Introduction
More than 90% of all malignant epithelial tumors
arising in the oral cavity are squamous cell carcinomas
(SCC)(1,2). Oral squamous cell carcinoma (OSCC) is
one of the most common malignancies in humans. However, the overall
survival rates have not substantially improved for decades
(3) and a significantly increased
incidence of OSCC in young subjects has been reported in recent
decades (4,5).
In general, varying degrees of inflammatory cell
infiltration are observed around malignant tumors containing SCC.
Cytokines or cytokine-related mediators have direct proliferative
and anti-proliferative effects on tumor cells, and influence the
cellular behavior of malignant cells (6–8).
Interleukin (IL)-22 is a newly discovered member of the IL-10
family, and is expressed mainly in activated T, mast and NK cells
(9,10). Additionally, a subset of helper T
cells abundantly produces IL-22, suggesting it plays a significant
role in skin homeostasis and pathology (11). IL-22 receptor is a heterodimeric
receptor of class II cytokine consisting of two chains, IL-22R1 and
IL-10R2. IL-22R1 is expressed in non-immune tissues, including the
skin, lungs, small intestine, liver, colon, kidneys, and pancreas
(12), unlike IL-10R2, which is
ubiquitously expressed in various organs and cells. Since IL-22
does not act between immune cells, but rather from immune cells to
the non-immune cell compartment, IL-22 appears to be unique among
cytokines.
Although a few studies have so far addressed the
roles of IL-22 in malignant cell proliferation and apoptosis, there
are inconsistencies in the findings. IL-22 induces the activation
of the major MAPK pathways in hepatoma cells (13), and increases the expression of many
anti-apoptotic and mitogenic proteins following the activation of
STAT3 (14). IL-22 can accelerate
inducible nitric-oxide synthase expression in human colon
adenocarcinoma cells (15). IL-22
protects human lung non-small cell carcinoma cells against
chemotherapy via the activation of anti-apoptotic proteins
(16). Conversely, IL-22 treatment
induces the cell cycle arrest of murine breast adenocarcinoma EMT6
cells through the inhibition of ERK1/2 and AKT phosphorylation
(17). The survival of mice with
IL-22-expressing Colon 26 cells significantly increased in
comparison to the control mice, suggesting that IL-22 might play a
protective role in hosts with tumors (18). Although IL-22 appears to act
variously in different carcinoma cells, there is little knowledge
on the potential roles of IL-22 in OSCCs.
This study analyzed the signal transduction and
genes induced in OSCC cells, to comprehensively evaluate the
potential biological activity of IL-22 in OSCC. Additionally, the
cell differentiation of OSCC cells by IL-22 was examined.
Materials and methods
Reagents
Recombinant human IL-6, IL-22 and TNF-α (Wako,
Osaka, Japan), and mouse EGF (Sigma-Aldrich, St. Louis, MO, USA)
were used for the study. Antibodies reactive to total STAT3,
phospho-STAT3 (pY705, pS727), ERK1, phospho-ERK1/2 (pT202/pY204),
p38α and phospho-p38 (pT180/pY182) were purchased from BD
Biosciences (Franklin Lakes, NJ, USA). The antibodies for total
STAT3 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and
IL-22R (Novus Biologicals, Littleton, CO, USA) were used for the
immunocytochemical and immunohistochemical studies,
respectively.
Samples and immunohistochemistry
Samples of primary OSCC and metastatic OSCC in the
cervical lymph node diagnosed in the Department of Oral and
Maxillofacial Surgery, Kyushu University Hospital in 2011 were
immunostained in this study. This study was approved by the local
research ethics committee.
Immunohistochemical staining was performed on 5
μm paraffin sections. The endogenous peroxide activity was
eliminated by treatment with 3% hydrogen peroxide in methanol for
20 min. Non-specific protein binding was blocked with 10% goat
serum for 20 min, and then the sections were reacted with the
primary antibody at 4°C overnight. The sections were incubated with
the Fab’ fragment of the secondary antibody conjugated with a
peroxidase-labeled amino acid polymer (Histofine Simple Stain MAX
PO, Nichirei, Japan) for 30 min at room temperature. After washing
with PBS, the immunoreactivity was visualized with a solution of 3,
3′-diaminobenzidine and <0.1% hydrogen peroxide (Nichirei).
Subsequently, the sections were counterstained with hematoxylin.
For the negative control, PBS was substituted for the primary
antibody.
Cell lines and culture conditions
Human OSCC cell lines, MISK81-5 (19), HSC-3, HSC-4 (Japanese Cancer
Research Resources Bank), SAS (20), and SQUU-B (21), a human keratinocyte cell line,
HaCaT, and a human erythroleukemia cell line, K562, were used.
MISK81-5, HSC-3, HSC-4 cells and K562 cells were grown in α-MEM
(Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum
(Filtron, Brooklyn, Australia). SAS and SQUU-B cells were incubated
in DMEM/F-12 (Invitrogen) with 10% serum. HaCaT cells were
maintained in DMEM (Invitrogen) with 10% serum.
Semiquantitative RT-PCR and real-time
quantitative PCR analyses
The total RNAs were isolated using the SV Total RNA
Isolation System (Promega, Madison, WI, USA), and cDNAs were
generated from isolated total RNA with the SuperScript VILO cDNA
Synthesis Kit (Invitrogen). Semiquantitative PCR was amplified with
Advantage 2 (Clontech, Mountain View, CA, USA). Real-time
quantitative PCR was performed using a Thermal Cycler
Dice® Real Time System with SYBR® Premix Ex
Taq™ II (Takara, Shiga, Japan).
A reference gene was determined among the various
housekeeping genes (Table I). The
relative expression level of each targeted gene was normalized
using the ΔΔCT comparative method, based on the
reference gene threshold cycle (CT) values (22).
 | Table I.Primer sets used in the present
study. |
Table I.
Primer sets used in the present
study.
| Target | | Sequence |
|---|
| For
semiquantitative RT-PCR | | |
| IL-22R1 | sense | 5′-CTC CAC AGC GGC
ATA GCC T-3′ |
| antisense | 5′-ACA TGC AGC TTC
CAG CTG G-3′ |
| IL-10R2 | sense | 5′-GGC TGA ATT TGC
AGA TGA GCA-3′ |
| antisense | 5′-GAA GAC CGA GGC
CAT GAG G-3′ |
| β-actin | sense | 5′-ATC TGG CAC CAC
ACC TTC TAC AAT GAG CTG CG-3′ |
| antisense | 5′-CGT CAT ACT CCT
GCT TGC TGA TCC ACA TCT GC-3′ |
| For quantitative
RT-PCR | | |
| Bcl-xL | sense | 5′-TAG GGT GGC CCT
TGC AGT TC-3′ |
| antisense | 5′-GTG AGG CAG CTG
AGG CCA TAA-3′ |
| Survivin | sense | 5′-TTC TCA AGG ACC
ACC GCA TC-3′ |
| antisense | 5′-GCC AAG TCT GGC
TCG TTC TC-3′ |
| c-Myc | sense | 5′-CGG ATT CTC TGC
TCT CCT CGA C-3′ |
| antisense | 5′-CCT CCA GCA GAA
GGT GAT CCA-3′ |
| Cyclin D1 | sense | 5′-GTG CAT CTA CAC
CGA CAA CTC CA-3′ |
| antisense | 5′-TGA GCT TGT TCA
CCA GGA GCA-3′ |
| SOCS3 | sense | 5′-CCC AAG GAC GGA
GAC TTC GAT-3′ |
| antisense | 5′-GAA ACT TGC TGT
GGG TGA CCA T-3′ |
| TFRC | sense | 5′-GCG AGC ACT GAC
CAG ATA AGA ATG-3′ |
| antisense | 5′-TCC CGA TAA TGT
GTT AGG ATT GTG A-3′ |
| β-actin | sense | 5′-TGG CAC CCA GCA
CAA TGA A-3′ |
| antisense | 5′-CTA AGT CAT AGT
CCG CCT AGA AGC A-3′ |
| GAPDH | sense | 5′-GCA CCG TCA AGG
CTG AGA AC-3′ |
| antisense | 5′-TGG TGA AGA CGC
CAG TGG A-3′ |
| B2M | sense | 5′-CGG GCA TTC CTG
AAG CTG A-3′ |
| antisense | 5′-GGA TGG ATG AAA
CCC AGA CAC ATA G-3′ |
| Loricrin | sense | 5′-TCA TGA TGC TAC
CCG AGG TTT G-3′ |
| antisense | 5′-TGC AAA TTT ATT
GAC TGA GGC ACT G-3′ |
| Involucrin | sense | 5′-TAA CCA CCC GCA
GTG TCC AG-3′ |
| antisense | 5′-ACA GAT GAG ACG
GGC CAC CTA-3′ |
| Keratin 1 | sense | 5′-AGA TCA CTG CTG
GCA GAC ATG G-3′ |
| antisense | 5′-TGA TGG ACT GCT
GCA AGT TGG-3′ |
| Keratin 5 | sense | 5′-GAT AGC ATC ATC
GCT GAG GTC AAG-3′ |
| antisense | 5′-AGC CTC TGG ATC
ATC CGG TTC-3′ |
| Keratin 10 | sense | 5′-AGG CTG GCA GCT
GAT GAC TTC-3′ |
| antisense | 5′-CAG GGT CAG CTC
ATC CAG CA-3′ |
| Keratin 14 | sense | 5′-ACT TCA AGA CCA
TTG AGG ACC TGA G-3′ |
| antisense | 5′-CAG GGT CAG TTC
GTC CAG CA-3′ |
| SERPINB3 | sense | 5′-GGC AGC AAT ACC
ACA TTG GTT C-3′ |
| antisense | 5′-TGT ATT GCC TCA
TCA TCT GTA TGG A-3′ |
| SERPINB4 | sense | 5′-GGG ACT ATT GGC
AAT GAT ACG ACA C-3′ |
| antisense | 5′-AGG ACC TTG GCC
TGT ACA TCC TC-3′ |
The mRNA expression of the STAT3 downstream genes,
keratinocyte differentiation-related genes and SERPINB3/4 (Squamous
Cell Carcinoma Antigen, SCCA1/2) genes, well-known SCC markers,
were examined in OSCC cells after IL-22 stimulation (Table I). The specificity of the PCR
products was determined using a melting curve and/or gel
electrophoresis.
Immunoblotting
Proteins were separated by 12% SDS-polyacrylamide
gel electrophoresis, and transferred to an Immun-Blot®
PVDF Membrane (Bio-Rad, Hercules, CA, USA). Antibodies bound to
proteins were visualized by the ECL plus detection system
(Amersham, Piscataway, NJ, USA). The protein concentration was
estimated using a Micro BCA Protein Assay Kit (Pierce
Biotechnology, Inc., IL, USA).
Immunocytochemistry for STAT3
Following incubation with the primary antibody, the
cells were incubated in Alexa Fluor® 568 goat
anti-rabbit IgG or 594 rabbit anti-mouse IgG (Invitrogen). The
nuclei were counterstained with DAPI (Dojindo, Kumamoto,
Japan).
Cell proliferation assay
The proliferation of IL-22-treated cells was
quantified using the CellTiter-Glo® Luminescent Cell
Viability Assay (Promega) and a Microplate Luminometer (Turner
Biosystems, Sunnyvale, CA, USA). The cells were stimulated with 20
ng/ml of IL-22 every 24 h during the 48 h culture period.
Construction of an NF-κB-responsive
Luciferase Reporter Vector and the luciferase assay
Four tandem copies of the NF-κB consensus sequence
were inserted upstream of the minimal promoter (minP) in pGL4.26
[luc2/minP/Hygro] (Promega). After clonal selection of
stably transfected MISK81-5 cells with hygromycin, MISK-pGL4-NF-κB
cells were generated. Luminescence was measured using the One-Glo
luciferase system (Promega) and the Microplate Luminometer.
Transient transfection of siRNA for
STAT3
siRNAs for human STAT3 (GenBank Accession Number:
NM_003150) and GAPDH, and a siRNA universal negative control
(Sigma-Aldrich) were used as a target and positive and negative
controls, respectively. The cells were transfected with siRNA (10
nM) using the Lipofectamine RNAiMAX (Invitrogen).
Statistical analysis
All experiments were independently repeated at least
three times. Statistical analysis was performed using the one-way
ANOVA with the Tukey-Kramer comparison test, Dunnett’s test or
Student’s t-test. A p-value <0.05 or <0.01 was considered to
indicate statistically significant differences.
Results
Human oral squamous cell carcinoma cell
lines express IL-22 receptor chains
First, we immunohistochemically examined the IL-22R
expression in OSCC. The immunostaining revealed that the intensity
increased in the OSCC cells, although weak IL-22R signals were also
observed throughout the normal oral mucosa (Fig. 1A). Significant staining was also
observed in the metastatic carcinoma cells present in the cervical
lymph node (Fig. 1B and C).
IL-22R1 and IL-10R2 were both detectable in all
tested OSCC cells (Fig. 1D),
although their expression intensity varied. HaCaT cells served as a
positive control (23). K562 cells
were analyzed as a negative control for IL-22R1. The mRNA
expression of IL-22R1 and IL-10R2 were also detectable in all the
OSCC cell lines under serum-free conditions (data not shown).
MISK81-5 squamous cell carcinoma cells
are responsive to IL-22
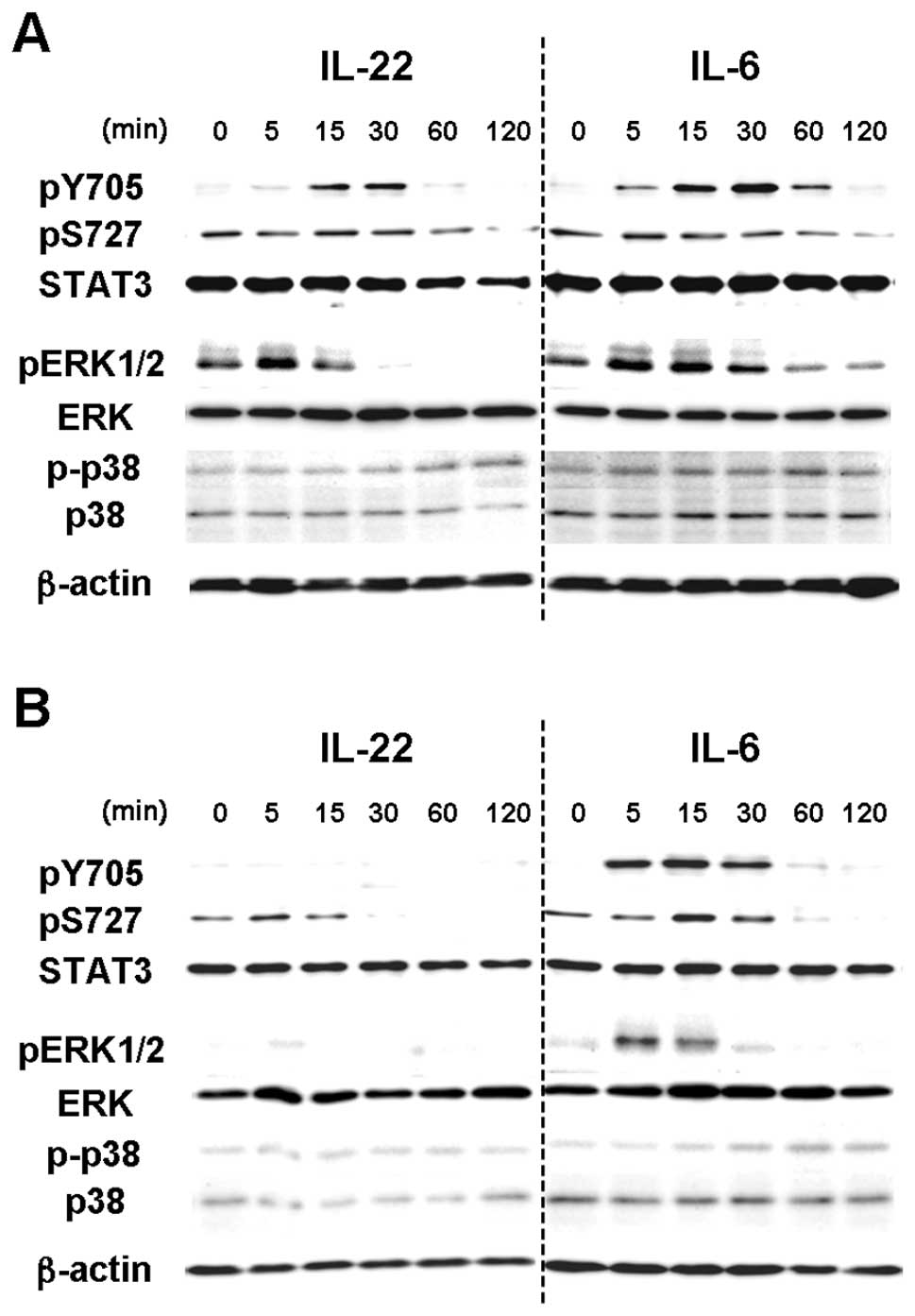
IL-22 induced the tyrosine phosphorylation of STAT3
(pY705-STAT3) in MISK81-5 cells within 15 min, peaking at 30 min
(Fig. 1B), as seen in other cell
lines by IL-22 (13,17,24–26).
This phosphorylation was transient, and decreased toward the
baseline until reaching barely detectable levels after 120 min. The
change in the serine phosphorylation of STAT3 (pS727-STAT3) in
MISK81-5 cells treated with IL-22 was subtle within the tested
periods. At the same time, pY705-STAT3 increased within 5 min and
still remained detectable in MISK81-5 cells at least 1 h after IL-6
stimulation. IL-6 treatment led to a subtle change in pS727-STAT3
within the tested periods. Conversely, IL-6 had a similar effect on
pY705-STAT3 in HSC-3 cells, but the activation of pY705-STAT3 by
IL-22 was not detectable during the tested periods (Fig. 2A).
IL-22 induced the phosphorylation of ERK1/2 in
MISK81-5 cells within 5 min, but the level slightly decreased at 15
min (Fig. 2B). This
phosphorylation decreased to below control levels after 30 min.
IL-22 also induced a delayed phosphorylation of p38 MAP kinase
after 60 min. Although the peak of pERK1/2 was noted at 15 min,
similar results were obtained in MISK81-5 cells treated with IL-6.
The activation of ERK1/2 and p38 MAP kinases was undetectable in
HSC-3 cells after IL-22 treatment (Fig. 2A). IL-6 showed similar activation
of ERK1/2 and p38 MAP kinases to that in MISK81-5 cells treated
with IL-22 or IL-6.
IL-22 induces the translocation of pSTAT3
into the nucleus of MISK81-5 cells
STAT3 expression was noted in both the nucleus and
cytoplasm of MISK81-5 cells before IL-6 stimulation, and was
observed in the nucleus of many MISK81-5 cells within 5 min after
IL-6 stimulation. STAT3 was again detectable in the cytoplasm after
30 min (Fig. 3A). The increased
signal for pSTAT3 in the nucleus of MISK81-5 cells was observed at
30 min after IL-22 treatment (Fig.
3B), whereas no nuclear translocation of pSTAT3 was detected in
HSC-3 cells treated with IL-22 (Fig.
3C).
IL-22 promotes the expression of
anti-apoptotic and mitogenic genes in MISK81-5 cells
Since cytokine stimulation can induce instability in
the housekeeping gene expression (27–29),
B2M was selected as the internal control among the various
housekeeping genes tested using the geNorm system (http://medgen.ugent.be/~jvdesomp/genorm).
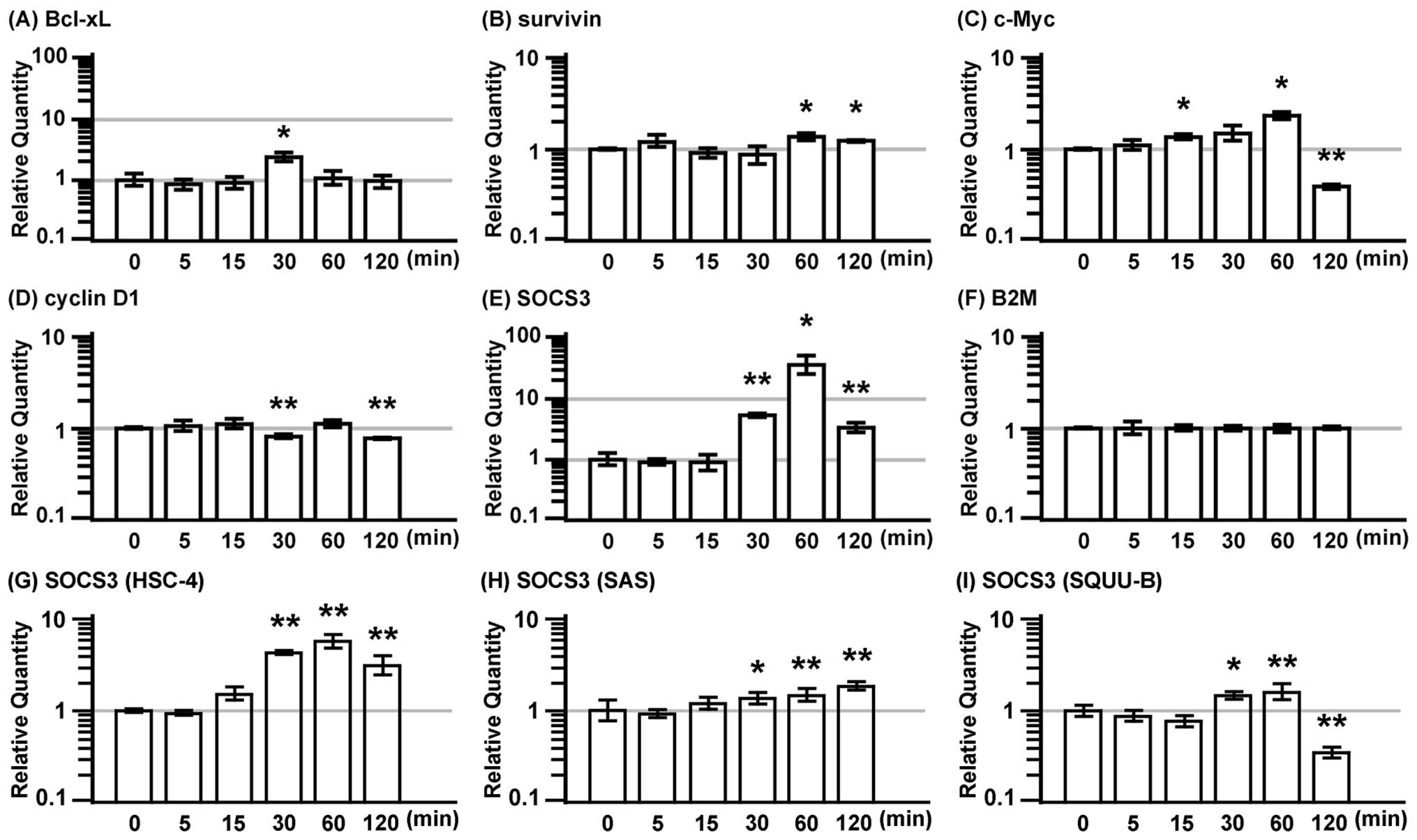
The expression of anti-apoptotic proteins, Bcl-xL
and survivin, and the mitogenic proteins, c-Myc and cyclin D1, and
the suppressor for STAT3, the SOCS3 gene, were examined in MISK81-5
cells treated with IL-22 (Fig. 4).
The expression of Bcl-xL and c-Myc genes exhibited a 2.4-fold
increase, and peaked at 30 and 60 min after IL-22 stimulation,
respectively (Fig. 4A and C).
However, the c-Myc gene expression dramatically decreased to 40% of
the basal level at 120 min. SOCS3 expression was markedly induced
at 30 min after IL-22 stimulation, it exhibited a 37-fold increase
at 60 min and subsequently decreased at 120 min (Fig. 4E). IL-22 significantly increased
the gene expression of survivin at 60 and 120 min. The expression
of cyclin D1 significantly decreased at 30 and 120 min (Fig. 4B and D).
SOCS3 expression was markedly induced at 30 min in
the HSC-4 cells after IL-22 stimulation, it peaked at 60 min, and
subsequently decreased at 120 min (Fig. 4G). Similar results were observed
for IL-22 stimulation in the SQUU-B cells. However, its expression
dramatically decreased to less than the baseline level at 120 min
(Fig. 4I). The SOCS3 expression in
the SAS cells treated with IL-22 was gradually increased from 30
min to 120 min (Fig. 4H).
IL-22 slightly induces tumor cell
proliferation in vitro and the cellular NF-κB activation
status
The effect of IL-22 on the proliferation of
MISK81-5, HSC-4, SAS and SQUU-B cells in vitro was examined.
MISK81-5 and SQUU-B cells treated with IL-22 showed 1.3- and
1.1-fold increase in viability compared with control samples,
respectively. A significant difference was demonstrated between the
IL-22-treated cells and controls (p<0.01). Although HSC-4 and
SAS cells were subtly increased by IL-22, there was no significant
difference in the viability of these cells between the
IL-22-treated cells and controls (Fig.
5A).
MISK-pGL4-NF-κB cells stimulated with 50 and 100
ng/ml TNF-α demonstrated significant 3.6-fold and 3.8-fold
increases in luciferase activity, respectively, compared with the
unstimulated cells (Fig. 5B).
However, the effects of IL-22 and IL-6 were subtle or negligible.
No significant difference was noted between the stimulated and
control samples (Fig. 5B).
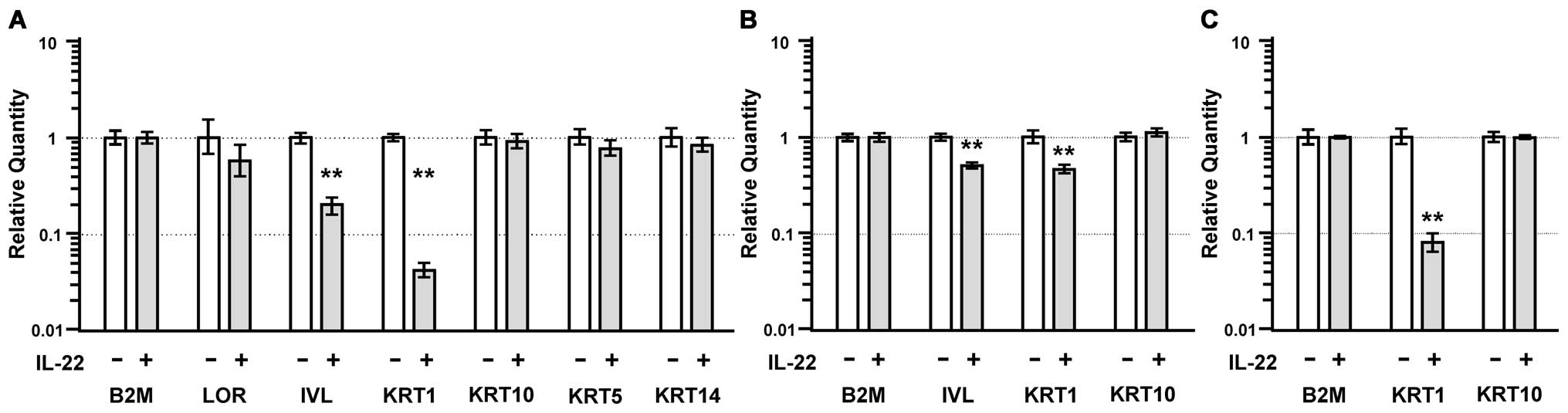
IL-22 reduces the expression of
keratinocyte differentiation-related genes
The expression of the involucrin (IVL) and keratin 1
(KRT1) genes significantly decreased to ∼20% and ∼5% of control
levels by IL-22 treatment, respectively (p<0.01; Fig. 6A). In addition, the expression of
these genes in HSC-4 cells significantly decreased to ∼50% after
IL-22 treatment (p<0.01) (Fig.
6B). The KRT1 expression in SAS cells significantly decreased
to ∼10% after IL-22 treatment (p<0.01) (Fig. 6C). The expression of keratin 10
(KRT10) was unchanged in the MISK81-5 (Fig. 6A), HSC-4 (Fig. 6B), SAS (Fig. 6C) and SQUU-B cells treated with
IL-22.
To examine whether IL-22 induces a reduction of the
KRT1 expression through STAT3, we used siRNA to selectively reduce
the STAT3 expression. STAT3 siRNA induced a significant
downregulation of the STAT3 mRNA and protein levels (Fig. 7A and B), and inhibited the
downregulation of KRT1 expression by IL-22 (Fig. 7C). Similarly, the transfection of
the SAS cells with a siRNA for STAT partially inhibited the
downregulation of the KRT1 expression by IL-22 (Fig. 7D). However, neither ERK nor pERK1/2
was affected by the STAT3 siRNA treatment (Fig. 7E).
 | Figure 7.STAT3 siRNA inhibits IL-22-induced
reduction of KRT1 expression, but it has little impact on pERK. (A)
siRNA selectively reduced the gene expression in the MISK81-5 cells
at 30 h after siRNA transfection. Significant differences in the
gene expression are indicated by double asterisks
(**p<0.01). B2M was used as a reference gene. (B) An
immunoblot analysis also revealed that GAPDH and STAT3 siRNAs cause
a depletion of the GAPDH and STAT3 protein levels in the MISK81-5
cells, respectively. (C) At 30 h before IL-22 stimulation, the
MISK81-5 cells were transfected with siRNA. The expression of B2M,
GAPDH, STAT3 and KRT1 was compared between MISK81-5 cells after
IL-22 stimulation for 24 h and unstimulated cells
(*p<0.05; **p<0.01). (D) The
downregulation of KRT1 expression by IL-22 was inhibited in the SAS
cells transfected with a siRNA for STAT3 and in unstimulated cells.
Significant differences in the gene expression are indicated by
single or double asterisks (*p<0.05;
**p<0.01). (E) At 30 h after siRNA transfection, the
MISK81-5 cells were treated with IL-22 for 10 min. Total cell
lysates (10 μg/sample) were analyzed by immunoblotting with
an antibody against pY705-STAT3. The membrane was repeatedly
reprobed and immunoblotted with an anti-pERK1/2, anti-total STAT3,
or an anti-ERK antibody and then with an anti-β-actin antibody.
w/o, sample without siRNA treatment; C, sample treated with
negative control siRNA; P, sample treated with GAPDH siRNA as a
positive control; S, sample treated with STAT3 siRNA. |
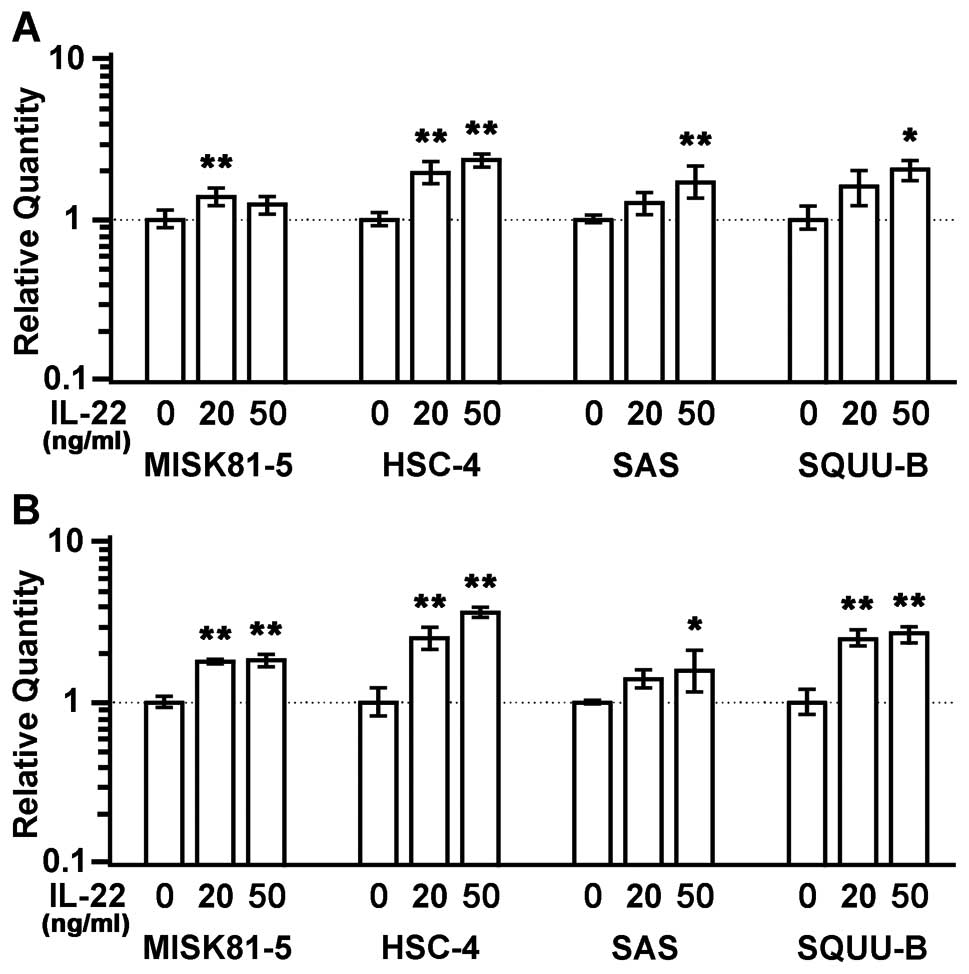
IL-22 upregulates the expression of
SERPINB3/4 (SCCA1/2) genes
Squamous cell carcinoma antigen (SCCA) 1 was
originally identified in squamous cell carcinoma (SCC) of the
uterine cervix (30). An elevated
expression of SCCA1 and its isoform, SCCA2, has been used as a
biomarker for aggressive SCC in the cervix, lung, head and neck
(31–33). SCCA belongs to the serine protease
inhibitor (Serpin) family of proteins, and SCCA1 and SCCA2 are
called SERPNB3 and SERPINB4, respectively. The SERPINB3 expression
showed respective 1.2-, 2.4-. 1.7- and 2.0-fold increases in the
MISK81-5, HSC-4, SAS and SQUU-B cells treated with IL-22 (50 ng/ml)
compared with control samples. The SERPINB4 expression also showed
1.8-, 3.6-. 1.6- and 3.0-fold increases, respectively, in the
MISK81-5, HSC-4, SAS and SQUU-B cells treated with IL-22 (50 ng/ml)
compared with control samples. A significant difference in the
expression levels of these genes was noted between all of the
IL-22-treated (50 ng/ml) cells and control cells, except for the
SERPINB3 expression in MISK81-5 cells (p<0.01 or p<0.05)
(Fig. 8).
Discussion
Immunostaining for IL-22R revealed that the
intensity was increased in the primary and metastatic OSCC cells.
IL-22 induced the transient phosphorylation of STAT3 and led to its
translocation into the nucleus. IL-22 activated the ERK and p38
MAPK pathways, but did not have a significant effect on NF-κB.
IL-22 mildly affected the proliferation of OSCC cells and
downregulated the expression of keratinocyte
differentiation-related genes. STAT3 siRNA inhibited the
IL-22-mediated downregulation of the keratinocyte
differentiation-related genes, but did not affect the activation of
the ERK pathway. The expression of the SERPINB3/4 genes in OSCC
cells was upregulated by IL-22 stimulation, thus suggesting that
IL-22 plays a key role in the biology of OSCC cells.
Immunohistochemical staining showed that IL-22R was
expressed in OSCC. The expression of both IL-22 receptor chains was
confirmed in MISK81-5, HSC-3, HSC-4, SAS and SQUU-B OSCC cell lines
by RT-PCR. In the immunoblotting analysis, MISK81-5 cells showed
the transient phosphorylation of STAT3 at Y705 by IL-22
stimulation. Similar results were reported for IL-22 stimulation in
other types of cells (13,23). In the immunocytochemistry
experiments, a transient translocation of STAT3 into the nucleus
was observed in MISK81-5 cells. When pY705-STAT3 decreased, STAT3
was again detected in the cytoplasm, similar to unstimulated cells.
These results suggest that pY705 mediated the translocation of
pSTAT3. This finding was supported by the study of Zhong et
al(34), in which the
phosphorylation of STAT3 at Y705 was shown to lead to the
translocation of STAT3 into the nucleus, thereby activating the
transcription of multiple target genes. Conversely, the change in
pS727-STAT3 was subtle in this study. The phosphorylation of STAT3
at S727 in OSCC cells was different from that in the study of
Lejeune et al(13) who
showed transient increases in pS727-STAT3 in hepatoma cells after
treatment with IL-22. Although pS727 is thought to play a
regulatory role in STAT3 activation, resulting in its maximal
transcriptional activity (35),
the function of pS727 remains unclear in this study. In addition,
STAT3 phosphorylation was not observed in HSC-3 cells following
IL-22 stimulation. This result indicates that the IL-22 receptors
were functional in MISK81-5 cells, but that not all squamous cell
carcinomas activate STAT3 signaling after exposure to IL-22.
The activity of MAP kinases such as ERK and p38
after IL-22 stimulation in this study (Fig. 2), is partly reminiscent of that in
hepatoma cells observed in other studies (13,14).
After IL-22 stimulation, ERK activation preceded that of STAT3. The
phosphorylation of ERK1/2 induced by IL-22 stimulation was not
affected by STAT3 siRNA (Fig. 7).
These results suggest that other STAT3-independent mechanisms are
acting on MISK81-5 cells under IL-22 stimulation. While IL-22
transiently activated ERK1/2 and induced a delayed phosphorylation
of p38 MAP kinase, ERK1/2 phosphorylation decreased to less than
the control level after 30 min. A similar result was seen in the
IL-22 treatment of murine breast adenocarcinoma EMT6, in which
ERK1/2 phosphorylation was inhibited by IL-22, thus leading to cell
cycle arrest (17). Additionally,
the transient activation of STAT3 also involved the transient
upregulation of SOCS3 expression in OSCC cells. The transient
upregulation of SOCS3 expression may affect the transient
activation of STAT3 and STAT3-associated factors in OSCC cells, as
SOCS3 acts as a suppressor of STAT signaling, while SOCS3 is one of
the downstream genes of STAT3. IL-22 may constitutively contribute
to the activation of STAT3 and the expression of anti-apoptotic and
mitogenic genes in OSCC cells under the suppression of SOCS3, since
SOCS3 causes growth inhibition in SCC cell lines (25,36).
Indeed, IL-22 mildly stimulated the cell proliferation of MISK81-5
and SQUU-B cells in this study. The proliferation of HSC-4 and SAS
cells was limited after IL-22 stimulation. This stimulation may be
due to a complicated synergistic effect among the transiently
increased activity of ERK1/2 and the expression of c-Myc and cyclin
D1 genes, the inhibition of ERK1/2 phosphorylation, and/or SOCS3
expression.
Keratinocytes are thought to show changes in their
expression and synthesis of cytoskeletal proteins after exposure to
proliferative or inflammatory cytokines (37). IVL, LOR, KRT1 and KRT10 are the
characteristic markers of normal suprabasal keratinocytes (38). IL-22 significantly reduced the
expression of the IVL and/or KRT1 genes in MISK81-5, HSC-4 and SAS
cells. Our results indicated that IL-22 could thus play a role in
regulating the terminal differentiation of OSCC cells through STAT3
activation similar to the effects in keratinocytes. Since these
factors play important roles during the terminal differentiation of
keratinocytes and are associated with apoptotic processes (39–42),
the control of the IL-22 function in OSCCs may therefore make it
possible to induce apoptosis in OSCC cells via differentiation.
In this study, IL-22 induced the upregulation of
SERPINB3 and SERPINB4 expression in OSCC cells. The downregulation
of SERPINB3 by an antisense method significantly increased the
cellular susceptibility to drug-induced apoptosis (43). Our previous study showed that
SERPINB3/B4 contributed, at least in part, to preventing TNF-α
induced cell death by impeding the cytochrome c release from
the mitochondria (44). Ahmed
et al(45) demonstrated
that squamous carcinoma cells promote cell survival through
activation of SERPINB3/B4 genes by activated STAT3. Thus, IL-22 may
play a role in the attenuation of drug-induced apoptosis by the
increasing the expression of SERPINB3/B4 in cancer cells.
Our present study shows that IL-22 affects several
important functions of OSCC cells via the STAT3-dependent and/or
-independent pathways, suggesting that IL-22 may play a role in
carcinoma cell differentiation and the upregulation of SERPINB3/B4,
well-known biomarkers for SCC. However, the response against IL-22
varies in OSCC cell lines. Further studies are required to
elucidate the mechanisms by which IL-22 is involved in the biology
of OSCC carcinogenesis. Elucidating the functions of IL-22 could
lead to the development of new perspectives on this disease, and
potentially new therapies with few side-effects, thereby improving
the treatment of patients with OSCC.
Acknowledgements
The present study was funded in part
by Grant-in-Aid from the Ministry of Education, Culture, Sports,
Science and Technology of Japan, #20390466, #23659880 (to H.S.) and
#80117077 (to S.O.).
References
|
1.
|
MM ChidzongaL MahomvaSquamous cell
carcinoma of the oral cavity, maxillary antrum and lip in a
Zimbabwean population: a descriptive epidemiological studyOral
Oncol42184189200610.1016/j.oraloncology.2005.07.01116256417
|
|
2.
|
MM ChidzongaOral malignant neoplasia: a
survey of 428 cases in two Zimbabwean hospitalsOral
Oncol42177183200610.1016/j.oraloncology.2005.07.00316256412
|
|
3.
|
IW DimeryWK HongOverview of combined
modality therapies for head and neck cancerJ Natl Cancer
Inst8595111199310.1093/jnci/85.2.958418313
|
|
4.
|
SP SchantzGP YuHead and neck cancer
incidence trends in young Americans, 1973–1997, with a special
analysis for tongue cancerArch Otolaryngol Head Neck
Surg128268274200211886342
|
|
5.
|
CD LlewellynK LinklaterJ BellNW JohnsonKA
WarnakulasuriyaSquamous cell carcinoma of the oral cavity in
patients aged 45 years and under: a descriptive analysis of 116
cases diagnosed in the South East of England from 1990 to 1997Oral
Oncol39106114200312509963
|
|
6.
|
ME DudleyJR WunderlichPF RobbinsJC YangP
HwuDJ SchwartzentruberSL TopalianR SherryNP RestifoAM HubickiCancer
regression and autoimmunity in patients after clonal repopulation
with antitumor
lymphocytesScience298850854200210.1126/science.107651412242449
|
|
7.
|
S NégrierB EscudierF GomezJY DouillardA
RavaudC ChevreauM BuclonD PérolC LassetPrognostic factors of
survival and rapid progression in 782 patients with metastatic
renal carcinomas treated by cytokines: a report from the Groupe
Français d’ImmunothérapieAnn Oncol1314601468200212196373
|
|
8.
|
SA RosenbergProgress in human tumour
immunology and
immunotherapyNature411380384200110.1038/3507724611357146
|
|
9.
|
K BonifaceE GuignouardN PedrettiM GarciaA
DelwailFX BernardF NauG GuilletG DagregorioH YsselA role for T
cell-derived interleukin 22 in psoriatic skin inflammationClin Exp
Immunol150407415200710.1111/j.1365-2249.2007.03511.x17900301
|
|
10.
|
K WolkR SabatInterleukin-22: a novel T-
and NK-cell derived cytokine that regulates the biology of tissue
cellsCytokine Growth Factor
Rev17367380200610.1016/j.cytogfr.2006.09.00117030002
|
|
11.
|
S TrifariCD KaplanEH TranNK CrellinH
SpitsIdentification of a human helper T cell population that has
abundant production of interleukin 22 and is distinct from T(H)-17,
T(H)1 and T(H)2 cellsNat
Immunol10864871200910.1038/ni.177019578368
|
|
12.
|
K WolkS KunzE WitteM FriedrichK AsadullahR
SabatIL-22 increases the innate immunity of
tissuesImmunity21241254200410.1016/j.immuni.2004.07.00715308104
|
|
13.
|
D LejeuneL DumoutierS ConstantinescuW
KruijerJJ SchuringaJC RenauldInterleukin-22 (IL-22) activates the
JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma
cell line. Pathways that are shared with and distinct from IL-10J
Biol Chem2773367633682200210.1074/jbc.M20420420012087100
|
|
14.
|
S RadaevaR SunHN PanF HongB GaoInterleukin
22 (IL-22) plays a protective role in T cell-mediated murine
hepatitis: IL-22 is a survival factor for hepatocytes via STAT3
activationHepatology3913321342200410.1002/hep.2018415122762
|
|
15.
|
E ZieschéM BachmannH KleinertJ
PfeilschifterH MühlThe interleukin-22/STAT3 pathway potentiates
expression of inducible nitric-oxide synthase in human colon
carcinoma cellsJ Biol Chem2821600616015200717438334
|
|
16.
|
W ZhangY ChenH WeiC ZhengR SunJ ZhangZ
TianAntiapoptotic activity of autocrine interleukin-22 and
therapeutic effects of interleukin-22-small interfering RNA on
human lung cancer xenograftsClin Cancer
Res1464326439200810.1158/1078-0432.CCR-07-440118927282
|
|
17.
|
GF WeberFC GaertnerW ErlKP JanssenB
BlechertB HolzmannH WeighardtM EsslerIL-22-mediated tumor growth
reduction correlates with inhibition of ERK1/2 and AKT
phosphorylation and induction of cell cycle arrest in the G2-M
phaseJ
Immunol17782668272200610.4049/jimmunol.177.11.826617114505
|
|
18.
|
H NagakawaO ShimozatoL YuY TakiguchiK
TatsumiT KuriyamaM TagawaExpression of interleukin-22 in murine
carcinoma cells did not influence tumour growth in vivo but did
improve survival of the inoculated hostsScand J
Immunol60449454200410.1111/j.0300-9475.2004.01504.x15541036
|
|
19.
|
K MatsuoY IshibashiI KobayashiS OzekiM
OhishiT TangeJ HirataT KiyoshimaH SakaiNew human oral squamous
carcinoma cell line and its tumorigenic subline producing
granulocyte colony-stimulating factorJpn J Cancer
Res8512571262199410.1111/j.1349-7006.1994.tb02938.x7531680
|
|
20.
|
K TakahashiH KanazawaY AkiyamaS TasakiM
TakaharaT MutoH TanzawaK SatoEstablishment and characterization of
a cell line (SAS) from poorly differentiated human squamous cell
carcinoma of the tongueJ Jpn Stomatol Soc3820281989
|
|
21.
|
M MorifujiS TaniguchiH SakaiY NakabeppuM
OhishiDifferential expression of cytokeratin after orthotopic
implantation of newly established human tongue cancer cell lines of
defined metastatic abilityAm J
Pathol15613171326200010.1016/S0002-9440(10)65002-X
|
|
22.
|
JS YuanA ReedF ChenCN Stewart
JrStatistical analysis of real-time PCR dataBMC
Bioinformatics785200610.1186/1471-2105-7-8516504059
|
|
23.
|
K WolkE WitteE WallaceWD DockeS KunzK
AsadullahHD VolkW SterryR SabatIL-22 regulates the expression of
genes responsible for antimicrobial defense, cellular
differentiation, and mobility in keratinocytes: a potential role in
psoriasisEur J
Immunol3613091323200610.1002/eji.20053550316619290
|
|
24.
|
K BonifaceFX BernardM GarciaAL GurneyJC
LecronF MorelIL-22 inhibits epidermal differentiation and induces
proinflammatory gene expression and migration of human
keratinocytesJ
Immunol17436953702200510.4049/jimmunol.174.6.369515749908
|
|
25.
|
TL LeeJ YehC Van WaesZ ChenEpigenetic
modification of SOCS-1 differentially regulates STAT3 activation in
response to interleukin-6 receptor and epidermal growth factor
receptor signaling through JAK and/or MEK in head and neck squamous
cell carcinomasMol Cancer
Ther5819200610.1158/1535-7163.MCT-05-0069
|
|
26.
|
LH WeiML KuoCA ChenCH ChouWF ChengMC
ChangJL SuCY HsiehThe anti-apoptotic role of interleukin-6 in human
cervical cancer is mediated by up-regulation of Mcl-1 through a
PI3-K/Akt
pathwayOncogene2057995809200110.1038/sj.onc.120473311593385
|
|
27.
|
H ZhongJW SimonsDirect comparison of
GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards
for quantifying RNA levels under hypoxiaBiochem Biophys Res
Commun259523526199910.1006/bbrc.1999.081510364451
|
|
28.
|
K HagiharaT NishikawaT IsobeJ SongY
SugamataK YoshizakiIL-6 plays a critical role in the synergistic
induction of human serum amyloid A (SAA) gene when stimulated with
proinflammatory cytokines as analyzed with an SAA isoform real-time
quantitative RT-PCR assay systemBiochem Biophys Res
Commun314363369200410.1016/j.bbrc.2003.12.096
|
|
29.
|
J VandesompeleK De PreterF PattynB PoppeN
Van RoyA De PaepeF SpelemanAccurate normalization of real-time
quantitative RT-PCR data by geometric averaging of multiple
internal control genesGenome
Biol30034200210.1186/gb-2002-3-7-research003412184808
|
|
30.
|
H KatoT TorigoeRadioimmunoassay for tumor
antigen of human cervical squamous cell
carcinomaCancer4016211628197710.1002/1097-0142(197710)40:4%3C1621::AID-CNCR2820400435%3E3.0.CO;2-I332328
|
|
31.
|
JM DukHW de BruijnKH GroenierH HollemaKA
ten HoorCancer of the uterine cervix: sensitivity and specificity
of serum squamous cell carcinoma antigen determinationsGynecol
Oncol39186194199010.1016/0090-8258(90)90430-S2227594
|
|
32.
|
JM DukKH GroenierHW de BruijnH HollemaKA
ten HoorAG van der ZeeJG AaldersPretreatment serum squamous cell
carcinoma antigen: a newly identified prognostic factor in
early-stage cervical carcinomaJ Clin Oncol1411111819968558185
|
|
33.
|
R MolinaX FilellaJM AugéR FuentesI BoverJ
RifaV MorenoE CanalsN ViñolasA MarquezTumor markers (CEA, CA 125,
CYFRA 21-1, SCC and NSE) in patients with non-small cell lung
cancer as an aid in histological diagnosis and prognosis.
Comparison with the main clinical and pathological prognostic
factorsTumour Biol24209218200310.1159/000074432
|
|
34.
|
Z ZhongZ WenJE Darnell JrStat3: a STAT
family member activated by tyrosine phosphorylation in response to
epidermal growth factor and
interleukin-6Science2649598199410.1126/science.81404228140422
|
|
35.
|
JJ SchuringaH SchepersE VellengaW
KruijerSer727-dependent transcriptional activation by association
of p300 with STAT3 upon IL-6 stimulationFEBS
Lett4957176200110.1016/S0014-5793(01)02354-711322950
|
|
36.
|
A WeberUR HenggeW BardenheuerI TischoffF
SommererA MarkwarthA DietzC WittekindA TannapfelSOCS-3 is
frequently methylated in head and neck squamous cell carcinoma and
its precursor lesions and causes growth
inhibitionOncogene2466996708200510.1038/sj.onc.1208818
|
|
37.
|
M Hernández-QuinteroW Kuri-HarcuchA
González RoblesF Castro-MuñozledoInterleukin-6 promotes human
epidermal keratinocyte proliferation and keratin cytoskeleton
reorganization in cultureCell Tissue Res3257790200616550359
|
|
38.
|
R EichnerP BonitzTT SunClassification of
epidermal keratins according to their immunoreactivity, isoelectric
point, and mode of expressionJ Cell
Biol9813881396198410.1083/jcb.98.4.13886201491
|
|
39.
|
E FuchsEpidermal differentiation: the bare
essentialsJ Cell
Biol11128072814199010.1083/jcb.111.6.28072269655
|
|
40.
|
RM PorterS LeitgebDW MeltonO SwenssonRA
EadyTM MaginGene targeting at the mouse cytokeratin 10 locus:
severe skin fragility and changes of cytokeratin expression in the
epidermisJ Cell Biol132925936199610.1083/jcb.132.5.9258603923
|
|
41.
|
E FuchsK WeberIntermediate filaments:
structure, dynamics, function, and diseaseAnnu Rev
Biochem63345382199410.1146/annurev.bi.63.070194.0020217979242
|
|
42.
|
E FuchsKeratins and the skinAnnu Rev Cell
Dev Biol11123153199510.1146/annurev.cb.11.110195.001011
|
|
43.
|
Y SuminamiS NagashimaA MurakamiS NawataT
GondoH HirakawaF NumaGA SilvermanH KatoSuppression of a squamous
cell carcinoma (SCC)-related serpin, SCC antigen, inhibits tumor
growth with increased intratumor infiltration of natural killer
cellsCancer Res6117761780200111280721
|
|
44.
|
K HashimotoT KiyoshimaK MatsuoS OzekiH
SakaiEffect of SCCA1 and SCCA2 on the suppression of
TNF-alpha-induced cell death by impeding the release of
mitochondrial cytochrome c in an oral squamous cell carcinoma cell
lineTumour Biol26165172200510.1159/00008694916006770
|
|
45.
|
ST AhmedJE Darnell JrSerpin B3/B4,
activated by STAT3, promote survival of squamous carcinoma
cellsBiochem Biophys Res
Commun378821825200910.1016/j.bbrc.2008.11.14719070595
|