Introduction
Non-small cell lung cancer (NSCLC) including
adenocarcinoma (ADC) and squamous cell carcinoma (SQCC) accounts
for approximately 80–90% of all diagnosed lung cancers (1,2).
Despite recent advances in understanding complex pathophysiology,
uncovering predictive/prognostic markers, and developing novel
technologies to aid diagnosis/treatment decision, NSCLC is still
the major determinant of overall cancer mortality (3–7). The
5-year survival rate was 15% across all stages of the
disease-ranging from 2.8% for patients with distant metastases to
nearly 50% for those presenting with local disease. Because NSCLC
is a heterogeneous disease entity, the accurate stratification of
the patients with high risk of developing recurrence or distant
metastasis would improve final prognosis.
While the brain is a major site of relapse
contributing to the unfavorable prognosis of the NSCLC, the
relevant molecular mechanisms are largely unknown. Lung ADC is
known to establish distant macrometastasis within months of initial
diagnosis (8–10). This short latency implies that
metastatic competence would result from early oncogenic events that
drive primary tumor growth rather than late-arising, rare genomic
alterations specific for metastasis (11). Therefore, defining consistent
chromosomal changes in the primary NSCLC could help to identify
metastasis-specific signatures. Molecular inversion probe (MIP)
technology is a high-throughput genotyping capable of providing
copy number measurements with high precision and specificity,
proven to be a valid array for analyzing copy number in
formaldehyde-fixed and paraffin-embedded (FFPE) samples (12–14).
In this study, to delineate prognostic markers to stratify NSCLC
patients at higher risk for developing brain metastasis (BM) and
novel therapeutic candidates targeting BM, the patterns of copy
number alterations (CNA) in primary lung NSCLC FFPE samples were
analyzed by MIP technology.
Materials and methods
Pathology samples from NSCLS patients
with brain metastases
Our study was reviewed and approved by the
Institutional Review Board of the Samsung Medical Center (Seoul,
Korea). All pathology samples and clinical data were obtained with
written informed consent according to the institutional
regulations. In the pathology database of the Samsung Medical
Center (1994.12–2010.7), 160 NSCLC brain metastasis patients were
identified. From them, 36 cases were selected; exclusion criteria,
the biopsy of primary lung cancer without brain metastasis sample;
inclusion criteria, harbouring visible tumors. A total of 37
formaldehyde-fixed and paraffin-embedded (FFPE) pathology samples
were analyzed by this MIP analysis including 3 normal lung and 5
brain controls as follows: 20 ADCs of 8 paired cases (lung cancer
and BM derived from same patient) and 4 only lung cancer cases; 9
SQCCs of 3 paired cases and 3 only lung cancers. Normal lung and
brain tissues were obtained from the patients with other benign
diseases.
All NSCLC patients were classified according to the
standard World Health Organization (WHO) histological typing of
lung carcinomas and the TNM (tumor-node-metastasis) staging system
of the International Union Against Cancer (UICC).
Clinicopathological data including age, gender, tumor stage and
treatment history were obtained by a review of the medical records.
Sites of distant metastasis and disease recurrence after treatment
were traced by serial computed tomography (CT), magnetic resonance
imaging (MRI) and positron emission tomography (PET). BM was
defined as synchronous when metastasis was detected within 3 months
of the initial diagnosis of primary tumor. The others were defined
as metachronous.
Isolation of genomic DNA
Genomic DNA (gDNA) was extracted from three
5-μm-thick sections per FFPE block for each pathology sample.
Deparaffinization and RNA extraction/purification was performed
using a QIAamp DNA FFPE\Tissue kit (Qiagen) according the proposed
protocol. The only change to the standard protocol was to increase
the proteinase K digestion time (overnight). Extracted gDNA was
stored at −20°C, and DNA quantity was analyzed by a Quant-iT
Picogreen dsDNA kit (Invitrogen).
Molecular inversion probe (MIP) assay and
data analysis
The MIP assay was performed using the OncoScan™ FFPE
Express 330K MIP platform (Affymetrix). The MIP assay was performed
as described previously (12,15).
Briefly, gDNA samples (75 ng per each sample) were annealed with
the 24,037 MIP probes (200 amol/μl per probe) in a 384-well plate
on ice at 20°C for 4 minutes (min), at 95°C for 5 min, and then at
58°C overnight. The mixtures were circularized with the addition of
4 μl of the appropriate nucleotide at 58°C for 10 min (gap-fill
ligation). Un-circularized MIP probes and gDNA were eliminated by
addition of 4 μl of exonucleases (37°C for 15 min followed by heat
inactivation). The circularized probes were linearized by
restriction enzyme digest at 37°C for 15 min, and then amplified
using a universal primer for 18 cycles [95°C for 20 seconds (sec),
64°C for 40 sec, and 72°C for 10 sec]. For the labeling reaction,
the amplified products were further amplified with the label
primers for 10 cycles. The MIP polymerase chain reaction (PCR)
products were mixed with hybridization cocktail, denatured, and
hybridized to 30K Universal Tag Arrays (Affymetrix) at 39°C for 16
h with two arrays for each allele. The hybridized arrays were
washed on a standard Affymetrix fluidic station and stained with
streptavidin-phycoerythrin (5 ng/ml, Invitrogen).
Copy number estimation was obtained from the barcode
hybridization signals as described previously (12). The copy number changes of the
NSCLCs were analyzed compared with the copy number of the normal
non-neoplastic lung and brain samples. Data analysis including copy
sum and copy contrast computation, allele ratio, sample
normalization, data smoothing was performed by Nexus copy number
analytics Version 5.1 Software (Biodiscovery), using algorithm
SNP-FASST2 with sensitivity p<0.05.
Results
Detailed clinicopathological data of the 18 NSCLC
patients with BM (12 ADCs and 6 SQCCs) are summarized in Table I. The median age of the NSCLC
patients at the initial diagnosis of the primary cancers was 55
years (range 33–72 years). Synchronous (metastasis within 3 months
of the initial diagnosis of primary NSCLC) and metachronous
(metastasis after 3 months of the initial diagnosis of primary
NSCLC) BM were 6 (33%) and 12 cases (67%), respectively. Median
time between initial diagnosis of primary tumors and onset of
overall BM was 257.5 days (8.6 months). The median lag (range) till
BM for synchronous tumors was 0 (0–88) while for metachronous
tumors the lag was 545 (180–2190) days.
 | Table ICliniopathological features of 18
non-small cell lung cancer (NSCLC) cases studied. |
Table I
Cliniopathological features of 18
non-small cell lung cancer (NSCLC) cases studied.
| Sample ID | Gender/age | Pattern of
metastasis | Subtype | Treatment for
PT | Treatment for
BM | Extracranial
metastasis | Interval between PT
and BM (days) | OSa (days) | PFSb (days) | KRAS mutation | EGFR mutation |
|---|
| 1 | M/62 | S | ADC | Lobectomy, RT | TR, RT, GKRS
(2) | None | 0 | 496 | 110 | G12V | wt |
| 2 | M/66 | S | | Lobectomy, RT | TR, RT | None | 0 | 1098 | 1098 | G12D | wt |
| 3 | F/55 | S | | Lobectomy, RT | TR, RT | None | 0 | 1590 | 1590 | wt | L858R |
| 4 | M/33 | S | | Neoadjuvant CCRT,
lobectomy | TR (2), RT, GKRS
(4) | None | 88 | 670 | 70 | wt | E746-A750 del |
| 5 | F/55 | M | | Lobectomy,
CCRT | TR, RT | None | 2190 | 99 | - | wt | wt |
| 6 | F/66 | M | | Lobectomy, RT | TR, RT | None | 1230 | 933 | 271 | wt | E746-A750 del |
| 7 | F/55 | M | | Lobectomy, RT | TR | Adrenal gland,
liver | 1240 | 30 | - | wt | E746-A750 del |
| 8 | F/46 | M | | Lobectomy,
CCRT | TR, GKRS, RT | T-spine | 780 | 32 | - | wt | wt |
| 9 | M/64 | M | | Neoadjuvant CCRT,
lobectomy | TR, GKRS | None | 1110 | 922 | - | wt | wt |
| 10 | M/51 | M | | Lobectomy, RT | TR, RT | None | 210 | 254 | - | wt | wt |
| 11 | M/45 | M | | Lobectomy | GKRS (2), TR | None | 310 | 922 | - | NA | NA |
| 12 | M/56 | M | | Lobectomy,
CCRT | TR, RT, GKRS | None | 830 | 405 | - | NA | NA |
| 13 | M/67 | S | SQCC | Lobectomy | TR | None | 0 | 24 | - | wt | wt |
| 14 | M/54 | S | | Lobectomy | TR (3), GKRS
(4) | None | 0 | 2633 | 1607 | wt | wt |
| 15 | F/47 | M | | Lobectomy,
CTx. | TR, RT | None | 180 | 69 | - | wt | wt |
| 16 | M/72 | M | | Lobectomy,
CTx. | TR, RT | None | 310 | 1210 | - | wt | wt |
| 16 | M/51 | M | | Lobectomy,
CCRT | TR (2), RT, GKRS
(2) | None | 210 | 470 | 390 | wt | wt |
| 17 | F/52 | M | | Lobectomy,
CTx. | TR, GKS (2),
RT | None | 305 | 136 | - | wt | wt |
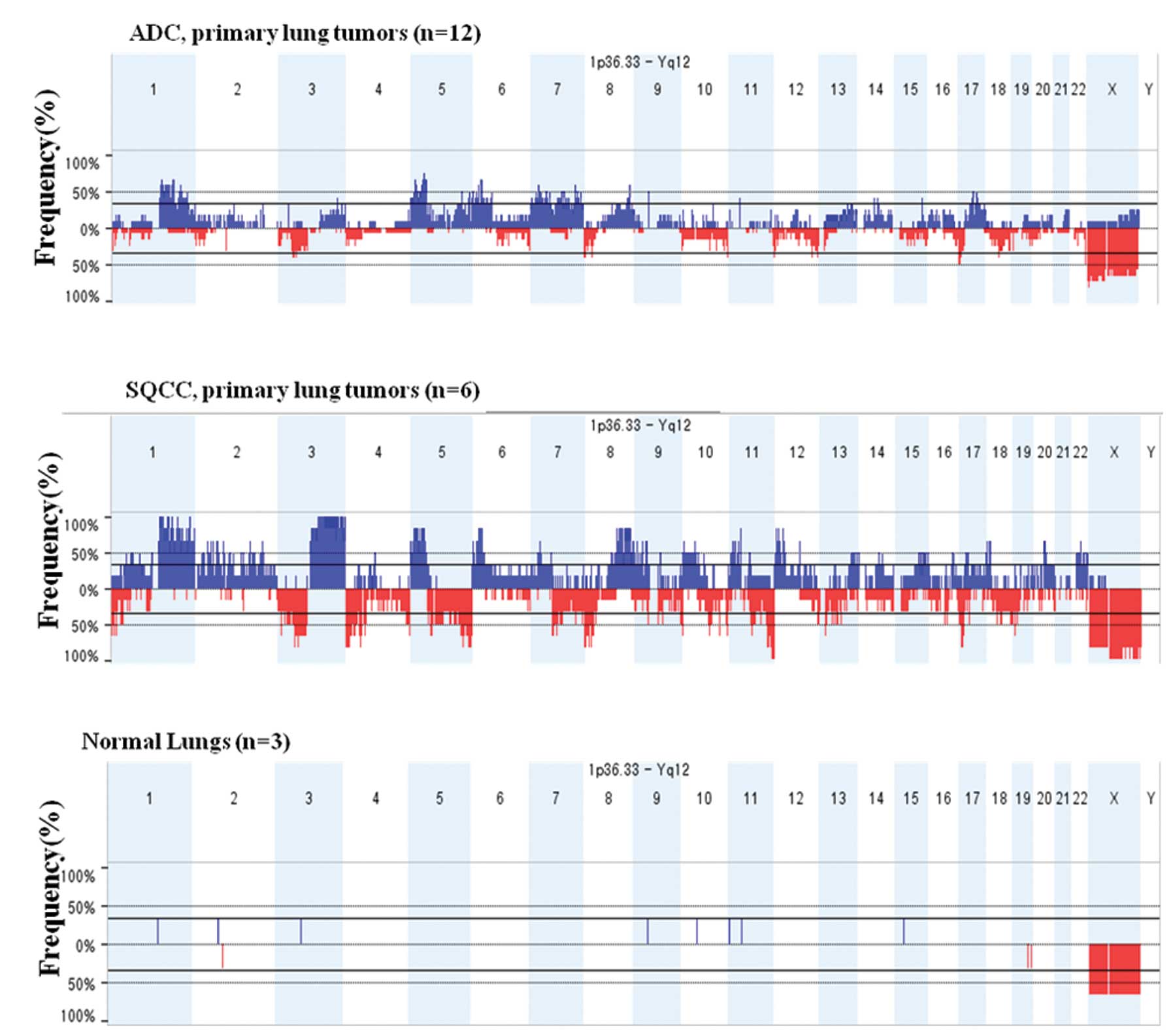
Specific copy number alterations
depending on distinct histologic-subtypes of non-small cell lung
cancer
In this study, chromosomal aberration of 18 NSCLC
lung samples (12 ADCs and 6 SQCCs) and 3 normal lung controls was
analyzed by the MIP technology to identify chromosomal aberrations
involved in the distinct pathogenesis. Common genomic aberrations
were defined as continuous genomic regions where the average signal
in tumors consistently and significantly differed from that of
normal lung samples. Since there is growing evidence that lung ADC
and SQCC have distinct oncogenic mutations and divergent
therapeutic responses (16,17),
12 ADCs and 6 SQCCs were analyzed separately. A full view of the
frequently detected significant chromosomal aberrations is
presented in Fig. 1. The most
frequent aberrations identified in primary ADCs (>40% of cases)
were 3q, 5p, 5q, 6p, 8q, 9p, 11p, 15q and 17q for copy number gains
and 10q and 22q for losses (Table
II). On the other hand, primary SQCCs revealed frequent copy
number gains in 4q and 12q and copy number loss in 9q (Table II). Previously, whole-genome array
comparative genomic hybridization (aCGH) identified significantly
different chromosomal signatures between two subtypes of the NSCLC
(18). Compared with the previous
results using different experimental techniques, similar patterns
of CNAs were observed in this study, confirming the validity of the
MIP technology (19–22). However, several studies
demonstrated additional CNAs including gains of 1q and losses of 9q
and 10p in ADC and gains of 3q, 7p, 12p and 20q as well as losses
of 2q, 3p, 16p and 17p in SQCC (21,23–25).
 | Table IIFrequent regions of
adenocarcinoma-specific and squamous cell carcinoma-specific copy
number alteration. |
Table II
Frequent regions of
adenocarcinoma-specific and squamous cell carcinoma-specific copy
number alteration.
| NSCLC subtype | Chromosomal
aberration | Region (start
site-end site) | Cytoband
location | Frequency % | P-value |
|---|
| Adenocarcinoma
(n=12) | CN gain |
chr3:173,271,977–173,427,105 | q26.31 | 41.7 | 0.011 |
| CN gain |
chr5:40,349,188–40,886,467 | p13.1 | 75 | 0.018 |
| CN gain |
chr5:150,438,972–150,523,583 | q33.1 | 50 | 0.001 |
| CN gain |
chr5:172,100,206–172,250,021 | q35.1–q35.2 | 50 | 0.001 |
| CN gain |
chr6:26,128,335–26,352,388 | p22.1 | 66.7 | 0.004 |
| CN gain |
chr8:134,026,676–134,089,398 | q24.22 | 58.3 | 0 |
| CN gain |
chr9:43,997,314–44,825,192 | p11.2 | 50 | 0 |
| CN loss |
chr10:133,727,624–135,374,737 | q26.3 | 41.7 | 0.026 |
| CN gain |
chr11:35,117,949–35,240,617 | p13 | 41.7 | 0 |
| CN gain |
chr15:83,849,162–83,995,470 | q25.3 | 41.7 | 0 |
| CN gain |
chr17:45,586,978–45,621,111 | q21.33 | 50 | 0.015 |
| CN gain |
chr17:55,139,289–55,290,700 | q23.1 | 50 | 0.015 |
| CN loss |
chr22:47,207,744–47,732,937 | q13.32 | 50 | 0 |
| Squamous cell
carcinoma (n=6) | CN gain |
chr4:87,398,382–87,491,412 | q21.3 | 50 | 0.028 |
| CN loss |
chr9:139,183,759–140,273,252 | q34.3 | 66.7 | 0.042 |
| CN gain |
chr12:74,190,395–74,668,717 | q21.2 | 66.7 | 0.026 |
Continuous somatic evolution eventually giving rise
to overt metastasis revealed the metastasis-specific related genes
that mediate or impede metastatic progression without affecting
primary malignancy (26). When
patterns of CNAs in 11 BM (8 ADCs and 3 SQCCs) were compared with
those of corresponding primary lung tumors, BM were found to carry
the majority of genetic alterations present in the corresponding
primary tumors (data not shown). However, in ADCs, BM harbored
specific CNAs; new 11p and 15q gain (data not shown). In SQCCs, no
BM specific CNAs were detected, likely because of the small number
(n=3) of the SQCC cases. Therefore, BM specific CNAs were further
analyzed in the ADCs.
Genetic signatures associated with early
brain metastasis of lung adenocarcinoma
Although comparing the genetic alterations between
primary and metastatic tumor would have functional implications,
BM-associated genetic signatures in the primary lung ADC would have
been more clinically relevant. Lung ADC is characterized by the
early development of BM and the incidence of BM based on autopsy
findings was as high as 50% in patients with lung ADC (8,27).
Given that BM significantly worsens prognosis of lung ADC patients,
lung ADC patients who are likely to develop BM need adjuvant
treatments for BM. Primary lung ADCs with early development of BM
(synchronous) would contain more CNAs predictive of metastatic
potential or aggressive transformation. To uncover complex genetic
‘signatures’ that can predict the risk of BM, the copy number
changes of 4 lung ADCs with synchronous BM were compared with those
of 8 lung ADCs with metachronous BM (Table III and Fig. 2). Amplification in 5q35.1–2,
10q23.31, 17q23.3–24.1 and 17q24.1 was detected in 100, 75, 100 and
75% of the cases with synchronous BM, respectively, significantly
more frequent than that of lung ADCs with metachronous BM (Table III). Differentially gained regions
between primary ADCs with synchronous and metachronous BM were
found to contain putative metastasis promoting genes, NeurL1B,
ACTA2, FAS and ICAM2 (Table
III).
 | Table IIIThe comparison of recurrent copy
number alterations in primary lung adenocarcinomas derived from the
patients with synchronous (n=4) and metachronous (n=8) brain
metastasis. |
Table III
The comparison of recurrent copy
number alterations in primary lung adenocarcinomas derived from the
patients with synchronous (n=4) and metachronous (n=8) brain
metastasis.
| Event | Chromosome
cytoband | Start | End | Region length | Genes | Frequency %
synchronous BM (Total cases = 8) | Frequency % BM
metachronous (Total cases = 4) | P-value |
|---|
| Gain | 5q35.1 | 172008530 | 172100206 | 91,676 | NeurL1B | 100 | 12.5 | 0.01 |
| 5q35.2 | 172250021 | 172277415 | 27394 | ERGIC1 | 100 | 12.5 | 0.01 |
| 10q23.31 | 90682710 | 90735753 | 53043 | ACTA2 | 75 | 0 | 0.018 |
| 10q23.31 | 90749267 | 90751025 | 1758 | FAS | 75 | 0 | 0.018 |
| 17q23.3 | 59424619 | 59455000 | 30381 | C17orf72,
ICAM2 | 100 | 0 | 0.002 |
| 17q24.1 | 60155862 | 60203841 | 47979 | LOC146880 | 100 | 12.5 | 0.01 |
| 17q24.1 | 60635140 | 60665382 | 30242 | RGS9 | 75 | 0 | 0.018 |
Discussion
Although the BM affecting up to 25% of NSCLC during
their lifetime negatively impacts survival, currently, there are no
standard practice measures to reduce BM risk in NSCLC (28,29).
Under the hypothesis that the genes residing in amplified or
deleted regions in each subtype play an important role in the
histology-specific pathogenesis of NSCLC, this study was designed
to identify ‘meta-signatures’ that stratify NSCLC patients at
higher risk for BM. The NSCLC harbours several histopathologically
and molecularly distinct subtypes including ADC and SQCC. However,
important molecular differences between primary lung ADC and SQCC
have been identified, suggesting that future targeted therapies
need to be histology-specific (8,16,17,30–36).
For example, KRAS and epidermal growth factor receptor (EGFR) gene
mutations are found almost exclusively in ADCs rather than SQCCs.
Therefore, primary lung ADCs were separately analyzed in this
study.
Due to increasing incidence and substantial relapse
rate, the pattern of CNAs was compared between primary ADCs derived
from patients with synchronous and metachronous BM to find
brain-specific meta-signature and putative targets associated with
BM in ADC subtype. Several putative genes reported to be involved
in tumorigenesis of various cancers were demonstrated in our study.
For example, Neuralized-1B is the E3 ubiquitin ligase that is
required for endocytosis regulating both the receptor and ligand
side in Notch signaling (37,38).
Relatively frequent deregulation of the Notch pathway and Notch
ligand Jagged2/miR-200-dependent pathway in NSCLC indicates the
significance and mechanisms underpinning Notch pathway activation
in lung cancer (39–41). In addition, Fas signalling exhibits
tumor-promoting effects by increasing proliferation and
invasiveness (42–46). Fas can promote lung cancer growth
by recruiting MDSC via cancer cell-derived PGE2 and signal for cell
invasion via the glycogen synthase kinase 3β pathway (47,48).
Human smooth muscle α-actin (SMA/ACTA2) has been used as one of
mesenchymal cell-specific markers showing the
epithelial-to-myofibroblast transition involving actin-skeleton
remodeling and myogenic reprogramming. The appearance of dot-like
α-SMA staining in cytokeratin positive cells may indicate the
initial phase of the epithelial to mesenchymal transition (EMT)
(49–54). Finally, a recent study reported
ICAM2 as a mediator for a survival signal sufficient to block
apoptosis by activation of the PI3K/AKT pathway (55). As tumor progression and response to
treatment are determined by numerous co-dependent prognostic
factors, a multi-genetic approach to determining the optimal
treatment for individual patients is more likely to be successful
rather than a single prognostic biomarker.
Evidence that highly metastatic clones from primary
tumor had a higher rate of genetic mutability than non-metastatic
clones from the same tumor provided an early link between
metastasis and intrinsic genetic instability such as mutations and
chromosomal rearrangements (56).
Although several high-throughput technologies have been developed
to detect genomic CNAs in a variety of cancers until recently, high
sensitivity to detect single copy number changes and the ability to
test FFPE samples in which DNA is known to be degraded to different
degrees is important (12).
Analyzing FFPE samples allows the utilization of the vast majority
of cancer samples since most cancer tissue samples are available as
FFPE. For our FFPE NSCLC samples, the MIP technology was used,
which has been validated to be able to substitute aCGH (12–15,57,58).
Because MIP uses probes that have a genomic footprint of ∼40 bp,
this high density platform only requires small amounts of DNA and,
consequently, is well-suited for the analysis of degraded DNA in
FFPE tissues (59). The results of
this study using the MIP technique showed differential genetic
alteration between lung ADCs and SQCCs, correlating well with
previous studies using aCGH. The similarities support the validity
of the technique and the results from it. However, due to
inadequate amount of extracted gDNA, quantitative-real-time-PCR
(qRT-PCR) to validate copy number alterations identified by the MIP
array could not be performed in our study.
In this study, we identified several genes
associated with early BM of lung ADCs. Although our data suggested
a new aspect of genomic alterations associated with BM of NSCLC,
further biological studies to validate the role of identified
candidates in brain-specific metastasis and testing the predictive
power of our meta-signature in larger NSCLC samples would be
required. In spite of several limitations of our study including
very small number of NSCLC samples and absence of validation
studies such as qRT-PCR, these genomic signatures may help to
generate useful markers to refine prognosis and guide therapeutic
decisions for improvement of prognosis and quality of life for
patients with NSCLC.
Acknowledgements
This study was supported by a grant of
the Korea Healthcare Technology R&D Project, Ministry for
Health and Welfare Affairs, Republic of Korea (A092255).
References
|
1.
|
Choi YW, Choi JS, Zheng LT, et al:
Comparative genomic hybridization array analysis and real time PCR
reveals genomic alterations in squamous cell carcinomas of the
lung. Lung Cancer. 55:43–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Travis WD, Brambilla E, Muller-Hermelink
HK and Harris CC: World Health Organization Classification of
Tumors. Pathology and Genetics of the Lung, Pleura, Thymus and
Heart. IARC Press; Lyon: 2004
|
|
3.
|
Spira A and Ettinger DS: Multidisciplinary
management of lung cancer. N Engl J Med. 350:379–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Salgia R, Hensing T, Campbell N, et al:
Personalized treatment of lung cancer. Semin Oncol. 38:274–283.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37(Suppl
8): S4–S66. 2001.PubMed/NCBI
|
|
6.
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
7.
|
Moran C: Importance of molecular features
of non-small cell lung cancer for choice of treatment. Am J Pathol.
178:1940–1948. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Hoffman PC, Mauer AM and Vokes EE: Lung
cancer. Lancet. 355:479–485. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Hess KR, Varadhachary GR, Taylor SH, et
al: Metastatic patterns in adenocarcinoma. Cancer. 106:1624–1633.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Feld R, Rubinstein LV and Weisenberger TH:
Sites of recurrence in resected stage I non-small-cell lung cancer:
a guide for future studies. J Clin Oncol. 2:1352–1358.
1984.PubMed/NCBI
|
|
11.
|
Bernards R and Weinberg RA: A progression
puzzle. Nature. 418:8232002. View
Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Wang Y, Moorhead M, Karlin-Neumann G, et
al: Allele quantification using molecular inversion probes (MIP).
Nucleic Acids Res. 33:e1832005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Johnson CE, Gorringe KL, Thompson ER, et
al: Identification of copy number alterations associated with the
progression of DCIS to invasive ductal carcinoma. Breast Cancer Res
Treat. 133:889–898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Schiffman JD, Wang Y, McPherson LA, et al:
Molecular inversion probes reveal patterns of 9p21 deletion and
copy number aberrations in childhood leukemia. Cancer Genet
Cytogenet. 193:9–18. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Wang Y, Moorhead M, Karlin-Neumann G, et
al: Analysis of molecular inversion probe performance for allele
copy number determination. Genome Biol. 8:R2462007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Travis WD and Rekhtman N: Pathological
diagnosis and classification of lung cancer in small biopsies and
cytology: strategic management of tissue for molecular testing.
Semin Respir Crit Care Med. 32:22–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Travis WD, Rekhtman N, Riley GJ, et al:
Pathologic diagnosis of advanced lung cancer based on small
biopsies and cytology: a paradigm shift. J Thorac Oncol. 5:411–414.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Kang JU, Koo SH, Kwon KC, Park JW and Kim
JM: Identification of novel candidate target genes, including
EPHB3, MASP1 and SST at 3q26.2–q29 in squamous cell carcinoma of
the lung. BMC Cancer. 9:2372009.PubMed/NCBI
|
|
19.
|
Yokoi S, Yasui K, Iizasa T, Imoto I,
Fujisawa T and Inazawa J: TERC identified as a probable target
within the 3q26 amplicon that is detected frequently in non-small
cell lung cancers. Clin Cancer Res. 9:4705–4713. 2003.PubMed/NCBI
|
|
20.
|
Ubagai T, Matsuura S, Tauchi H, Itou K and
Komatsu K: Comparative genomic hybridization analysis suggests a
gain of chromosome 7p associated with lymph node metastasis in
non-small cell lung cancer. Oncol Rep. 8:83–88. 2001.PubMed/NCBI
|
|
21.
|
Kang JU, KS, Kwon KC, Park JW, Shin SY,
Kim JM and Jung SS: High frequency of genetic alterations in
non-small cell lung cancer detected by multi-target fluorescence in
situ hybridization. J Korean Med Sci. 22:47–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Dehan E, Ben-Dor A, Liao W, et al:
Chromosomal aberrations and gene expression profiles in non-small
cell lung cancer. Lung Cancer. 56:175–184. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Chujo M, Noguchi T, Miura T, Arinaga M,
Uchida Y and Tagawa Y: Comparative genomic hybridization analysis
detected frequent overrepresentation of chromosome 3q in squamous
cell carcinoma of the lung. Lung Cancer. 38:23–29. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kang JU, Koo SH, Kwon KC, Park JW and Jung
SS: Gain of the EGFR gene located on 7p12 is a frequent and early
event in squamous cell carcinoma of the lung. Cancer Genet
Cytogenet. 184:31–37. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Massion PP, Kuo WL, Stokoe D, et al:
Genomic copy number analysis of non-small cell lung cancer using
array comparative genomic hybridization: implications of the
phosphatidylinositol 3-kinase pathway. Cancer Res. 62:3636–3640.
2002.
|
|
26.
|
Kikuchi T, Daigo Y, Ishikawa N, et al:
Expression profiles of metastatic brain tumor from lung
adenocarcinomas on cDNA microarray. Int J Oncol. 28:799–805.
2006.PubMed/NCBI
|
|
27.
|
Robnett TJ, Machtay M, Stevenson JP,
Algazy KM and Hahn SM: Factors affecting the risk of brain
metastases after definitive chemo-radiation for locally advanced
non-small-cell lung carcinoma. J Clin Oncol. 19:1344–1349.
2001.PubMed/NCBI
|
|
28.
|
Grinberg-Rashi H, Ofek E, Perelman M, et
al: The expression of three genes in primary non-small cell lung
cancer is associated with metastatic spread to the brain. Clin
Cancer Res. 15:1755–1761. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Oh Y, Taylor S, Bekele BN, et al: Number
of metastatic sites is a strong predictor of survival in patients
with nonsmall cell lung cancer with or without brain metastases.
Cancer. 115:2930–2938. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Tam IY, Chung LP, Suen WS, et al: Distinct
epidermal growth factor receptor and KRAS mutation patterns in
non-small cell lung cancer patients with different tobacco exposure
and clinicopathologic features. Clin Cancer Res. 12:1647–1653.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Mok TS, Wu YL, Thongprasert S, et al:
Gefitinib or carboplatinpaclitaxel in pulmonary adenocarcinoma. N
Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Shaw AT, Yeap BY, Mino-Kenudson M, et al:
Clinical features and outcome of patients with non-small-cell lung
cancer who harbor EML4-ALK. J Clin Oncol. 27:4247–4253. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Scagliotti G, Brodowicz T, Shepherd FA, et
al: Treatment-byhistology interaction analyses in three phase III
trials show superiority of pemetrexed in nonsquamous non-small cell
lung cancer. J Thorac Oncol. 6:64–70. 2011.PubMed/NCBI
|
|
34.
|
Chansky K, Sculier JP, Crowley JJ, Giroux
D, Van Meerbeeck J and Goldstraw P: The International Association
for the Study of Lung Cancer Staging Project: prognostic factors
and pathologic TNM stage in surgically managed non-small cell lung
cancer. J Thorac Oncol. 4:792–801. 2009. View Article : Google Scholar
|
|
35.
|
Pisters KM, Evans WK, Azzoli CG, et al:
Cancer Care Ontario and American Society of Clinical Oncology
adjuvant chemotherapy and adjuvant radiation therapy for stages
I–IIIA resectable non-small-cell lung cancer guideline. J Clin
Oncol. 25:5506–5518. 2007.PubMed/NCBI
|
|
36.
|
Bass AJ, Watanabe H, Mermel CH, et al:
SOX2 is an amplified lineage-survival oncogene in lung and
esophageal squamous cell carcinomas. Nat Genet. 41:1238–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Rullinkov G, Tamme R, Sarapuu A, et al:
Neuralized-2: expression in human and rodents and interaction with
Delta-like ligands. Biochem Biophys Res Commun. 389:420–425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Song R, Koo BK, Yoon KJ, et al:
Neuralized-2 regulates a Notch ligand in cooperation with Mind
bomb-1. J Biol Chem. 281:36391–36400. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Collins BJ, Kleeberger W and Ball DW:
Notch in lung development and lung cancer. Semin Cancer Biol.
14:357–364. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Westhoff B, Colaluca IN, D’Ario G, et al:
Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci
USA. 106:22293–22298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Yang Y, Ahn YH, Gibbons DL, et al: The
Notch ligand Jagged2 promotes lung adenocarcinoma metastasis
through a miR-200-dependent pathway in mice. J Clin Invest.
121:1373–1385. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Peter ME, Budd RC, Desbarats J, et al: The
CD95 receptor: apoptosis revisited. Cell. 129:447–450. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Peter ME, Legembre P and Barnhart BC: Does
CD95 have tumor promoting activities? Biochim Biophys Acta.
1755:25–36. 2005.PubMed/NCBI
|
|
44.
|
Mitsiades CS, Poulaki V, Fanourakis G, et
al: Fas signaling in thyroid carcinomas is diverted from apoptosis
to proliferation. Clin Cancer Res. 12:3705–3712. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Barnhart BC, Legembre P, Pietras E, Bubici
C, Franzoso G and Peter ME: CD95 ligand induces motility and
invasiveness of apoptosis-resistant tumor cells. EMBO J.
23:3175–3185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Chen L, Park SM, Tumanov AV, et al: CD95
promotes tumour growth. Nature. 465:492–496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Zhang Y, Liu Q, Zhang M, Yu Y, Liu X and
Cao X: Fas signal promotes lung cancer growth by recruiting
myeloid-derived suppressor cells via cancer cell-derived PGE2. J
Immunol. 182:3801–3808. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Kleber S, Sancho-Martinez I, Wiestler B,
et al: Yes and PI3K bind CD95 to signal invasion of glioblastoma.
Cancer Cell. 13:235–248. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Masszi A, Speight P, Charbonney E, et al:
Fate-determining mechanisms in epithelial-myofibroblast transition:
major inhibitory role for Smad3. J Cell Biol. 188:383–399. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Wang J, Zohar R and McCulloch CA: Multiple
roles of alpha-smooth muscle actin in mechanotransduction. Exp Cell
Res. 312:205–214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Han M, Liu M, Wang Y, et al: Re-expression
of miR-21 contributes to migration and invasion by inducing
epithelial-mesenchymal transition consistent with cancer stem cell
characteristics in MCF-7 cells. Mol Cell Biochem. 363:427–436.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Owens GK and Thompson MM: Developmental
changes in isoactin expression in rat aortic smooth muscle cells in
vivo. Relationship between growth and cytodifferentiation. J Biol
Chem. 261:13373–13380. 1986.PubMed/NCBI
|
|
53.
|
Pirozzi G, Tirino V, Camerlingo R, et al:
Epithelial to mesenchymal transition by TGFbeta-1 induction
increases stemness characteristics in primary non-small cell lung
cancer cell line. PLoS One. 6:e215482011. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Valcz G, Sipos F, Krenacs T, et al:
Increase of alpha-SMA(+) and CK (+) cells as an early sign of
epithelial-mesenchymal transition during colorectal carcinogenesis.
Pathol Oncol Res. 18:371–376. 2012.
|
|
55.
|
Perez OD, Kinoshita S, Hitoshi Y, et al:
Activation of the PKB/AKT pathway by ICAM-2. Immunity. 16:51–65.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Fidler IJ: The pathogenesis of cancer
metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003.
|
|
57.
|
Ji H, Kumm J, Zhang M, et al: Molecular
inversion probe analysis of gene copy alterations reveals distinct
categories of colorectal carcinoma. Cancer Res. 66:7910–7919. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Lapuk A, Volik S, Vincent R, et al:
Computational BAC clone contig assembly for comprehensive genome
analysis. Genes Chromosomes Cancer. 40:66–71. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Wang Y, Carlton VE, Karlin-Neumann G, et
al: High quality copy number and genotype data from FFPE samples
using Molecular Inversion Probe (MIP) microarrays. BMC Med
Genomics. 2:82009. View Article : Google Scholar : PubMed/NCBI
|