Introduction
The trefoil factor (TFF) family is a group of
protease-resistant peptides characterized by a conserved three-loop
domain, designated as a TFF domain (1–3). The
TFF family consists of pS2 (TFF1), spasmolytic polypeptide SP
(TFF2) and intestinal trefoil factor (TFF3) (4), which are all widely expressed in the
gastrointestinal epithelium. TFF1 was initially described as an
estrogen-inducible gene in the hormone-sensitive breast cancer cell
line MCF-7 (5), but was later
found to be spontaneously expressed at a high level in gastric
epithelial cells (6). TFF1
interacts with mucin core proteins to stabilize the surface mucus
gel layers of the gastric epithelium and plays an important role in
the protection and regeneration of the gastric mucosa (2,7,8). On
the contrary, TFF1-deficient mice develop antral neoplasms of the
stomach (9), thus suggesting that
TFF1 acts as a tumor-suppressor gene in the stomach. Moreover,
several investigators have reported that TFF1 expression is related
to carcinogenesis and metastasis in various types of cancers, such
as prostate, breast, pancreatic and upper gastrointestinal cancer
(10–14).
The mechanisms regulating TFF1 expression were
previously addressed. It has been demonstrated that several similar
sequences at the 5′-flanking regions of human TFF genes (motifs
I–IV) play an important role in the stomach-specific expression of
TFF1 (15,16). GATA-6 activates TFF1 expression in
the stomach via motif III (17)
and HNF-3 activates TFF1 transcription via motif IV, located close
to the TATA box (18). Hernández
et al(19) also indicated
that hypoxia inducible factor (HIF)-1 mediates the induction of
TFF1 expression in gastric epithelial cells under hypoxic
conditions. However, few studies have so far examined the clinical
significance of TFF1 expression and its regulatory mechanism in
gastric cancer patients.
The present study evaluated the immunohistochemical
expression of TFF1 in 182 gastric cancer patients and examined
whether or not TFF1 is associated with the clinicopathological
factors and patient survival. In addition, to clarify the mechanism
regulating TFF1 expression, we focused on the methylation status of
the TFF1 promoter region.
Materials and methods
Patients
One hundred and eighty-two patients with advanced
gastric cancer who underwent curative surgery at our institution
between June 2000 and December 2008 were enrolled. None of these
patients had hepatic, peritoneal, or distant metastasis, nor any
tumor cells in the peritoneal fluid. The stage classification was
performed according to the guidelines of the Japanese Gastric
Cancer Association (20). Informed
consent to use specimens was obtained from all patients and the
study protocol was approved by the Ethics Committee of Saga
University Faculty of Medicine.
Cell culture
Eight gastric cancer cell lines (MKN1, MKN7, MKN28,
MKN45, MKN74, HSC45, HSC57 and KATO-III) were used for the study.
HSC45 and HSC57 were provided by Dr K. Yanagihara (National Cancer
Center Research Institute, Tokyo, Japan) (21,22)
and the remaining six cell lines were purchased from Cell Bank,
RIKEN BioResource Center (Ibaraki, Japan). The cells were
maintained under either normoxic conditions (containing 20%
O2 and 5% CO2 in air) or hypoxic conditions
(containing 1% O2, 5% CO2 and 94%
N2).
Immunohistochemistry
Paraffin-embedded sections (4-μm-thick) were
incubated with an anti-TFF1 antibody (1:100 dilution; sc-28925,
Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at room
temperature and with the corresponding secondary antibody for 30
min. The slides were washed in PBS and incubated with a
diaminobenzidine substrate kit (Nichirei Corp., Tokyo, Japan). The
level of cytoplasmic staining for TFF1 was classified as high
expression and low expression, according to the percentage of
stained cancer cells. Tumors showing 30% or more positive cancer
cells were considered to have high expression, while <30%
staining was judged as the low expression.
Total-RNA extraction and real-time
RT-PCR
RT-PCR was performed using the Light Cycler
instrument system (Roche Diagnostics GmbH, Mannheim, Germany)
according to the manufacturer’s instructions. The primers were
designed as follows: TFF1, 5′-TTGTGGTTTTCCTGGTGTCA-3′, 5′-GGGACG
TCGATGGTATTAGG-3′ (111 bp) and β-actin, 5′-TTAAGG
AGAAGCTGTGCTACG-3′, 5′-GTTGAAGGTAGTTTCGTG GAT-3′ (206 bp). A
melting curve analysis was used to control for the specificity of
the amplification products. The quantitative value was normalized
to the β-actin expression level, which was used as an internal
control.
DNA extraction and bisulfite
sequencing
The frozen tissue specimens from six representative
patients were cut into 8 μm-thick sections and cancer foci
were macrodissected. Genomic DNA from gastric cancer cell lines and
macrodissected cancer foci obtained from gastric cancer tissues was
extracted using a DNeasy Blood & Tissue kit (Qiagen, Hilden,
Germany). Bisulfite modification was then carried out using an
EpiTect Plus Bisulfite kit (Qiagen) with 500 ng of genomic DNA.
Bisulfite-specific primers were designed as follows:
5′-TTTGTTTTGGTTTTTTAAAGTGTTG-3′, 5′-TTCTCC ATAATAACCATTACCTCCT-3′
(611 bp) and 5′-GAGGAG GTAATGGTTATTATGGAGA-3′, 5′-AAACCCTACCAC
CCTAAATTACTCT-3′ (283 bp). Bisulfite PCR was performed using the
Takara ExTaq Hot Start Version (Takara, Shiga, Japan). The PCR
products were purified using a QIAquick PCR Purification kit
(Qiagen) and then ligated into the pT7Blue-T-vector (Novagen,
Madison, WI) using a DNA Ligation kit Ver.2.1 (Takara). E.
coli DH5α competent cells (Toyobo Co., Ltd., Osaka, Japan) were
transformed by inserting the vector described above and were seeded
on LB plates containing the appropriate antibiotics and incubated
overnight at 37°C. For each sample, at least 10 clones were
sequenced by a dye terminator method on an Applied Biosystems 3130
Genetic Analyzer (Applied Biosystems, Inc., Foster City, CA) using
a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems).
The results were analyzed using the Sequence Scanner Software
program, v1.0 (Applied Biosystems).
Knockdown of TFF1
siRNA oligos for the human TFF1 gene
(SASI_Hs01_00121167) and non-silencing (scrambled) siRNA (siRNA
Universal Negative Control #1) were purchased from Sigma-Aldrich,
Inc. The siRNA oligos were transiently transfected into HSC45 and
HSC57 cells using a MicroPorator-mini (MP-100) (Digital Bio
Technology, Seoul, Korea) in combination with the Neon™
100-μl kit (Invitrogen Corp., Carlsbad, CA), according to
the manufacturer’s instructions. The transfected cells were
cultured in complete medium and used on 1 to 4 days of culture.
Western blot analysis confirmed that the silencing effect on the
gene expression continued for 144 h after the transfection.
Western blot analysis
Whole-cell lysates from cultured cells were prepared
using lysis buffer as described in a previous report (23). Aliquots containing 50 μg of
protein were subjected to 4 to 12% Bis-Tris gel electrophoresis
(NuPAGE; Invitrogen) and were transferred onto Amersham Hybond-ECL
membranes (GE Healthcare, Buckinghamshire, UK) in transfer buffer.
After blocking with 5% skim milk for 60 min, the membranes were
incubated with primary antibodies overnight at 4°C. The primary
antibodies were anti-TFF1 (1:1000 dilution; 2801-1, Epitomics,
Burlingame, CA) and anti-β-actin (1:10,000 dilution; Sigma-Aldrich,
Inc.). After incubation with the corresponding secondary
antibodies, the signals were developed using an Amersham ECL Prime
Western Blotting Detection System (GE Healthcare).
In vitro cell invasion assay
Polycarbonate filters (6.5-mm diameter, 8-μm
pore size) of the Falcon Transwell™ chemo-taxis chambers
(Becton-Dickinson, Franklin Lakes, NJ) were coated with 50
μl (1 mg/ml) of Matrigel biomatrix (Becton-Dickinson) in
cold RPMI-1640 medium and dried overnight. Suspensions of
1×105 (HSC45) or 5×105 (HSC57) cells in 200
μl of complete RPMI-1640 medium were placed in the upper
compartments of the chamber, whereas the lower compartments were
filled with 800 μl of conditioned medium from MRC5
fibroblasts. These culture units were incubated for 48 h at 37°C
under normoxia. The number of viable invasive cells, which had
infiltrated onto the lower surface of the filter, was counted.
Cell proliferation assay
Cell proliferation was analyzed by the MTT assay
using a Cell-Titer 96™ non-radioactive cell proliferation assay kit
(Promega, Madison, WI). In brief, 1×103 (HSC45) or
5×103 (HSC57) cells per well were seeded in triplicate
onto 96-well plates and incubated at 37°C in a humidified
atmosphere. After 24 h, the numbers of viable cells were measured
in triplicate every day for four days. The proliferation curves
were then constructed by calculating the mean value of the optical
density measurements at 590 nm using a 96-well plate reader
(Immuno-mini NJ2300, Nalge Nunc International K.K., Tokyo,
Japan).
Statistical analysis
The statistical analysis was performed using the
computer software program SPSS 15.0J for windows (Chicago, IL). The
χ2 test was used to compare categorical data and
differences in the mean values were evaluated by Student’s t-test
and the Mann-Whitney U test. Both the univariate analysis and
multivariate analysis for survival were performed using Cox’s
proportional hazards model. The survival curves were generated
using the Kaplan-Meier method and the statistical significance of
differences was compared using the log-rank test. Values of
p<0.05 were considered to be statistically significant.
Results
Patient characteristics
The 182 patients included 120 males and 62 females,
ranging from 26 to 91 years old (median, 67.9 years). The median
follow-up period was 46.3 months (range, 0.3–120.0 months). Among
the 182 patients, 74 received 5-FU-based adjuvant chemotherapy
(adjuvant group), while the remaining 108 did not receive adjuvant
treatment (surgery group) because of the diagnosis of early stage
disease, advanced age and/or various complications.
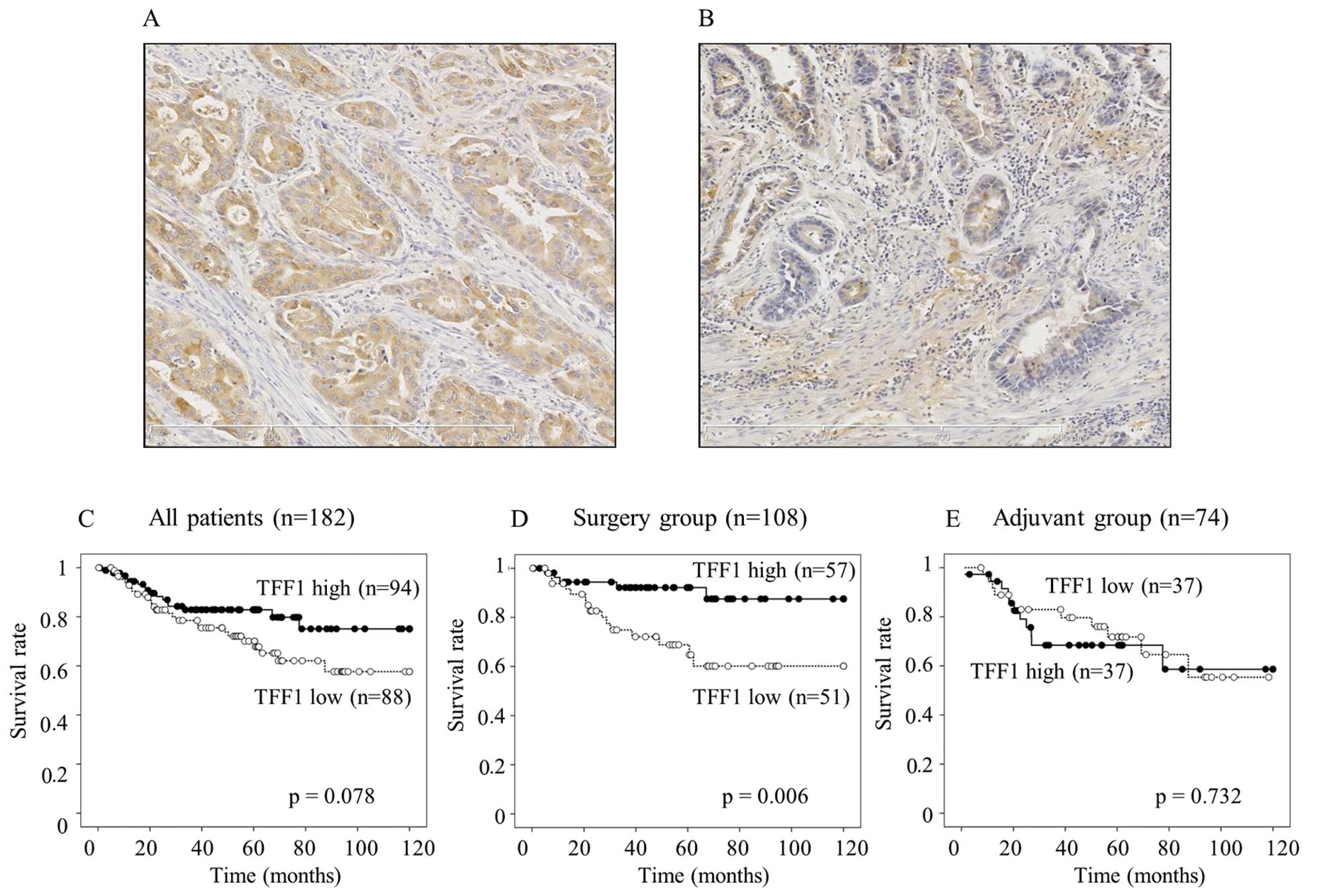
Immunohistochemical staining of TFF1
The staining of TFF1 was predominantly distributed
in the cytoplasm. High expression of TFF1 (Fig. 1A) was observed in 94 gastric cancer
patients, whereas the remaining 88 patients showed low expression
(Fig. 1B). Low expression of TFF1
was significantly correlated with deeper invasion of the tumor
(p=0.037) (Table I). However, no
significant correlations were observed between the TFF1 expression
and other factors including age, gender, lymph node metastasis (N),
lymphatic invasion (ly), vascular invasion (v), or tumor stage.
 | Table ICorrelation between TFF1 expression
and clinicopathological features. |
Table I
Correlation between TFF1 expression
and clinicopathological features.
| TFF1 expression
| |
|---|
|
Characteristics | High (n=94) | Low (n=88) | p-value |
|---|
| Age (mean ±
SD) | 68.0±11.2 | 68.4±12.3 | 0.824 |
| Gender | | | |
| Male | 61 | 59 | 0.760 |
| Female | 33 | 29 | |
| Histology | | | |
|
Differentiated | 42 | 30 | 0.144 |
|
Undifferentiated | 52 | 58 | |
| Ta | | | |
| 2 | 31 | 17 | 0.037 |
| 3/4 | 63 | 71 | |
| Nb | | | |
| − | 32 | 33 | 0.627 |
| + | 62 | 55 | |
| lyc | | | |
| − | 22 | 16 | 0.463 |
| + | 72 | 72 | |
| vd | | | |
| − | 54 | 42 | 0.212 |
| + | 40 | 46 | |
| Stage | | | |
| I/II | 54 | 38 | 0.054 |
| III | 40 | 50 | |
| Adjuvant
chemotherapy | | | |
| − | 57 | 51 | 0.764 |
| + | 37 | 37 | |
TFF1 expression and patient survival
In all the 182 patients, the patients with a low
expression of TFF1 tended to have a worse survival than those with
a high expression (p=0.078, Fig.
1C). A low expression of TFF1 significantly associated with a
worse survival in the surgery group (p=0.006, Fig. 1D), whereas the TFF1 expression did
not contribute to the patient survival in the adjuvant group
(p=0.732, Fig. 1E). A univariate
analysis demonstrated that the depth of tumor invasion (T), lymph
node metastasis (N), lymphatic invasion (ly), tumor stage and TFF1
expression were significantly associated with the disease-specific
survival in the surgery group. A multivariate analysis confirmed
both TFF1 expression and lymph node metastasis to be independent
predictive factors for disease-specific survival (p=0.026, 0.009,
respectively) (Table II).
 | Table IIUnivariate and multivariate analysis
for disease-specific survival in 108 patients in the surgery
group. |
Table II
Univariate and multivariate analysis
for disease-specific survival in 108 patients in the surgery
group.
| | Univariate
| Multivariate
|
|---|
|
Characteristics | No. | HR (95% CI) | p-value | HR (95% CI) | p-value |
|---|
| TFF1 | | | 0.010 | | 0.026 |
| High | 57 | 1 | | 1 | |
| Low | 51 | 3.774
(1.370-10.417) | | 3.268
(1.151-9.259) | |
| Age (years) | | | 0.411 | | |
| <70 | 43 | 1 | | | |
| ≥70 | 65 | 1.473
(0.585-3.704) | | | |
| Gender | | | 0.828 | | |
| Male | 74 | 1 | | | |
| Female | 34 | 1.107
(0.442-2.778) | | | |
| Histology | | | 0.750 | | |
|
Differentiated | 50 | 1 | | | |
|
Undifferentiated | 58 | 1.157
(0.470-2.849) | | | |
| Ta | | | 0.016 | | 0.266 |
| 2 | 43 | 1 | | 1 | |
| 3/4 | 65 | 6.061
(1.404-26.316) | | 2.353
(0.521-10.638) | |
| Nb | | | 0.001 | | 0.009 |
| − | 58 | 1 | | 1 | |
| + | 50 | 27.027
(3.636-200.000) | | 16.393
(2.041-142.857) | |
| lyc | | | 0.029 | | 0.386 |
| − | 33 | 1 | | 1 | |
| + | 75 | 9.434
(1.266-71.429) | | 2.494
(0.316-19.608) | |
| vd | | | 0.344 | | |
| − | 65 | 1 | | | |
| + | 43 | 1.527
(0.635-3.676) | | | |
| Stage | | | 0.001 | | |
| I/II | 78 | 1 | | | |
| III | 30 | 13.158
(4.367-40.000) | | | |
Correlation between promoter methylation
and the expression of TFF1
The expression of TFF1 mRNA was detected in the
order of KATO-III > HSC45 > HSC57 > MKN45 > MKN7 >
MKN28 > MKN74 > MKN1 (Fig.
2A). Based on the results, we classified the cell lines into a
high-expression group (KATO-III, HSC45 and HSC57) and a
low-expression group (MKN1, MKN28 and MKN74). The hypoxia-mediated
induction of mRNA was evaluated in the high-expression group and
the degree of the induction was observed in the order of KATO-III
> HSC45 > HSC57 (Fig.
2B).
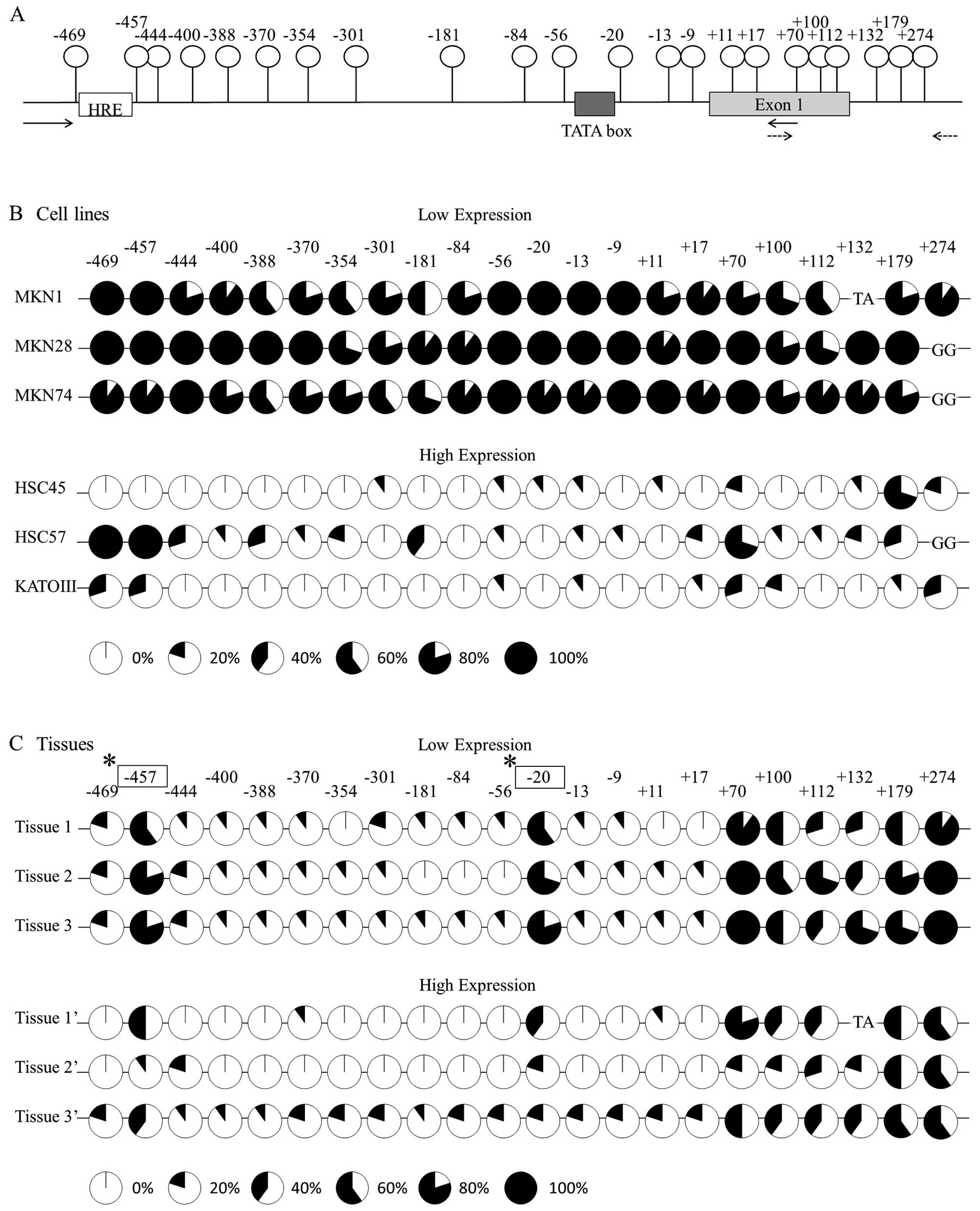
A diagram of the promoter region of the TFF1 gene is
shown in Fig. 3A. There are 22 CpG
sites between 274 and −469 bp. Fig.
3B indicates the methylation status of each CpG site in the six
cell lines. The cell lines classified into the low-expression group
showed dense methylation; in contrast, the cell lines in the
high-expression group showed sparse methylation. In sharp contrast,
in cancer tissues with low TFF1 expression, two specific CpG sites,
which are located at −457 and −20 from the transcript initiation
site, showed a significantly denser methylation in comparison to
the high-expression tissues (Fig.
3C).
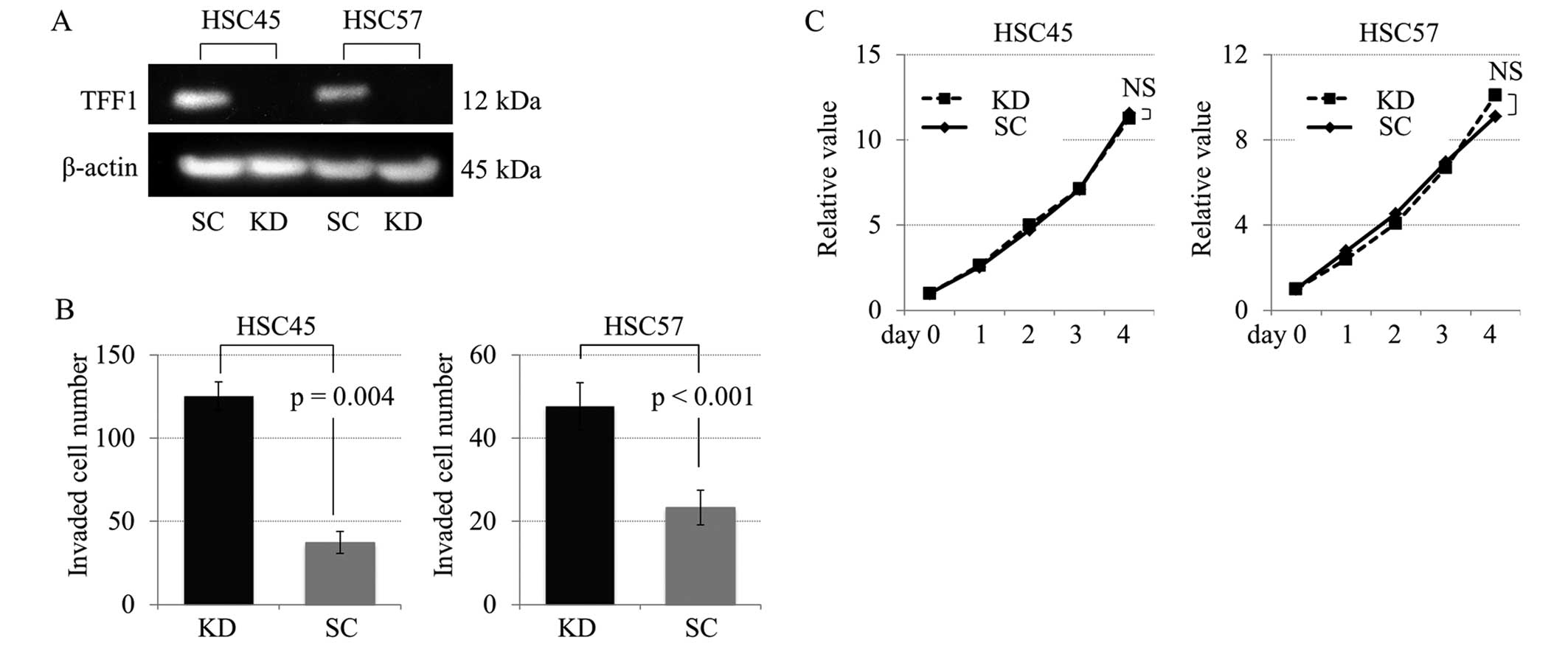
Characterization of TFF1 knockdown
cells
TFF1 knockdown study was performed in order to
investigate the role of TFF1 in cell proliferation and invasion
using gastric cancer cell lines HSC45 and HSC57 with TFF1 positive
expression. The TFF1 knockdown by siRNA was validated by a western
blot analysis (Fig. 4A). As
demonstrated in Fig. 4B, the
invasive capacity was significantly increased in the TFF1 knockdown
cells compared with the control cells, while there were no
significant differences in the proliferation curves between the
knockdown and control cells (Fig.
4C).
Discussion
The loss of TFF1 expression in gastric mucosa leads
to the initiation of gastric cancer (9). However, it has been under discussion
whether changes in TFF1 expression might affect the progression of
gastric cancer. The present study showed a significant correlation
between the depth of cancer invasion and the immunohistochemical
expression of TFF1 in resected tissue samples from patients with
gastric cancer (p=0.037, Table I).
Furthermore, in an in vitro analysis using TFF1 knockdown
cells established from HSC45 and HSC57, the invasive activity of
gastric cancer cells increased significantly in TFF1-deficient
cells compared with the control cells (Fig. 4B). These results indicated that
TFF1 acts not only as a tumor suppressor gene during gastric
carcinogenesis, but also as an inhibitor of tumor invasion during
gastric cancer progression. Hence, it is speculated that loss of
TFF1 at carcinogenesis might sequentially promote cancer invasion
in the stomach.
In a series of survival analysis, low expression of
TFF1 tended to be associated with a worse survival (p=0.078,
Fig. 1C) in all the patients.
Furthermore, the expression of TFF1 was an independent predictive
factor of patient survival in the surgery group (p=0.006, Fig. 1D). On the contrary, TFF1 expression
did not contribute to the survival of patients who received
adjuvant therapy (p=0.732, Fig.
1E). One of the reasons for these results might be due to the
differences in lymph node metastasis between the two groups. In
this series, 67 of 74 patients (90.5%) in the adjuvant group had
lymph node metastasis, whereas only 50 patients (46.3%) in the
surgery group had metastatic lymph nodes. TFF1 expression might not
influence lymph node metastasis during gastric cancer progression
and therefore resulted in no correlation with survival in the
adjuvant group, with high level of lymph node metastasis, compared
with the surgery group. Furthermore, in vitro analysis
revealed no effect of TFF1 expression on the drug sensitivity to
5-FU (data not shown). Taken together, it is suggested that TFF1
expression is a suitable biomarker predicting tumor invasion of
gastric cancer.
TFF peptides have been reported to stimulate the
migration of epithelial cells to promote the repair of mucosal
wounds (2,7,8).
Moreover, the oral intake of recombinant human TFF peptides
protects the gastric mucosa against ethanol- and
indomethacin-induced mucosal injuries in rats (24). Transgenic mice that overexpress
human TFF1 in the jejunum demonstrated an increased resistance to
indomethacin-induced mucosal damage (25). Clinically, phase II studies have
currently demonstrated that oral spray formulation of recombinant
human TFF prevented chemotherapy induced oral mucositis in patients
with colorectal cancer (26).
Based on these studies, oral intake or local administration of TFF1
peptide might suppress tumor invasion of gastric cancer.
The present study further addressed the regulatory
mechanism of TFF1 expression. Previous studies have demonstrated
the genetic regulation of TFF1, in which the loss of TFF1
expression in gastric cancer was explained by LOH and mutations
(27–31). Yio et al demonstrated that
TFF1 mutations enhance the invasion of gastric cancer cells in
vitro(31). Park et al
also indicated that the allelic loss of TFF1 plays an important
role in gastric carcinogenesis and detected somatic missense
mutations of TFF1 in 7 of 43 gastric cancers (27,28).
In addition to these studies, a low-oxygen environment induced the
expression of TFF1 in gastric epithelial cells via the HIF-1
pathway (19). The present study
investigated the epigenetic regulation of this gene by means of
bisulfate sequencing. In the experiment using six gastric cancer
cell lines, the cells with low TFF1 expression showed dense
methylation in the promoter region of this gene, while those with
high expression levels revealed sparse methylation (Fig. 3B). These results indicate that TFF1
expression in gastric cancer cells is strongly dependent on the DNA
methylation of the promoter region. The same experiment approached
the methylation status in the resected cancer tissues.
Interestingly, cancer tissues with the low expression showed a
significantly higher methylation at two specific CpG sites, located
at −20 and −457 from the transcript initiation site, compared to
the high-expression tissues (Fig.
3C). The result indicated that TFF1 expression is dependent on
the site-specific methylation in gastric cancer tissues. A TATA box
(5′-tataaaa-3′) is located upstream of the −20 bp CpG site and a
consensus sequence of hypoxia response element [HRE (5′-gCGtg-3′)]
is located upstream of the −457 bp CpG site.
A TATA box is one of the core elements in a promoter
region, which is necessary to direct the initiation of
transcription, cooperatively acting with other molecules (32). Thus, hypermethylation in the CpG
site at −20 bp downstream of a TATA box might inhibit the binding
of basal transcription complex, resulting in suppression of the
basal expression of TFF1. HRE is a binding site of the HIF-1
complex, which induces the expression of hypoxia-target genes under
hypoxic conditions (33). In the
present study, TFF1 expression was induced by a hypoxic environment
in KATO-III and HSC45 cells, whereas little induction was observed
in the HSC57 cells (Fig. 2B). The
CpG site of −457 bp in the HSC57 cells showed dense methylation in
bisulfite sequencing analysis, thus we speculated that epigenetic
modifications at this region possibly inhibited the binding of
HIF-1 to HRE and thus led to the decreased hypoxic induction in
HSC57 cells. Methylation status of these CpG sites could
potentially be a promising biomarker predicting TFF1 expression in
gastric cancer tissues.
In conclusion, this is the first report
demonstrating that a low expression of TFF1 in gastric cancer is
not only associated with the depth of tumor invasion, but also with
the poor survival in the patients who undergo curative surgery
without adjuvant therapy. Furthermore, epigenetic analysis revealed
that TFF1 expression in gastric cancer tissues is significantly
dependent on DNA methylation of two specific CpGs at −20 bp and
−457 bp in the promoter region. The expression or methylation
status of TFF1 in gastric cancer tissues might therefore be useful
marker predicting patient survival. Moreover, a novel treatment by
TFF1 peptide should be selectively performed in gastric cancer
patients with low TFF1 expression to improve the survival by
inhibiting tumor invasion.
Acknowledgements
This study was supported by a
Grant-in-Aid for Young Scientists (B) (23791548) from the Japan
Society for the Promotion of Science. We also thank Mr. Fumihiro
Mutoh for his helpful contributions to cutting the embedded frozen
specimens for the analyses.
References
|
1
|
Wong WM, Poulsom R and Wright NA: Trefoil
peptides. Gut. 44:890–895. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoffmann W and Jagla W: Cell type specific
expression of secretory TFF peptides: colocalization with mucins
and synthesis in the brain. Int Rev Cytol. 213:147–181. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Taupin D and Podolsky DK: Trefoil factors:
initiators of mucosal healing. Nat Rev Mol Cell Biol. 4:721–732.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thim L: Trefoil peptides: from structure
to function. Cell Mol Life Sci. 53:888–903. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Masiakowski P, Breathnach R, Bloch J,
Gannon F, Krust A and Chambon P: Cloning of cDNA sequences of
hormone-regulated genes from the MCF-7 human breast cancer cell
line. Nucleic Acids Res. 10:7895–7903. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rio MC, Bellocq JP, Daniel JY, et al:
Breast cancer-associated pS2 protein: synthesis and secretion by
normal stomach mucosa. Science. 241:705–708. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomasetto C, Masson R, Linares JL,
Wendling C, Lefebvre O, Chenard MP and Rio MC: pS2/TFF1 interacts
directly with the VWFC cysteine-rich domains of mucins.
Gastroenterology. 118:70–80. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hoffmann W, Jagla W and Wiede A: Molecular
medicine of TFF-peptides: from gut to brain. Histol Histopathol.
16:319–334. 2001.PubMed/NCBI
|
|
9
|
Lefebvre O, Chenard MP, Masson R, et al:
Gastric mucosa abnormalities and tumorigenesis in mice lacking the
pS2 trefoil protein. Science. 274:259–262. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Colombel M, Dante R, Bouvier R, Ribieras
S, Pangaud C, Marechal JM and Lasne Y: Differential RNA expression
of the pS2 gene in the human benign and malignant prostatic tissue.
J Urol. 162:927–930. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Buache E, Etique N, Alpy F, et al:
Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity of
human breast cancer cells and mammary tumor development in
TFF1-knockout mice. Oncogene. 30:3261–3273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arumugam T, Brandt W, Ramachandran V, et
al: Trefoil factor 1 stimulates both pancreatic cancer and stellate
cells and increases metastasis. Pancreas. 40:815–822. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McChesney PA, Aiyar SE, Lee OJ, Zaika A,
Moskaluk C, Li R and El-Rifai W: Cofactor of BRCA1: a novel
transcription factor regulator in upper gastrointestinal
adenocarcinomas. Cancer Res. 66:1346–1353. 2006. View Article : Google Scholar
|
|
14
|
Milne AN, Carvalho R, Morsink FM, Musler
AR, de Leng WW, Ristimäki A and Offerhaus GJ: Early-onset gastric
cancers have a different molecular expression profile than
conventional gastric cancers. Mod Pathol. 19:564–572. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gött P, Beck S, Machado JC, Carneiro F,
Schmitt H and Blin N: Human trefoil peptides: genomic structure in
21q22.3 and coordinated expression. Eur J Hum Genet. 4:308–315.
1996.PubMed/NCBI
|
|
16
|
Beck S, Sommer P, Blin N and Gött P:
5′-flanking motifs control cell-specific expression of trefoil
factor genes (TFF). Int J Mol Med. 2:353–361. 1998.
|
|
17
|
Al-azzeh ED, Fegert P, Blin N and Gött P:
Transcription factor GATA-6 activates expression of
gastroprotective trefoil genes TFF1 and TFF2. Biochim Biophys Acta.
1490:324–332. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beck S, Sommer P, dos Santos Silva E, Blin
N and Gött P: Hepatocyte nuclear factor 3 (winged helix domain)
activates trefoil factor gene TFF1 through a binding motif adjacent
to the TATAA box. DNA Cell Biol. 18:157–164. 1999. View Article : Google Scholar
|
|
19
|
Hernández C, Santamatilde E, McCreath KJ,
et al: Induction of trefoil factor (TFF) 1, TFF2 and TFF3 by
hypoxia is mediated by hypoxia inducible factor-1: implications for
gastric mucosal healing. Br J Pharmacol. 156:262–272.
2009.PubMed/NCBI
|
|
20
|
Japanese Gastric Cancer Association:
Japanese classification of gastric carcinoma: 3rd English ed.
Gastric Cancer. 14:101–112. 2011. View Article : Google Scholar
|
|
21
|
Yanagihara K, Ito A, Toge T and Numoto M:
Antiproliferative effects of isoflavones on human cancer cell lines
established from the gastrointestinal tract. Cancer Res.
53:5815–5821. 1993.PubMed/NCBI
|
|
22
|
Yanagihara K, Tanaka H, Takigahira M, et
al: Establishment of two cell lines from human gastric scirrhous
carcinoma that possess the potential to metastasize spontaneously
in nude mice. Cancer Sci. 95:575–582. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nakamura J, Kitajima Y, Kai K, et al:
Expression of hypoxic marker CA IX is regulated by site-specific
DNA methylation and is associated with the histology of gastric
cancer. Am J Pathol. 178:515–524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Babyatsky MW, deBeaumont M, Thim L and
Podolsky DK: Oral trefoil peptides protect against ethanol- and
indomethacin-induced gastric injury in rats. Gastroenterology.
110:489–497. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Playford RJ, Marchbank T, Goodlad RA,
Chinery RA, Poulsom R and Hanby AM: Transgenic mice that
overexpress the human trefoil peptide pS2 have an increased
resistance to intestinal damage. Proc Natl Acad Sci USA.
93:2137–2142. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peterson DE, Barker NP, Akhmadullina LI,
et al: Phase II, randomized, double-blind, placebo-controlled study
of recombinant human intestinal trefoil factor oral spray for
prevention of oral mucositis in patients with colorectal cancer who
are receiving fluorouracil-based chemotherapy. J Clin Oncol.
27:4333–4338. 2009. View Article : Google Scholar
|
|
27
|
Park WS, Oh RR, Park JY, et al: Mapping of
a new target region of allelic loss at 21q22 in primary gastric
cancers. Cancer Lett. 159:15–21. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park WS, Oh RR, Park JY, et al: Somatic
mutations of the trefoil factor family 1 gene in gastric cancer.
Gastroenterology. 119:691–698. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carvalho R, Kayademir T, Soares P, et al:
Loss of heterozygosity and promoter methylation, but not mutation,
may underlie loss of TFF1 in gastric carcinoma. Lab Invest.
82:1319–1326. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Beckler AD, Roche JK, Harper JC, et al:
Decreased abundance of trefoil factor 1 transcript in the majority
of gastric carcinomas. Cancer. 98:2184–2191. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yio X, Diamond M, Zhang JY, Weinstein H,
Wang LH, Werther L and Itzkowitz S: Trefoil factor family-1
mutations enhance gastric cancer cell invasion through distinct
signaling pathways. Gastroenterology. 130:1696–1706. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lifton RP, Goldberg ML, Karp RW and
Hogness DS: The organization of the histone genes in Drosophila
melanogaster: functional and evolutionary implications. Cold
Spring Harb Symp Quant Biol. 42:1047–1051. 1978.
|
|
33
|
Semenza GL, Jiang BH, Leung SW, Passantino
R, Concordet JP, Maire P and Giallongo A: Hypoxia response elements
in the aldolase A, enolase 1, and lactate dehydrogenase A gene
promoters contain essential binding sites for hypoxia-inducible
factor 1. J Biol Chem. 271:32529–32537. 1996. View Article : Google Scholar
|