Introduction
Post-translational protein modification by
poly(ADP-ribosyl) ation is involved in a variety of biological
processes including chromatin structural regulation, transcription,
DNA repair, DNA replication, telomere homeostasis, cell division,
cell proliferation, cell death and other physiological and
pathological functions (1–5). The reactions are catalytically
mediated by poly(ADP-ribose) polymerases(PARPs). Recent studies
indicated that there are 17 members of the PARP superfamily
(6).
By comparing the catalytic domain structures of
these 17 members, three different classes of PARP proteins have
been defined (7). The first
subfamily consists of enzymes that have been demonstrated or
structurally predicted to catalyze poly(ADP-ribosyl)ation. PARP1,
PARP2, PARP3, PARP4, PARP5a and PARP5b belong to this subfamily.
The second subfamily members (PARP6, PARP7, PARP8, PARP10, PARP11,
PARP12, PARP14, PARP15 and PARP16) have a considerably shorter
nicotinamide-ribose-binding site than PARP1, thus these enzymes
have been structurally predicted to catalyze
mono(ADP-ribosyl)ation. Indeed, the first analyzed enzyme belonging
to this subfamily, PARP10, has been demonstrated to be a
mono(ADP-ribosyl) transferase with auto-mono(ADP-ribosyl)ation
activity (7). The third subfamily
members (PARP9 and PARP13) lack the β-NAD+
cofactor-binding domain.
Among PARPs, the members capable of mediating
poly(ADP-ribosyl)ation have been extensively examined
physiologically, but the biological functions of the
mono(ADP-ribosyl) transferase subfamily members are largely
unexplored. Several limited studies suggested the involvement of
mono(ADP-ribosyl) transferases in growth control (7–13),
opening new areas for investigation. PARP10 was identified as a
novel protein that interacts with Myc oncoproteins (13). When expressed in rat embryo
fibroblasts, oncogenic transformation induced by c-Myc plus Ha-Ras
expression was inhibited (13).
Also, the growth rate was reduced by PARP10 expression in HeLa
cells (7). PARP10 phosphorylation
by cyclin-dependent kinases was suggested to be its cell cycle
regulation mechanism (10). PARP14
was identified as a novel modification protein that stabilizes
autocrine motility factor (AMF)/phosphoglucose isomerase (12). PARP14 is also known as a regulator
of Stat6 transcription activity, which leads to the transduction of
cell survival signals (8,9,11).
We analyzed PARP6 and found that it is involved in
negative regulation of cell cycle progression. When expressed in
HeLa cells, cell proliferation was inhibited depending on the PARP6
catalytic domain, which is highly conserved among vertebrates.
Immunohistochemical analysis of human colorectal cancer specimens
demonstrated that PARP6 protein expression was inversely correlated
with Ki-67 positivity and was linked to a good prognosis. To our
knowledge, this is the first demonstration that PARP6 controls
cancer cell growth.
Materials and methods
Cell lines and cell culture
The human cervical cancer cell line, HeLa, was
provided by the late Professor Masakatsu Horikawa, Faculty of
Pharmaceutical Sciences, Kanazawa University (Kanazawa, Japan)
(14). HEK293FT cells were
purchased from Life Technologies, Japan. Cells were cultured in
Dulbecco’s modified Eagle’s medium containing 10% fetal bovine
serum, penicillin (50 U/ml) and streptomycin (50 μg/ml), at 37°C in
a humidified atmosphere of 5% CO2 and 95% air.
Patients and tissue samples
A total of 126 advanced colorectal carcinomas (72
men and 54 women), including 62 cases with lymph node metastasis
and 24 cases with distant metastasis, were obtained from the
archive of Hiroshima University Hospital during 1984–2001 after
surgical resection. The age range was 37–84 years (mean, 64.3
years). Tissues were fixed in 10% buffered formalin and embedded in
paraffin. The study protocol followed the ethical guidelines of
Hiroshima University and Prefectural University of Hiroshima.
Informed consent was obtained from all subjects.
Plasmids and transfection
A full-length cDNA clone encoding PARP6 (BC110902)
was subcloned into a pEGFP vector (pEGFP-PARP6) to produce an
enhanced green fluorescent protein (EGFP)-tagged protein. cDNA
clones encoding two alternative splicing forms (AB499727 and
AB499728) were also subcloned into a pEGFP vector (pEGFP-PARP6-SP1
and pEGFP-PARP6-SP2). An N-terminal deletion mutant (deletion of
410 amino acid residues) was also constructed
[pEGFP-ΔN(1–410)PARP6]. Transfection was performed using
Lipofectamine 2000 (Life Technologies) according to the
manufacturer’s guidelines.
Cell growth assay
Proliferation activity was measured by the
water-soluble tetrazolium-1 reagent assay (WST-1, Roche Applied
Science) in transiently transfected cells, according to the
manufacturer’s protocol. HeLa cells were transfected with
pEGFP-empty, pEGFP-PARP6, pEGFP-PARP6-SP1 or pEGFP-PARP6-SP2.
Transfection efficiencies were >80% as confirmed by EGFP
expression under a fluorescence microscope. After 24 h, transfected
cells were replated in 96-well plates. WST-1 assay was performed at
each time point (1, 2 and 3 days) and the values were expressed as
a percentage change.
DNA histograms
Cells were transfected with pEGFP-empty, pEGFP-PARP6
or pEGFP-ΔN(1–410)PARP6. After 24 h, the transfected cells were
fixed with 20% ethanol and incubated with 0.1% RNase (Type II-A,
Sigma-Aldrich, Japan) for 30 min at 37°C. The cells were stained
with propidium iodide (50 μg/ml), and green (for EGFP) and red (for
propidium iodide) fluorescence from individual cells were measured
using a FACSort flow cytometer (BD Biosciences).
Immunoblot analysis
Transfected cells were lysed with ice-cold Laemmli
sodium dodecyl sulfate (SDS)-sample buffer (pH 6.8), consisting of
25 mM Tris-HCl, 5% glycerol, 2.5% 2-mercaptoethanol and 1% SDS,
containing protease inhibitor cocktail (Sigma-Aldrich). The lysates
were sonicated three times for 10 sec on ice and centrifuged at
15,000 rpm for 1 min at 4°C. The supernatant was collected and the
protein concentration was determined using the Bio-Rad protein
assay kit (Bio-Rad Laboratories). Equal amounts of protein (20 μg
per lane) were loaded on a 10% SDS-polyacrylamide gel. The cell
lysates were resolved by electrophoresis and transferred to
Immobilon-P membranes (Merck Millipore). Membranes were blocked
with skim milk, probed with primary antibodies, washed and then
incubated with secondary antibody. Anti-GFP antibody (JL-8,
Clontech Laboratories), anti-α-tubulin (CLT9002, Cedarlane
Laboratories) and anti-β-actin (A1978, Sigma-Aldrich) were used as
primary antibodies. Horseradish peroxidase-conjugated anti-mouse
antibody (GE Healthcare, Japan) was used as secondary antibody.
Proteins were visualized on X-ray film using ECL Western Blotting
Detection Reagent (GE Healthcare).
Immunohistochemical analysis
For immunohistochemical examination, serial 4-μm
sections were stained with hematoxylin and eosin and used for
immunohistochemical analysis. Immunohistochemical staining was
carried out with anti-PARP6 antibody (HPA026991, Sigma-Aldrich),
raised against the PARP6 catalytic domain (497–589), after antigen
retrieval by microwave treatment in citrate buffer (pH 6.0) and
detection was performed by the streptavidin-biotin peroxidase
system (Universal LSAB™2 kit, Dako, Japan). This PARP6 antibody is
specific to PARP6, but does not recognize PARP6-SP1 and PARP6-SP2.
In addition, to determine the proliferative cell activity and
correlate it with PARP6 expression, we examined Ki-67 expression
using anti-Ki-67 monoclonal antibody (MIB-1, Dako). The sections
were incubated with primary antibodies at 4°C overnight. The
immunostaining was defined as positive when >20% of the tumor
cells were stained for PARP6 in the cytoplasm. The
immunohistochemistry grade was defined as - to +++ according to the
number of cells stained and to the intensity of the reaction in
individual cells. Grades were defined as follows: -, mostly no
positive cells; +, 5–20% of tumor cells showed weak to moderate
immunoreactivity; ++, 20–50% of tumor cells showed moderate
immunoreactivity; +++, over 50% of tumor cells showed intense
immunoreactivity. Cases with grade ++ and +++ were regarded as
positive cases. A labeling index percentage of Ki-67-positive cells
was determined by examining at least 1,000 tumor cells at x200
magnification in five representative areas.
Statistical analysis
The Statcel software package (KaleidaGraph Version
4.1) was used for analysis. The α2 test and Fisher’s and
t-test (Statcel - The useful Addin Forms on Excel - 2nd edition)
were used for comparison of data between two groups. Survival
analysis was conducted according to the Kaplan-Meier method and
survival characteristics and were compared using log-rank tests. A
P<0.05 was considered to indicate statistical significance.
Results
PARP6 expression inhibits cell
growth
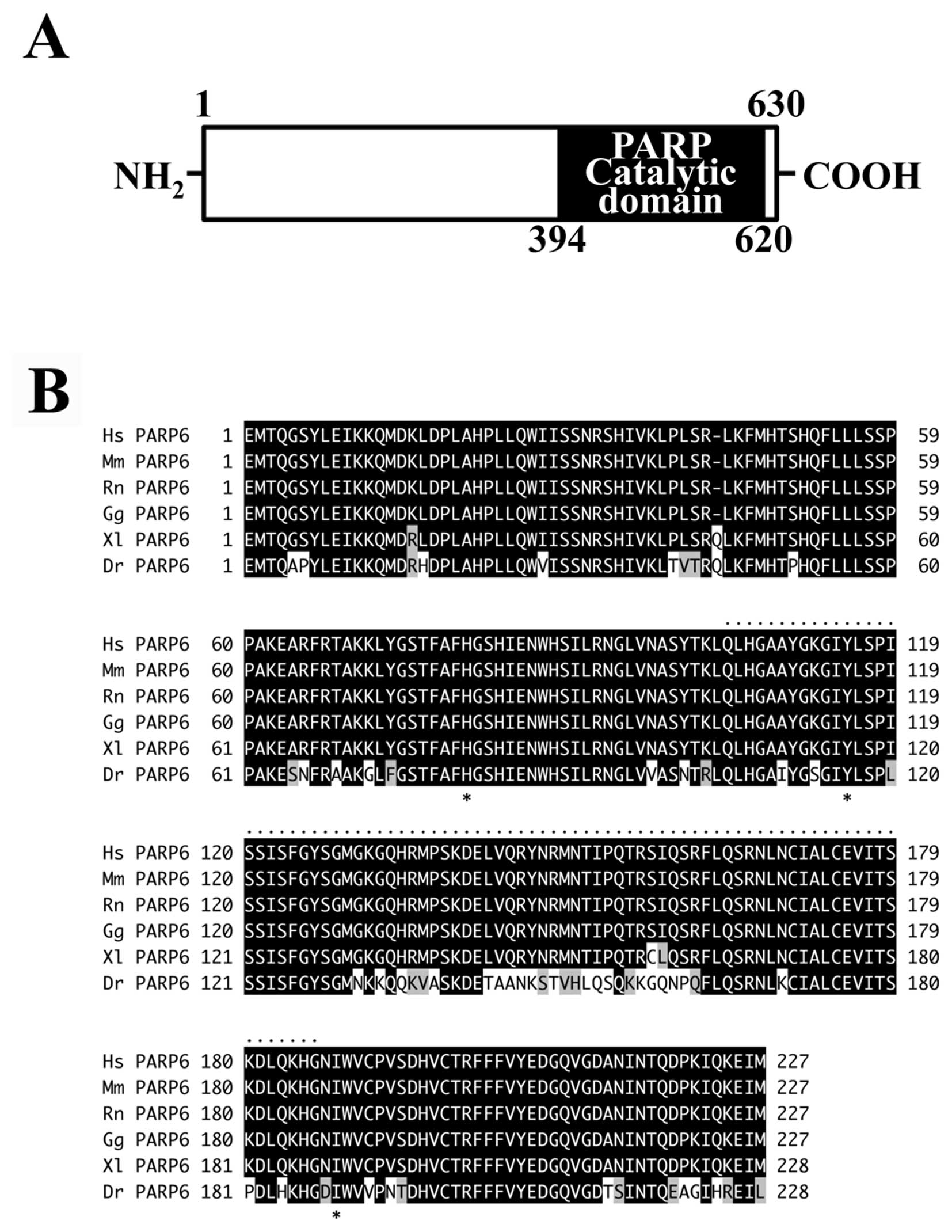
Full-length PARP6 consists of 630 amino acids with a
molecular mass of approximately 71 kDa. The C-terminal region
(residues 394–620) contains the PARP catalytic domain (Fig. 1A). Database analysis revealed the
presence of a putative PARP6 in vertebrates but not in
invertebrates. Within vertebrates, the catalytic domain of PARP6 is
highly conserved; in human, mouse, rat and chicken it is completely
identical, and in frog and fish it is 98 and 78% identical,
respectively (Fig. 1B). The
residues of the conserved ‘HYI’ triad, which is critical for the
mono(ADP-ribosyl)ation catalytic activity, are widely present in
vertebrates (Fig. 1B).
During the PARP6 cDNA screening in cDNA libraries
from HeLa cells and SW480 cells, we noticed that the full-length
PARP6 expression was extremely low, and there were two
alternatively spliced forms in these libraries. Both these forms,
PARP6-SP1 (accession number AB499727) and PARP6-SP2 (accession
number AB499728), lack the critical structure of the PARP catalytic
domain (Fig. 2A). To clarify the
effects of PARP6 expression on cell growth, HeLa cells were
transfected with plasmids (pEGFP-PARP6, pEGFP-PARP6-SP1 or
pEGFP-PARP6-SP2) to express EGFP-tagged PARP6 proteins (Fig. 2B). The data clearly revealed that
cell growth was inhibited by full-length PARP6 expression (Fig. 2C). On the other hand,
EGFP-PARP6-SP1 and EGFP-PARP6-SP2, which are catalytically
inactive, had no effect on cell growth (Fig. 2C). These results clearly indicate
that the PARP6 catalytic domain is essential for PARP6-mediated
cell growth inhibition.
PARP6 expression inhibits S-phase
progression
Next, we examined whether PARP6 blocks cell cycle
progression at a specific cell cycle phase. In PARP proteins, the
N-terminal region plays a role in the regulation of the catalytic
activity. Therefore, we constructed an expression vector encoding
an N-terminal deletion form of PARP6 (Fig. 3A). HEK293FT cells, which like HeLa
cells express extremely low levels of PARP6, were transfected with
plasmids [pEGFP-PARP6 or pEGFP-ΔN(1–410)PARP6] to express
EGFP-tagged full-length and N-terminal-deleted PARP6 proteins.
Cells were harvested 24 h after transfection, and cell cycle
distribution and expression level of the transfected plasmid were
analyzed by flow cytometry (Fig.
3B). In low-level-expressing cells (Gate A in Fig. 3C), the S-phase cell populations
were 29.79, 34.40 and 44.76% for EGFP-empty-expressing cells,
EGFP-PARP6-expressing cells and EGFP-ΔN (1–410)PARP6-expressing
cells, respectively. In moderate-level-expressing cells (Gate B in
Fig. 3C), over 90% of EGFP-ΔN
(1–410) PARP6-expressing cells were accumulated in the S-phase. In
high-level-expressing cells (Gate C in Fig. 3C), over 60% of
EGFP-PARP6-expressing cells were accumulated in the S-phase. Thus,
the expression of PARP6 induced S-phase arrest. The magnitude of
S-phase accumulation was greater in EGFP-ΔN (1–410)
PARP6-expressing cells than in EGFP-PARP6-expressing cells. Thus,
the PARP6 catalytic domain is likely to have an important role in
inhibition of S-phase progression.
PARP6 expression correlates with a good
prognosis in colorectal cancer
Because PARP6 functioned as a cell growth inhibitor,
we decided to explore the possibility that PARP6 may act as a tumor
suppressor in human cancer. The expression level of PARP6 was
examined in 126 colorectal cancer cases by immunohistochemistry,
using PARP6 antibody against the catalytic domain. We observed that
57 (45.6%) cases were positive. Among these 126 cases, the
frequency of proliferation marker Ki-67-positive cells was higher
in PARP6-negative cases than in PARP6-positive cases (Fig. 4). Thus, we confirmed that PARP6
negatively regulates cell growth in colorectal cancerous
tissues.
We also observed PARP6 positivity especially in the
cytoplasm of well-differentiated and moderately differentiated
adenocarcinoma, but hardly any in poorly differentiated
adenocarcinoma and mucinous carcinoma (Fig. 5). PARP6 positivity was inversely
correlated with loss of histological differentiation (P<0.01;
Table I). The correlation between
PARP6 positivity and other clinicopathological factors was also
examined, and we found that in primary colorectal cancer with
distant metastasis (P<0.01) and with stage D (P<0.01) the
PARP6-negative cases were significantly higher than the
PARP6-positive cases (Table
I).
 | Table ICorrelation between PARP6 expression
and clinico-pathological factors in colorectal cancer. |
Table I
Correlation between PARP6 expression
and clinico-pathological factors in colorectal cancer.
| PARP6 expression
| |
|---|
| Clinicopathological
factor | Negative 69 | Positive57 | P-value |
|---|
| Tumor size (mm) | | | |
| >50 | 32 | 24 | |
| ≤50 | 37 | 33 | |
| Histological
differentiation | | | |
| Por/Muca | 15 | 1 | <0.01 |
| W/Mb | 54 | 56 | |
| Lymph node
metastasis | | | |
| Negative | 30 | 34 | 0.071 |
| Positive | 39 | 23 | |
| Lymphatic
invasion | | | |
| Negative | 10 | 12 | |
| Positive | 59 | 45 | |
| Venous invasion | | | |
| Negative | 25 | 23 | |
| Positive | 44 | 34 | |
| Metastasis | | | |
| Negative | 49 | 53 | <0.01 |
| Positive | 20 | 4 | |
| Tumor stage | | | |
| B, C | 46 | 51 | <0.01 |
| D | 23 | 6 | |
To determine whether PARP6 expression is emerging as
a prognostic biomarker for survival in colorectal cancer patients,
we examined the survival rate of those 126 cases by the
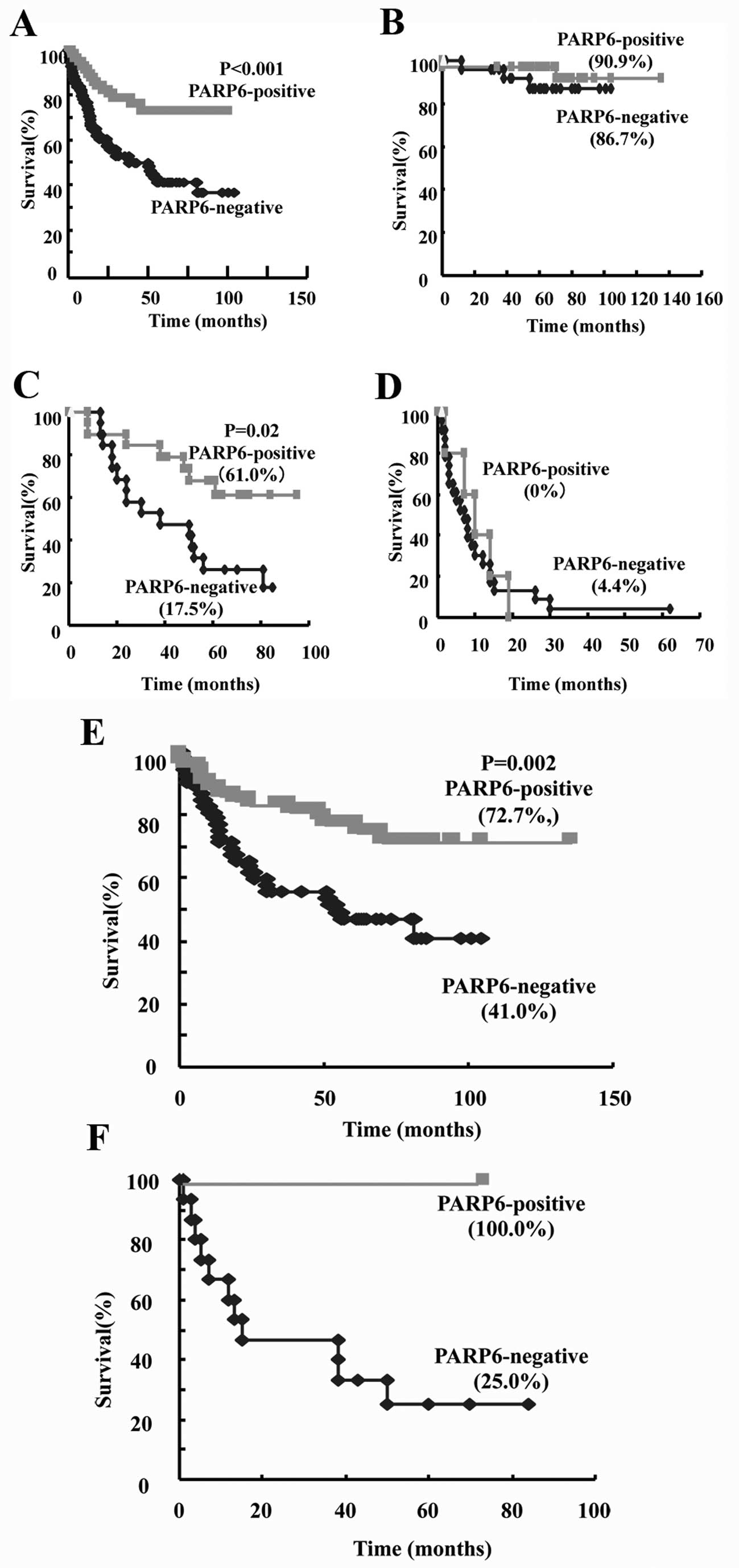
Kaplan-Meier method. The patient survival curves indicated that the
survival rate of PARP6-positive cases was higher than that of the
negative cases (P<0.001; Fig.
6A). When the survival rate within patients with B, C or D
stage was examined, we found that the 5-year-survival rate of
PARP6-positive cases was higher than that of negative cases within
stage C patients, and that it was not significantly different
within stage B or D patients (Fig. 6B,
C and D). When the survival rate within patients with
differentiated or undifferentiated type cancers was examined, we
found that the 5-year-survival rate of PARP6-positive cases was
higher than that of negative cases within the differentiated type
cases (Fig. 6E). A similar pattern
was observed within the undifferentiated type cases (Fig. 6F). In conclusion, PARP6 expression
may become a prognostic biomarker for colorectal cancer patient
survival.
Discussion
To date, the 17 identified PARPs share a PARP domain
and thus may contain polymerase activity (3,6,15,16).
The structure- and sequence-based analyses of the PARP catalytic
core motifs and the loop length (between β-sheets 4 and 5)
have indicated that these PARPs can be divide into three
functionally distinct subgroups according to their catalytic
activity: (i) poly(ADP-ribosyl) polymerase activity, (ii)
mono(ADP-ribosyl) transferase activity and (iii) catalytically
inactive (7). PARP6 has a
histidine (H473) and tyrosine (Y508) that are involved in
NAD+ binding, and an isoleucine (I581) that replaces the
catalytic glutamate found in PARP1 (E988). Moreover, the loop
between β-4 and β-5 is only two residues long in
PARP6. These features are characteristic of mono (ADP-ribosyl)
transferases. This is the first report demonstrating that PARP6 has
a physiological function.
An extremely low level of PARP6 expression was found
in cultured cancer cells such as HeLa cells and HEK293FT cells. We
were also unable to detect PARP6 expression in cultured colorectal
cancer cell lines, including SW480 cells and HCT116 cells (data not
shown). On the other hand, alternatively spliced forms of PARP6
lacking parts of the catalytic domain (PARP6-SP1 and PARP6-SP2)
were found in HeLa cells as well as in other cell lines including
colorectal cancer cells. It is not clear whether these
alternatively spliced species are actually translated in the cells.
However, forced expression of these forms had no effect on cell
growth. In contrast, full-length PARP6 and an N-terminal-deleted
catalytic domain inhibited cell growth, leading to S-phase
accumulation. In previous studies, PARP10, a mono(ADP-ribosyl)
transferase, has been demonstrated to have a growth inhibitory
effect (7,10,13).
It has been suggested that the direct interaction between PARP10
and Myc oncoprotein is the mechanism by which PARP10 functionally
inhibits c-Myc- and Ha-Ras-induced transformation of rat embryo
fibroblasts (8). Although the
anti-oncogenic effect of PARP10 has been considered to be
independent of PARP activity (13), mutational analysis of the PARP10
catalytic domain has implicated the catalytic activity of PARP10 in
growth inhibition (13). In
analogy to PARP10, the data presented in this study suggest that
the catalytic activity of PARP6 is required for negative regulation
of S-phase progression. Furthermore, the expression of
alternatively spliced forms lacking the catalytic domain in cancer
cells suggests dominant-negative effects on growth inhibition if
these forms are translated.
The mechanism by which PARP6 functionally inhibits
cell growth due to S-phase accumulation remains unclear. In the
case of PARP10, protein expression has been detected in both the
cytoplasm and nucleus, and the nuclear PARP10 is likely to be
critical for its function (10).
PARP14, another mono (ADP-ribosyl) transferase, has been reported
as both a cytoplasmic negative regulator of the cancer
metastasis-related protein, AMF (12), and as a nuclear transcription
switch of Stat6-dependent gene activation (8,9,11).
PARP6 was predominantly distributed in the cytoplasm of
well-differentiated adenocarcinoma (Fig. 5), suggesting it has cytoplasmic
target(s). Although elucidation of these molecular target(s)
remains challenging, PARP6 may function in a distinct subcellular
localization from PARP10 and PARP14.
In colorectal cancer, PARP6 expression was detected
in 57 (45.6%) of the 126 cases. Compared to Ki-67 expression, we
confirmed that PARP6 is a possible negative regulator of cell
growth in vivo as well as in vitro. The absence of
PARP6 would be expected to contribute to cancer progression, and
indeed, our analyses indicated that its reduced expression was
associated with a poor prognosis. The different incidence of PARP6
positivity between well-differentiated adenocarcinoma and poorly
differentiated adenocarcinoma suggests that PARP6 functions in
differentiated cells. PARP6 may serve as a novel biomarker for
colorectal cancer, and our present results may provide crucial
information for the creation of a novel therapeutic strategy using
selective PARP inhibitors, which is now becoming a promising
approach in cancer therapy.
Acknowledgements
This study was supported in part by
the Important Research Grant from the Prefectural University of
Hiroshima, Japan. We thank Saki Tomita, Hidehiko Kawai, Kenta
Watanabe, Shou Miyawaki, Takahiko Tsuno, Masato Hori, Mikiko Fujii,
Sanae Koya, Shou Kato, Tomohiro Doi, Satomi Koga, Tomoharu Miki and
Yuki Takeshita for their technical assistance.
References
|
1.
|
D’Amours D, Desnoyers S, D’Silva I and
Poirier GG: Poly(ADP-ribosyl)ation reactions in the regulation of
nuclear functions. Biochem J. 342:249–268. 1999.
|
|
2.
|
Kim MY, Zhang T and Kraus WL: Poly
(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a
nuclear signal. Genes Dev. 19:1951–1967. 2005.
|
|
3.
|
Schreiber V, Dantzer F, Ame JC and de
Murcia G: Poly(ADP-ribose): novel functions for an old molecule.
Nat Rev Mol Cell Biol. 7:517–528. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Hsiao SJ and Smith S: Tankyrase function
at telomeres, spindle poles, and beyond. Biochimie. 90:83–92. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kraus WL: Transcriptional control by
PARP-1: chromatin modulation, enhancer-binding, coregulation, and
insulation. Curr Opin Cell Biol. 20:294–302. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Hakme A, Wong HK, Dantzer F, et al: The
expanding field of poly(ADP-ribosyl)ation reactions. ‘Protein
Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep.
9:1094–1100. 2008.PubMed/NCBI
|
|
7.
|
Kleine H, Poreba E, Lesniewicz K, et al:
Substrate-assisted catalysis by PARP10 limits its activity to
mono-ADP-ribosylation. Mol Cell. 32:57–69. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Cho SH, Ahn AK, Bhargava P, et al:
Glycolytic rate and lymphomagenesis depend on PARP14, an ADP
ribosyltransferase of the B aggressive lymphoma (BAL) family. Proc
Natl Acad Sci USA. 108:15972–15977. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Cho SH, Goenka S, Henttinen T, et al:
PARP-14, a member of the B aggressive lymphoma family, transduces
survival signals in primary B cells. Blood. 113:2416–2425. 2009.
View Article : Google Scholar
|
|
10.
|
Chou HY, Chou HT and Lee SC: CDK-dependent
activation of poly (ADP-ribose) polymerase member 10 (PARP10). J
Biol Chem. 281:15201–15207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Mehrotra P, Riley JP, Patel R, et al:
PARP14 functions as a transcriptional switch for Stat6-dependent
gene activation. J Biol Chem. 286:1767–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Yanagawa T, Funasaka T, Tsutsumi S, et al:
Regulation of phosphoglucose isomerase/autocrine motility factor
activities by the poly(ADP-ribose) polymerase family-14. Cancer
Res. 67:8682–8689. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Yu M, Schreek S, Cerni C, et al: PARP-10,
a novel Myc-interacting protein with poly(ADP-ribose) polymerase
activity, inhibits transformation. Oncogene. 24:1982–1993. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Scherer WF, Syverton JT and Gey GO:
Studies on the propagation in vitro of poliomyelitis viruses. IV.
Viral multiplication in a stable strain of human malignant
epithelial cells (strain HeLa) derived from an epidermoid carcinoma
of the cervix. J Exp Med. 97:695–710. 1953. View Article : Google Scholar
|
|
15.
|
Ame JC, Spenlehauer C and de Murcia G: The
PARP superfamily. Bioessays. 26:882–893. 2004. View Article : Google Scholar
|
|
16.
|
Otto H, Reche PA, Bazan F, et al: In
silico characterization of the family of PARP-like poly
(ADP-ribosyl) transferases (pARTs). BMC Genomics.
6:1392005.PubMed/NCBI
|