Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common malignant tumor and the third leading cause of
cancer-related mortality worldwide (1). Considerable progress has been made
over the past few decades for diagnosing and treating HCC. However,
HCC is still associated with a high rate of mortality, and its
recurrence is often problematic and even lethal (2). Accumulating evidence suggests that
tumor maintenance and growth are sustained by a minority population
of cancer stem cells (CSCs) or tumor-propagating cells (TPCs)
(3–6). CSCs are posited to be responsible for
tumor initiation and for the generation of distant metastasis and
relapse after therapy (7). Despite
the current progress in understanding the contribution of CSCs to
tumorigenesis, it remains elusive whether CSCs are derived from
tissue-derived stem cells, bone marrow stem cells or differentiated
mature cells that have undergone a de-differentiation or a
trans-differentiation process (3–7).
The development of HCC usually follows a multistep
sequence and the carcinogenic sequence of chronic hepatitis,
cirrhosis, dysplastic nodule (DN), and HCC has been well
established. Nodular lesions that differ from the surrounding liver
parenchyma and that are characterized by cytological or structural
atypia are termed DNs. DNs are classified as low-grade (LGDN) or
high-grade (HGDN) depending on the degree of atypia (8). If CSCs in HCC arise from hepatic
progenitor cells (HPCs), the progenitors would be expected to be
already present in DN, a well-known precancerous lesion of HCC.
Clarifying the histogenesis of CSCs is very important, because it
may provide a rationale for novel therapeutic approaches to
HCC.
Epithelial cell adhesion molecule (EpCAM) is a
transmembrane intercellular cell-adhesion molecule that is
expressed in many human epithelial cells (9). EpCAM has been identified as a marker
of human hepatic stem/progenitor cells in the liver that is absent
in mature hepatocytes (10–13).
EpCAM is frequently expressed in most epithelial cell tumors,
including HCC (9). For this
reason, EpCAM has attracted major attention as a potential
therapeutic target for cancer patients. Indeed, the use of the
EpCAM-specific monoclonal antibody has been successful in treatment
of malignant tumors associated with EpCAM positive carcinomas
patients (14,15). Recent studies have suggested that
the role of EpCAM is not limited to cell adhesion, but it is also
involved in cellular signaling, cell differentiation,
proliferation, and migration (12,14,16).
Treatment of EpCAM-positive human breast cancer cells with
EpCAM-specific small interfering RNA (siRNA) reduces cell
proliferation, migration and invasion (17). Increased expression of EpCAM is
associated with tumor angiogenensis and poor prognosis of HCC
(13,18,19).
However, the function and clinical significance of EpCAM in HCC is
largely unknown.
In the present study, we examined the location and
expression of EpCAM in surgical specimens of DNs and HCCs, the
relationship between EpCAM expression and clinicopathologic factors
in HCCs, and whether EpCAM silencing by siRNA affects cell growth,
migration, and invasiveness in HCC cells. We also investigated
whether EpCAM expression affects tumor angiogenesis in HCC.
Materials and methods
Patients
To investigate the location and expression pattern
of EpCAM, we used 28 DNs (13 LGDNs and 15 HGDNs) and their
corresponding cirrhotic nodules, and 79 HCC specimens collected
from September 2004 to August 2008 at the Chonbuk National
University Hospital. This study was approved by the ethics
committees of Chonbuk National University. Written informed consent
was exempted by the board due to the retrospective nature of the
study. Representative 4-μm blocks were prepared from 10%
formalin-fixed, paraffin-embedded tissue sections for
immunohistochemical staining. In each case, clinicopathological
features including patient age at diagnosis, gender, etiology,
serological data including serum albumin level, α-fetoprotein
(AFP), presence of ascites, tumor size, Edmonson-Steiner grade,
microvessel invasion, presence of intrahepatic metastasis and
follow-up data were obtained from hospital records. Tumors were
staged according to the 2010 American Joint Committee on Cancer
tumor-node-metastasis classification (20). The follow-up period was determined
from the date of initial surgery until the date of the last
follow-up or death. A previous existing tissue microarray (TMA)
comprising 132 HCC cases was used to compare the concordance rates
of EpCAM expression in HCC between whole tissue and TMA (21).
HCC cell lines
Human HCC cell lines HLE, HLF and Huh-7 were
purchased from the Health Science Research Resources Bank (Osaka,
Japan). HepG2 cell line was obtained from the American Type Culture
Collection (Manassas, VA, USA). In addition, we used the
sarcomatoid HCC cell line, designated SH-J1, which was established
in our laboratory (22). All HCC
cell lines were cultured according to the recommendations of the
cell banks.
Immunohistochemistry
Immunohistochemical staining was performed by
polymer intense detection system using the Bond-Max Automatic
stainer (Leica, Bannockburn, IL, USA) in accordance with the
manufacturer’s instructions. Briefly, after antigen retrieval
(microwave at high power for 10 min in 0.01 M citrate buffer, pH
6), the samples were incubated with anti-EpCAM antibody (Abcam,
Cambridge, UK) for 30 min. Peroxidase activity was detected with
the enzyme substrate 3-amino-9-ethyl carbazole. For negative
controls, sections were treated the same way, except they were
incubated with citrate buffered saline instead of the primary
antibody. The samples subjected to immunostaining were rated
according to a score calculated by adding the cancer area of the
stain to the intensity of the stain. The area of staining was
scored as 0 (<10%), 1 (11–30%), 2 (31–60%) and 3 (≥61%). The
intensity of cell staining was grouped into four categories: 0, no
immunostaining; 1, weak; 2, moderate and 3, strong. The maximum
combined score was 6 and the minimum score was 0. If the combined
score was ≥3, the tumor was considered positive, otherwise the
tumor was considered negative. The cut-off score for determining
positive expression for EpCAM was determined by receiver-operating
characteristic (ROC) curve analysis. To study the relationship
between EpCAM expression and tumor angiogenesis in HCC, we also
examined the expression of CD34 (Dako, Carpinteria, CA, USA, for
sinusoidal capillarization) and α-smooth muscle actin (Dako, for
unpaired arteries) in 79 HCC specimens. The sinusoidal
capillarization and number of unpaired arteries in HCC was measured
as described previously (23).
Western blot analysis
Western blot analysis of EpCAM in HCC cell lines was
performed as described previously (24). Briefly, cell lysates were subjected
to denaturating sodium dodecyl sulfatepolyacrylamide gel
electrophoresis followed by electroblotting and immunoblotting for
anti-EpCAM (Abcam). Blots were developed using secondary antibody,
and immune complexes were visualized using an enhanced
chemiluminescence detection system (Amersham Biosciences,
Buckinghamshire, UK). They were then analyzed using a LAS-3000
luminescent image analyzer (Fuji Film, Tokyo, Japan).
Small interfering RNA transfection
Small interfering RNA (siRNA) sequences were used to
silence EpCAM expression. EpCAM siRNA and negative control were
purchased from Bioneer Corporation (Daejeon, Korea). Sequences for
EpCAM specific siRNAs and negative control siRNA were as follows:
EpCAM: sense 5′-GUGAGAACCUACUGGAUCA(dTdT)-3′, antisense
5′-UGAUCCAGUAGGUUCUCAC(dTdT)-3′, and negative control: sense
5′-CCUACGCCACCAAUUUCGU (dTdT)-3′, antisense antisense
5′-ACGAAAUUGGUGGCGUA GG(dTdT)-3′. Transfection of siRNA was
performed with Lipofectamine RNAiMAX transfection reagent
(Invitrogen, Carlsbad, CA, USA) following the manufacturer’s
instructions.
Cell growth and proliferation assay
Cell growth was determined by the colorimetric
tetrazolium derived XTT (sodium
3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis
(4-methoxy-6-nitro) benzene sulfonic acid hydrate) assay (Roche
Applied Science, Mannheim, Germany). DNA synthesis of cells was
assessed by the bromodeoxyuridine (BrdU) incorporation assay (Roche
Applied Science). For the cell growth and proliferation assay, 48 h
after transfection of siRNA the cells of each group were reseeded
in 96-well plates at a density of 0.3–1×104 cells per
well. After 24–48 h, XTT and incorporated BrdU were measured
colorimetrically using a microtiter plate reader (Bio-Rad,
Hercules, CA, USA) at a wavelength of 450 nm.
In vitro migration and invasion
assays
The migration assay was performed using a
24-transwell migration chamber (Corning Life Sciences, Acton, MA,
USA) and the cell invasion assay was performed using a 24-transwell
BioCoat Matrigel invasion chamber (BD Biosciences, San Jose, CA,
USA) with an 8 μm-pore size polyvinyl-pyrrolidone-free
polycarbonate membrane following the manufacturer’s protocol. The
cells that migrated or invaded to the lower surface of the filter
were counted under a light microscope at ×200 magnification in 10
randomly selected fields per well.
Statistical analyses
Comparisons between EpCAM expression and
clinicopathological factors were assessed by the χ2
test. Survival analyses were performed using the Kaplan-Meier
method, and differences in survival between different clinical
groups were determined by the log-rank test. A Cox proportional
hazards regression analysis was performed to estimate the impact of
clinicopathological factors on patient survival. P-values <0.05
were considered statistically significant. SPSS version 15.0
statistical software program (SPSS, Chicago, IL, USA) was used for
the statistical analyses.
Results
Clinical features
The 175 patients with HCC were 25–79 years of age
and consisted of 147 males and 28 females. A total of 126 patients
were positive for hepatitis B virus surface antigen, 19 were
alcohol related, 11 were positive for anti-hepatitis C virus
antibody and 19 patients were of unknown etiology (Table I). The 175 HCCs were composed of 35
small HCCs (≤2 cm) and 140 advanced HCCs (>2 cm). Among the 35
small HCCs, six were vaguely nodular, 27 were distinctly nodular
and two were infiltrative types.
 | Table IAssociation between pathological
features and EpCAM-positive patients with hepatocellular carcinoma
(HCC). |
Table I
Association between pathological
features and EpCAM-positive patients with hepatocellular carcinoma
(HCC).
| Overall HCC (n=175)
| T1 HCC (n=65)
|
|---|
|
Characteristics | Total |
EpCam+ | P-value | Total |
EpCam+ | P-value |
|---|
| Sex | | | | | | |
| Male | 147 | 56 | 0.060 | 51 | 20 | 0.230 |
| Female | 28 | 16 | | 14 | 8 | |
| Age (years) | | | | | | |
| <55 | 66 | 29 | 0.559 | 18 | 5 | 0.123 |
| ≥55 | 109 | 43 | | 47 | 23 | |
| Etiology | | | | | | |
| HBV | 126 | 52 | 0.111 | | | |
| HCV | 11 | 1 | | | | |
| Alcohol | 19 | 9 | | | | |
| Others | 19 | 10 | | | | |
| Etiology | | | | | | |
| Viral | 137 | 53 | 0.210 | 50 | 19 | 0.131 |
| Non-viral | 38 | 19 | | 15 | 9 | |
| Liver
cirrhosis | | | | | | |
| Absence | 86 | 33 | 0.464 | 24 | 12 | 0.388 |
| Presence | 89 | 39 | | 41 | 16 | |
| Ascites | | | | | | |
| Absence | 159 | 66 | 0.756 | 59 | 26 | 0.613 |
| Presence | 16 | 6 | | 6 | 2 | |
| Albumin, ng/dl | | | | | | |
| <3.5 | 22 | 9 | 0.981 | 13 | 7 | 0.381 |
| ≥3.5 | 153 | 63 | | 52 | 21 | |
| Microvessel
invasion | | | | | | |
| Absence | 72 | 31 | 0.667 | | | |
| Presence | 103 | 41 | | | | |
| Preoperative AFP,
ng/ml | | | | | | |
| ≥100 | 115 | 40 | 0.018 | 51 | 21 | 0.555 |
| >100 | 60 | 32 | | 14 | 7 | |
| Intrahepatic
metastasis | | | | | | |
| Absence | 118 | 49 | 0.882 | 60 | 27 | 0.278 |
| Presence | 57 | 23 | | 5 | 1 | |
| Histologic
grade | | | | | | |
| 1 and 2 | 110 | 34 | <0.001 | 48 | 15 | 0.001 |
| 3 and 4 | 65 | 38 | | 17 | 13 | |
| pT stage | | | | | | |
| 1 | 65 | 28 | 0.894 | | | |
| 2 | 76 | 31 | | | | |
| 3 and 4 | 34 | 13 | | | | |
Immunohistochemical results
Hepatocellular EpCAM expression in regenerating
nodules showed strong cytoplasmic and/or membranous staining in all
cirrhotic livers adjacent to DNs with the immunoreactivity
depending on the degree of hepatocellular differentiation. Reactive
ductular cells surrounding inflamed portal tract and periseptal
areas showed stronger positivity for EpCAM (Fig. 1A and B). However, the expression of
EpCAM was lost in the center of regenerating nodules, indicating
the differentiation towards mature hepatocytes (Fig. 1C). Of 28 DNs, only one LGDN showed
EpCAM expression. EpCAM expression in LGDN showed a geographic
staining with weak intensity and accentuation in cells around the
portal tracts (Fig. 1D and E). In
79 HCCs whole tissue sections, 31 were EpCAM-positive (39%). The
pattern of EpCAM expression in HCC was more homogeneous and
diffused than that of DN or regenerating nodules (Fig. 1F–I). Of the 175 HCC sections, EpCAM
expression was detected in 72 (41%) HCCs.
Among 35 small HCCs, EpCAM expression was detected
in 19 (54%) HCCs. Nineteen EpCAM-positive small HCCs were composed
of one vaguely nodular, 17 distinct nodular and one infiltrative
type. In the validation study between whole tissue section and TMA
samples, the concordance rate for EpCAM staining in HCC was 92% (33
of 36). Two EpCAM-positive HCCs in whole tissue sections changed to
negative cases in TMA samples, whereas one EpCAM-negative case
changed to positive case in TMA samples.
Correlation between immunohistochemical
results and clinicopathological features
To elucidate the significance of EpCAM in HCCs, a
correlation between EpCAM and the major clinicopathological
features was evaluated (Table
I).
The clinicopathological analysis revealed that
EpCAM-positive HCC was significantly associated with high
histological grade (P<0.001) and serum AFP level (P=0.018).
Other factors, including age, gender, etiology, background liver
disease, albumin level, presence of intrahepatic metastasis,
microvessel invasion and the presence of ascites were not
correlated with EpCAM expression. No significant differences were
observed between EpCAM expression and the sinusoidal
capillarization or number of unpaired arteries in HCC (Table II).
 | Table IIAssociation with EpCAM expression and
tumor angiogenesis in hepatocellular carcinoma. |
Table II
Association with EpCAM expression and
tumor angiogenesis in hepatocellular carcinoma.
| Overall HCC (n=79)
| T1 HCC (n=65)
|
|---|
|
Characteristics | Total |
EpCam+ | P-value | Total |
EpCam+ | P-value |
|---|
| SMA | | | | | | |
| 1+, 2+ | 31 | 14 | 0.953 | 14 | 6 | 0.769 |
| 3+, 4+ | 48 | 22 | | 23 | 11 | |
| CD334 | | | | | | |
| 1+, 2+ | 9 | 5 | 0.523 | 5 | 2 | 0.774 |
| 3+, 4+ | 70 | 31 | | 32 | 15 | |
Outcome
Follow-up intervals ranged from 1–142 months.
Sixty-one patients died during the follow-up period. In univariate
analysis, intrahepatic metastasis, serum albumin levels,
microvessel invasion and T stage were significantly associated with
poor patient survival (P<0.001, P=0.011, P=0.018 and P= 0.010,
respectively). Multivariate analysis revealed that T stage,
intrahepatic metastasis and serum albumin levels were independent
prognostic indicators (P=0.012, P=0.005 and P<0.001,
respectively) (Table III). Among
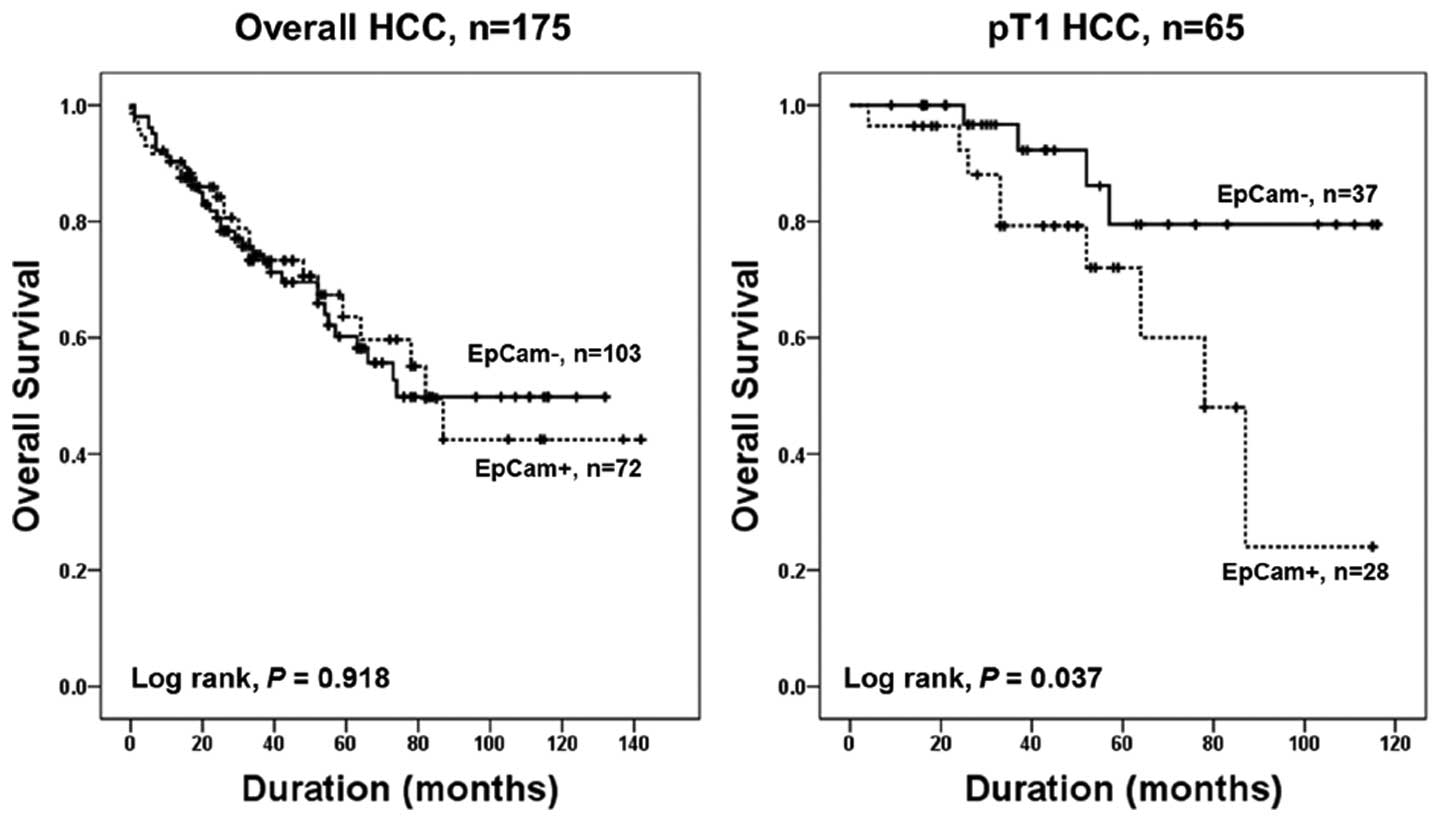
T1 HCC patients, mean survival of patients with EpCAM-positive HCC
was 74.4 months, and EpCAM-negative HCC was 101.7 months. EpCAM
expression was significantly associated with patient survival in T1
HCC patients in univariate and multivariate Cox survival analyses
(P= 0.049 and P= 0.023, respectively) (Table III, Fig. 2).
 | Table IIICox proportional hazard analyses of
factors associated with hepatocellular carcinoma (HCC) in 175
patients. |
Table III
Cox proportional hazard analyses of
factors associated with hepatocellular carcinoma (HCC) in 175
patients.
| Overall HCC (n=175)
| | T1 HCC (n=65)
|
|---|
|
Characteristics | HR | 95%CI | P-value | | HR | 95%CI | P-value |
|---|
| Univariate Cox
regression analysis | Univariate Cox
regression analysis |
| Intrahepatic
metastasis | 2.474 | 1.490–4.106 | <0.001 | EpCAM | 3.284 | 1.003–10.759 | 0.049 |
| Albumin | 0.426 | 0.220–0.825 | 0.011 | | | | |
| Microvessel
invasion | 1.969 | 1.122–3.453 | 0.018 | | | | |
| pT stage | | | 0.010 | | | | |
| 2.673 | 1.401–5.100 | 0.003 | | | | |
| 2.371 | 1.125–4.994 | 0.023 | | | | |
| Multivariate Cox
regression analysis | Multivariate Cox
regression analysisa |
| pT stage | | | 0.012 | EpCAM | 4.008 | 1.215–13.219 | 0.023 |
| 2.756 | 1.359–5.589 | 0.005 | | | | |
| 1.627 | 0.695–3.808 | 0.262 | | | | |
| Intrahepatic
metastasis | 2.255 | 1.271–4.002 | 0.005 | | | | |
| Albumin | 0.267 | 0.013–0.538 | <0.001 | | | | |
Expression of EpCAM in HCC cell
lines
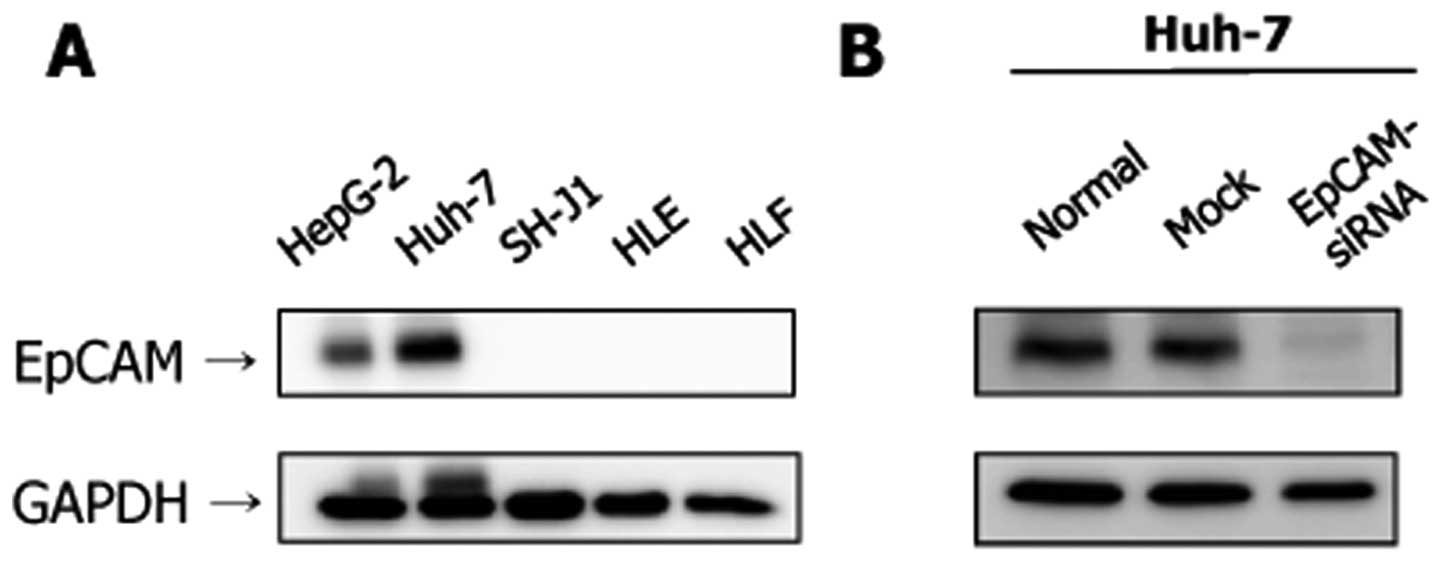
The expression level of the EpCAM protein was higher
in the Huh-7 and HepG2 cell lines (Fig. 3A). However, the expression of EpCAM
was not evident in the SH-J1, HLE and HLF cell lines. Transfection
with EpCAM siRNA resulted in a marked decrease of EpCAM protein
expression at 48 h post-transfection in Huh-7 cells (Fig. 3B).
Effects of EpCAM silencing on cell
growth, proliferation, migration, and invasion
Silencing EpCAM gene expression in Huh-7 cells by
EpCAM siRNA resulted in significant inhibition of cell growth
compared to those of the control (P<0.001) (Fig. 4A). Silencing EpCAM gene expression
also significantly decreased cell proliferation compared to those
of the control (P<0.001) (Fig.
4B). Silencing EpCAM gene expression dramatically inhibited the
migration and invasion ability of Huh-7 cells (Fig. 5).
Discussion
This study demonstrated that: i) EpCAM expression is
very rare in DNs but predominates in a distinctly nodular type of
small HCC; ii) EpCAM expression in HCC correlates with tumor cell
de-differentiation and serum AFP levels; iii) EpCAM silencing
induces significant inhibition in the growth and proliferation of
HCC cells; and iv) EpCAM silencing decreases cell migration and
invasion of HCC cells. We also found that EpCAM expression is an
independent prognostic indicator of T1 HCC. However, we could not
discern a significant association between EpCAM expression in HCC
cells and tumor angiogenesis. These data strongly suggest that
EpCAM expression occurs in the small nodular stage of HCC in
hepatocarcinogenesis and indicate important roles of EpCAM in HCC
progression.
We found that a strong expression of EpCAM in
proliferating ductular cells and regenerating hepatocellular cells
of regenerating nodules. On maturation of these regenerating
hepatocytes, EpCAM expression was lost. There was a transient loss
of hepatocellular EpCAM expression in DNs in regenerating nodules.
EpCAM expression reappeared in distinctly nodular HCCs. Small HCCs
(≤2 cm in diameter) can be subcategorized further into vaguely
nodular and distinctly nodular HCCs based on macroscopic features.
Vaguely nodular HCC is an early HCC, and distinctly nodular HCC is
a small progressed HCC (8).
Contrary to the notion that EpCAM-positive HCC may originate from
HPCs, our finding of the dominant reappearance of EpCAM in distinct
nodular HCC indicates that HCCs could obtain the EpCAM phenotype
during a small progressed stage of HCC. Our findings are consistent
with the results of Breuhahn et al, who reported the rarity
of EpCAM expression in DN, the earliest known premalignant lesion
in human HCC (11). The CSC model
is essentially synonymous with the hierarchy model of
carcinogenesis (25). However, the
expression of stemness-related markers exists as a functional
phenotype in the de-differentiation model and is evident by any
member of the malignant population in the presence of the
appropriate endogenous and exogenous factors (5). In HCC, EpCAM expression is regulated
by Wnt/β-catenin signaling and tumorigenicity, invasiveness, and
differentiation capabilities of EpCAM-positive HCC are regulated by
Wnt/β-catenin signaling (12).
Thus, EpCAM appears to be a common gene expressed in HCC with
activated Wnt/β-catenin signaling (12). Taken together, these findings
suggest that EpCAM expression is an acquired phenotype of cancer
cells during HCC progression, although CSCs might be another
contributor of EpCAM-positive HCC.
Presently, a proportion of HCCs expressed EpCAM and
EpCAM expression correlated with the grade of malignancy. Moreover,
EpCAM silencing resulted in a significant decrease in the rate of
cell proliferation of HCC cells. These findings agree with previous
studies demonstrating that the number of EpCAM-positive cells and
expression levels of EpCAM correlate with de-differentiation and
are associated with the proliferative activity of tumor cells
(17,26,28).
EpCAM is overexpressed in breast carcinoma and silencing of EpCAM
gene expression with siRNA decreases proliferation of breast cancer
cells (17). EpCAM blockage via
siRNA also inhibits spheroid formation and tumorigenicity of Huh-1
cells (12). Taken together, these
observations suggest that EpCAM is required for tumor cell
de-differentiation and increased proliferative activity in HCC.
This notion is supported by the fact that EpCAM has a direct impact
on cell cycle and the ability to rapidly upregulate the
proto-oncogene c-myc as well as cyclin A and E in human epithelial
kidney 293 cells (27).
Additionally, it has been shown that proteolytic cleavage of EpCAM
releases an intracellular domain, which forms a complex with
components of Wnt pathway and regulates gene transcription,
resulting in cell proliferation and tumor formation (16).
We also found that EpCAM expression in HCC was
significantly associated with high serum AFP level. A close
relationship between EpCAM expression and high AFP levels has been
demonstrated (12,13). Gene expression profiles have
revealed that EpCAM+/AFP+ HCCs have
progenitor features with poor prognosis, whereas
EpCAM−/AFP− HCCs have adult hepatocyte
features with good prognosis (13). The latter study confirmed that
EpCAM+ HCC cells are highly invasive and tumorigenic,
and activate Wnt/β-catenin signaling. The prognosis of patients
with EpCAM-positive HCC is thought to be worse than those with pure
HCC (13,18,19).
In this study, EpCAM expression was not associated with the overall
survival rate in all patients with HCC; however, we found that
EpCAM expression was an independent prognostic indicator of T1 HCC.
Because EpCAM expression was presently associated with high tumor
grade and high AFP levels-factors that are well known unfavorable
prognostic factors in HCC-the finding that EpCAM appears to be a
poor prognostic factor for HCC is reasonable. The collective
findings suggest that EpCAM expression in HCC plays an important
role in facilitating tumor cell proliferation, leading to high
grade HCC with high AFP level.
This study demonstrates that EpCAM silencing by
siRNA dramatically decreases cell migration and invasion of HCC
cells. This is in agreement with previous studies showing that
expression of EpCAM is related to the degree of invasion and/or
metastasis in breast (29), lung
(30) and pancreas cancers
(31). Silencing of EpCAM gene
expression decreased cell migration and invasion in a breast cancer
cell line (17). Similarly, EpCAM
blockage by siRNA decreases the population of EpCAM-positive cells
and significantly inhibited cellular invasion of Huh-1 cells
(12). Based on the above
observations, it is clear that EpCAM is an important player in
invasion and metastasis of tumor cells. However, the mechanisms of
the promoting role of EpCAM in tumor invasion and metastasis are
still not fully understood. EpCAM is a transmembrane glycoprotein
that has been proposed to mediate homophilic adhesive interactions,
thereby preventing cell scattering (9,10,26).
From this, one might expect that EpCAM prevents cancer metastasis.
However, in several tumor types, high expression of EpCAM has been
inversely correlated with metastasis and poor clinical outcome
(32–34). EpCAM is able to abrogate
E-cadherin-mediated cell-to-cell adhesion by disrupting the link
between α-catenin and cytoskeleton, resulting in promoting cell
motility, proliferation and metastasis (32,35).
EpCAM also interacts directly with CD44v4-v7, a tumor
metastasis-promoting cell adhesion molecule, and with claudin-7, a
tight cell junction protein (36,37).
These complexes can influence cell-to-cell adhesion and cell matrix
adhesion, and they appear to be involved in processes that promote
metastasis. Another potential mechanism involves the possible links
between EpCAM expression and activation of Wnt signaling. This
contention is supported by the observation that EpCAM
downregulation leads to a significant decrease in cytoplasmic
β-catenin through an increase in its association with the
E-cadherin adhesion complex (17).
Consequently, the decreased available β-catenin for Wnt signaling
leads to the shut-down of the activation of its target genes
involved in tumor progression.
EpCAM has been targeted in clinical trials using
monoclonal antibodies in various cancers (14,15,38)
and we believe that EpCAM represents a novel target for gene
therapy in HCC. In support of this hypothesis, we found that EpCAM
was overexpressed in a proportion of HCCs. Furthermore, silencing
EpCAM gene expression significantly decreased the proliferative
capacity and invasive potential of HCC cells. siRNA can be used
successfully for gene silencing in vivo (39,40).
Since EpCAM antibody and/or siRNA can be easily synthesized, this
study may provide a rationale for therapeutic approaches to
HCC.
Our study did not find a statistically significant
correlation between EpCAM expression and the number of unpaired
arteries, or the degree of sinusoidal capillarization in HCC. On
the other hand, some other studies have reported that high
expression of EpCAM correlates with tumor angiogenesis in HCC
(18,19). Further analysis of the EpCAM
expression and tumor angiogenesis in clinical HCC tumor samples
might provide useful information regarding prognosis and
treatment.
In conclusion, our data indicate that EpCAM
expression is very rare in DNs, but can reappear predominantly in
distinctly nodular small HCC. Although we cannot conclude whether
the EpCAM-positive HCCs originated from pre-existing CSC or from
de-differentiation of HCC cells, our study suggests that the EpCAM
phenotype might be an acquired feature of cancer cells during HCC
progression. EpCAM expression is associated with tumor cell
de-differentiation and serum AFP level, and is a negative
prognostic factor in HCC. Moreover, silencing EpCAM gene expression
significantly inhibits tumor cell proliferation and decreases the
migration and invasiveness of HCC cells. These findings strongly
suggest that EpCAM plays an important role in development and
progression of HCC.
Acknowledgements
This study was supported by the
National Research Foundation of Korea Grant funded by the Korean
Government (no. 2011-0028223).
References
|
1.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy KR: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Visvader JE: Cells of origin in cancer.
Nature. 469:314–322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Clarke MF, Dick JE, Dirks PB, et al:
Cancer stem cells - perspectives on current status and future
directions: AACR Workshop on cancer stem cells. Cancer Res.
66:9339–9344. 2006. View Article : Google Scholar
|
|
5.
|
Bomken S, Fiser K, Heidenreich O and
Vormoor J: Understanding the cancer stem cell. Br J Cancer.
103:439–445. 2010. View Article : Google Scholar
|
|
6.
|
Bjerkvig R, Tysnes BB, Aboody KS, Najbauer
J and Terzis AJ: Opinion: the origin of the cancer stem cell:
current controversies and new insights. Nat Rev Cancer. 5:899–904.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Marquardt JU, Factor VM and Thorgeirsson
SS: Epigenetic regulation of cancer stem cells in liver cancer:
current concepts and clinical implications. J Hepatol. 53:568–577.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Park YN: Update on precursor and early
lesions of hepatocellular carcinomas. Arch Pathol Lab Med.
135:704–715. 2011.PubMed/NCBI
|
|
9.
|
Litvinov SV, Balzar M, Winter MJ, et al:
Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell
interactions mediated by classic cadherins. J Cell Biol.
139:1337–1348. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Winter MJ, Nagtegaal ID, van Krieken JH
and Litvinov SV: The epithelial cell adhesion molecule (Ep-CAM) as
a morpho-regulatory molecule is a tool in surgical pathology. Am J
Pathol. 163:2139–2148. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Breuhahn K, Baeuerle PA, Peters M, et al:
Expression of epithelial cellular adhesion molecule (Ep-CAM) in
chronic (necro-) inflammatory liver diseases and hepatocellular
carcinoma. Hepatol Res. 34:50–56. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Yamashita T, Ji J, Budhu A, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Yamashita T, Forgues M, Wang W, et al:
EpCAM and alpha-fetoprotein expression defines novel prognostic
subtypes of hepatocellular carcinoma. Cancer Res. 68:1451–1461.
2008. View Article : Google Scholar
|
|
14.
|
Patriarca C, Macchi RM, Marschner AK and
Mellstedt H: Epithelial cell adhesion molecule expression (CD326)
in cancer: a short review. Cancer Treat Rev. 38:68–75. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Schmidt M, Scheulen ME, Dittrich C, et al:
An open-label, randomized phase II study of adecatumumab, a fully
human anti-EpCAM antibody, as monotherapy in patients with
meta-static breast cancer. Ann Oncol. 21:275–282. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Maetzel D, Denzel S, Mack B, et al:
Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell
Biol. 11:162–171. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Osta WA, Chen Y, Mikhitarian K, et al:
EpCAM is over-expressed in breast cancer and is a potential target
for breast cancer gene therapy. Cancer Res. 64:5818–5824. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Yang XR, Xu Y, Yu B, et al: High
expression levels of putative hepatic stem/progenitor cell
biomarkers related to tumour angiogenesis and poor prognosis of
hepatocellular carcinoma. Gut. 59:953–962. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Shan YF, Huang YL, Xie YK, et al:
Angiogenesis and clinicopathologic characteristics in different
hepatocellular carcinoma subtypes defined by EpCAM and
α-fetoprotein expression status. Med Oncol. 28:1012–1016.
2011.PubMed/NCBI
|
|
20.
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th ed.
Springer; New York, NY: 2010
|
|
21.
|
Bae JS, Choi HN, Noh SJ, et al: Expression
of K19 and K7 in dysplastic nodules and hepatocellular carcinoma.
Oncol Lett. 4:213–220. 2012.PubMed/NCBI
|
|
22.
|
Kim DG, Park SY, Kim H, Chun YH, Moon WS
and Park SH: A comprehensive karyotypic analysis on a newly
established sarcomatoid hepatocellular carcinoma cell line SH-J1 by
comparative genomic hybridization and chromosome painting. Cancer
Genet Cytogenet. 132:120–124. 2002. View Article : Google Scholar
|
|
23.
|
Park YN, Kim YB, Yang KM and Park C:
Increased expression of vascular endothelial growth factor and
angiogenesis in the early stage of multistep hepatocarcinogenesis.
Arch Pathol Lab Med. 124:1061–1065. 2000.PubMed/NCBI
|
|
24.
|
Kwon CY, Kim KR, Choi HN, et al: The role
of serum response factor in hepatocellular carcinoma: implications
for disease progression. Int J Oncol. 37:837–844. 2010.PubMed/NCBI
|
|
25.
|
Shackleton M, Quintana E, Fearon ER and
Morrison SJ: Heterogeneity in cancer: cancer stem cells versus
clonal evolution. Cell. 138:822–829. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Litvinov SV, van Driel W, van Rhijn CM, et
al: Expression of Ep-CAM in cervical squamous epithelia correlates
with an increased proliferation and the disappearance of markers
for terminal differentiation. Am J Pathol. 148:865–875. 1996.
|
|
27.
|
Münz M, Kieu C, Mack B, Schmitt B, Zeidler
R and Gires O: The carcinoma-associated antigen EpCAM upregulates
c-myc and induces cell proliferation. Oncogene. 23:5748–5758.
2004.PubMed/NCBI
|
|
28.
|
Yamashita T, Budhu A, Forgues M and Wang
XW: Activation of hepatic stem cell marker EpCAM by
Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res.
67:10831–10839. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Cimino A, Halushka M, Illei P, Wu X,
Sukumar S and Argani P: Epithelial cell adhesion molecule (EpCAM)
is overexpressed in breast cancer metastases. Breast Cancer Res
Treat. 123:701–708. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Piyathilake CJ, Frost AR, Weiss H, Manne
U, Heimburger DC and Grizzle WE: The expression of Ep-CAM (17-1A)
in squamous cell cancers of the lung. Hum Pathol. 31:482–487. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Scheunemann P, Stoecklein NH, Rehders A,
et al: Occult tumor cells in lymph nodes as a predictor for tumor
relapse in pancreatic adenocarcinoma. Langenbecks Arch Surg.
393:359–365. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
van der Gun BT, Melchers LJ, Ruiters MH,
de Leij LF, McLaughlin PM and Rots MG: EpCAM in carcinogenesis: the
good, the bad or the ugly. Carcinogenesis. 31:1913–1921.
2010.PubMed/NCBI
|
|
33.
|
Went P, Dirnhofer S, Salvisberg T, et al:
Expression of epithelial cell adhesion molecule (EpCam) in renal
epithelial tumors. Am J Surg Pathol. 29:83–88. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Ensinger C, Kremser R, Prommegger R,
Spizzo G and Schmid KW: EpCAM overexpression in thyroid carcinomas:
a histopathological study of 121 cases. J Immunother. 29:569–573.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Winter MJ, Nagelkerken B, Mertens AE,
Rees-Bakker HA, Briaire-de Bruijn IH and Litvinov SV: Expression of
Ep-CAM shifts the state of cadherin-mediated adhesions from strong
to weak. Exp Cell Res. 285:50–58. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Schmidt DS, Klingbeil P, Schnölzer M and
Zöller M: CD44 variant isoforms associate with tetraspanins and
EpCAM. Exp Cell Res. 297:329–347. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Ladwein M, Pape UF, Schmidt DS, et al: The
cell-cell adhesion molecule EpCAM interacts directly with the tight
junction protein claudin-7. Exp Cell Res. 309:345–357. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Chaudry MA, Sales K, Ruf P, Lindhofer H
and Winslet MC: EpCAM an immunotherapeutic target for
gastrointestinal malignancy: current experience and future
challenges. Br J Cancer. 96:1013–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
McCaffrey AP, Meuse L, Pham TT, Conklin
DS, Hannon GJ and Kay MA: RNA interference in adult mice. Nature.
418:38–39. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Lewis DL, Hagstrom JE, Loomis AG, Wolff JA
and Herweijer H: Efficient delivery of siRNA for inhibition of gene
expression in postnatal mice. Nat Genet. 32:107–108. 2002.
View Article : Google Scholar : PubMed/NCBI
|