected head-and-neck cancer cell
lines in comparison to a non-HPV-infected cell line and
relationship to signaling through AKT. Int J Radiat Oncol Biol
Phys. 74:928–933. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Y, McPhie DL, Hirschberg J and Neve
RL: The amyloid precursor protein-binding protein APP-BP1 drives
the cell cycle through the S-M checkpoint and causes apoptosis in
neurons. J Biol Chem. 275:8929–8935. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Boer MA, Jordanova ES, Kenter GG, et
al: High human papillomavirus oncogene mRNA expression and not
viral DNA load is associated with poor prognosis in cervical cancer
patients. Clin Cancer Res. 13:132–138. 2007.
|
|
14
|
Garrison SP, Jeffers JR, Yang C, et al:
Selection against PUMA gene expression in Myc-driven B-cell
lymphomagenesis. Mol Cell Biol. 28:5391–5402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Franken NA, Rodermond HM, Stap J, Haveman
J and van Bree C: Clonogenic assay of cells in vitro. Nat Protoc.
1:2315–2319. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abramoff MD, Magelhaes PJ and Ram SJ:
Image processing with ImageJ. Biophotonics Int. 11:36–42. 2004.
|
|
17
|
Rasband WS: ImageJ. National Institutes of
Health; Bethesda, MA: http://rsb.info.nih.gov/ij/,
1997–2008.
|
|
18
|
Kozarsky KF, McKinley DR, Austin LL, Raper
SE, Stratford-Perricaudet LD and Wilson JM: In vivo correction of
low density lipoprotein receptor deficiency in the Watanabe
heritable hyperlipidemic rabbit with recombinant adenoviruses. J
Biol Chem. 269:13695–13702. 1994.
|
|
19
|
Lassen P: The role of Human papillomavirus
in head and neck cancer and the impact on radiotherapy outcome.
Radiother Oncol. 95:371–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abida WM, Nikolaev A, Zhao W, Zhang W and
Gu W: FBXO11 promotes the Neddylation of p53 and inhibits its
transcriptional activity. J Biol Chem. 282:1797–1804. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim HJ, Kim SH, Shim SO, et al:
Drosophila homolog of APP-BP1 (dAPP-BP1) interacts
antagonistically with APPL during Drosophila development.
Cell Death Differ. 14:103–115. 2007. View Article : Google Scholar
|
|
22
|
Nakayama KI and Nakayama K: Ubiquitin
ligases: cell-cycle control and cancer. Nat Rev Cancer. 6:369–381.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia L, Yang J, Hao X, et al: Validation of
SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and
radiosensitizing target. Clin Cancer Res. 16:814–824. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Watson IR, Blanch A, Lin DC, Ohh M and
Irwin MS: Mdm2-mediated NEDD8 modification of TAp73 regulates its
transactivation function. J Biol Chem. 281:34096–34103. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Quiet CA, Weichselbaum RR and Grdina DJ:
Variation in radiation sensitivity during the cell cycle of two
human squamous cell carcinomas. Int J Radiat Oncol Biol Phys.
20:733–738. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tell R, Heiden T, Granath F, Borg AL, Skog
S and Lewensohn R: Comparison between radiation-induced cell cycle
delay in lymphocytes and radiotherapy response in head and neck
cancer. Br J Cancer. 77:643–649. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Milhollen MA, Traore T, Adams-Duffy J, et
al: MLN4924, a NEDD8-activating enzyme inhibitor, is active in
diffuse large B-cell lymphoma models: rationale for treatment of
NF-κB-dependent lymphoma. Blood. 116:1515–1523. 2010.PubMed/NCBI
|
|
28
|
Soucy TA, Smith PG, Milhollen MA, et al:
An inhibitor of NEDD8-activating enzyme as a new approach to treat
cancer. Nature. 458:732–736. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ang KK, Harris J, Wheeler R, et al: Human
papillomavirus and survival of patients with oropharyngeal cancer.
N Engl J Med. 363:24–35. 2010. View Article : Google Scholar : PubMed/NCBI
|