Introduction
Metastasis of tumor cells to distant organs is the
most common cause of death arising from human carcinomas. During
the metastatic cascade, carcinoma cells often initiate a
transdifferentiation step known as epithelial-mesenchymal
transition (EMT), one dynamic cellular process deemed to underlie
metastasis by promoting acquisition of migratory and invasive
abilities (1,2). EMT is characterized by loss of
E-cadherin that is proposed to be a critical switch of EMT,
accompanied with increased expression of mesenchymal cell phenotype
markers (Vimentin, N-cadherin and fibronectin) (3), which contribute to disruption of cell
adherens junction and the ensuing loss of cell polarity.
Subsequently, epithelial cells acquire a fibroblastic phenotype,
dissociate from the epithelium and migrate to distant organs.
Therefore, EMT underlying metastasis remains an important aspect of
cancer research.
During the acquisition of EMT phenotype, several
transcription factors including the Snail-family members Snail1,
Snail2 (formerly Slug), Twist1, E47 and the Zeb-family members Zeb1
and Zeb2 (Sip1) are known to potently modulate epithelial cell
plasticity and induce EMT phenotype by repressing E-cadherin
expression (4–7). The transcription factor
overexpression in human carcinomas binds to E-box elements within
the promoter region of E-cadherin, leading to transcription
inhibition of E-cadherin and triggering EMT in cancer cells, which
suggest that transcription factors play a key role in controlling
the induction of EMT phenotype.
Members of the Forkhead box protein family of
transcription factors (Fox-factors) have been shown to be involved
in regulating EMT of epithelial cancer cells (8). In humans, this transcription factor
family characterized by the presence of a DNA-binding domain called
the Forkhead box or winged helix domain comprises 17 subfamilies
(FoxA-FoxR) with 43 known members (9,10).
FOXC2 and FOXM1 have been recently implicated in aggressive
basal-like breast cancer with a role in regulating EMT and
metastasis (11,12). FOXQ1 belongs to the human Forkhead
box gene family (13). It has been
recognized that FOXQ1 as a potent modulator of epithelial cell
plasticity and is involved in epithelial differentiation and cell
proli feration which regulates the developmental function of many
organs in the body (14).
Moreover, overexpression of FOXQ1 has been reported in several
cancers including lung carcinoma cell lines, pancreatic ductal
adenocarcinomas (15) and in the
transition from normal intestinal epithelium to adenoma and
carcinomas in an APC min/t mouse (16). Interestingly, higher expression of
FOXQ1 in human colorectal carcinomas has been reported to directly
control the expression of p21Waf1/Cip1 and enhance
tumorigenicity and tumor progression and found to be associated
with poor prognosis of colorectal cancer patients (17). Furthermore, emerging evidence
suggests that FOXQ1 was identified as transcriptionally induced in
a TGF-β1 responsive cell culture model of cytokine-induced EMT-like
progression (18), suggesting a
potential impact of FOXQ1 expression in the regulation of EMT.
FOXQ1 has been confirmed to be increased in expression in a
Smad4-dependent manner in response to TGF-β1 treatment (19), demonstrating that the expression of
FOXQ1 is regulated by TGF-β1 which is profoundly known as a major
inducer of EMT processes and plays an important role in the outcome
of TGF-β1 signaling. These results strongly suggest that FOXQ1 may
have a crucial role in the development and progression of human
cancers. However, the precise mechanism for FOXQ1 inducing EMT
phenotype and tumor progression in the transitional cell carcinoma
(TCC) has not been elucidated. No previous study has examined the
effect of targeted inhibition or downregulation of FOXQ1 in bladder
cancer cells.
In this research, to study the functional
significance of EMT in initiating metastasis of bladder cancer, we
used short hairpin RNA (shRNA) against FOXQ1 gene to implement gene
silencing by means of RNA interference in the human bladder cancer
cell line T24 which has high metastatic potential. Then
investigated the regulatory effect of FOXQ1 on gene expression of
EMT molecular markers and further inspected its role in cell
proliferation, cell migration and invasion with in vitro
transfection assays. We showed that silencing of the FOXQ1 gene
induced changes in markers of epithelial and mesenchymal phenotypes
and the morphology of cells, displaying a mesenchymal-epithelial
transition (MET). We further demonstrated that the inhibition of
the FOXQ1 expression significantly depressed motility, subsequent
migration and invasion of bladder cancer cells.
Materials and methods
Cell line culture and maintenance
The human bladder cancer cell line T24 was chosen
for knockdown expression of FOXQ1 by stable transfection of FOXQ1
shRNA especially because T24 cells showed overexpression of FOXQ1
compared with BIU-87 cells. T24 cells are a cell line derived from
a grade III bladder carcinoma, which are poorly differentiated and
possess a higher potential of metastasis (20). T24 cells were maintained in our
laboratory. They were cultured in RPMI-1640 (Gibco, Rockville, MD)
with 10% fetal bovine serum (FBS) (Hyclone, Logan, OR) at 37°C
under 5% CO2 and 95% humidified air.
Tissue samples
This study was approved by the institutional Health
Care/Hospital Ethics Committees. Informed consent was obtained from
all patients prior to the study. Bladder tissue samples were
obtained from 65 patients (mean age 65.2 years, range 45–78) with
primary histologically-proven urothelial bladder cancer. For each
specimen, the cancer tissue, paracarcinoma tissue >1.5–2 cm from
cancer tissue and its remote normal mucosa were analyzed and
compared. These patients underwent transurethral resection
(TUR-Bt), partial cystectomy, or radical cystectomy between 2009
and 2011 at the Department of Urology, Union Hospital of Tongji
Medical College, Huazhong University of Science and Technology.
None of the patients had received preoperative treatment. The
samples collected displayed papillary or sessile appearance during
the endoscopic procedure. The tumors were stratified as non-muscle
invasive bladder cancer (NMIBC) or muscle invasive bladder cancer
(MIBC) according to the 2002 UICC TNM classification for the stage
(21), as shown in their
respective cystectomy specimens. Grade was assigned using 2004
WHO/ISUP classification (22)
(LMP, low malignant potential; LG, low grade; HG, high grade) which
showed LMP in 18 cases, LG in 27 cases and HG in 20 cases. Tissue
specimens from surgically resected bladder tumors were confirmed by
pathology and immunostaining was evaluated by 2 independent
pathologists to validate the diagnosis. The corresponding clinical
information was obtained from the Union Hospital of Tongji Medical
College.
Transient transfection
The short hairpin RNA eukaryotic expression vector
targeting FOXQ1 (referring to GenBank NM_033260.3) proceeded by
method of chemical synthesis according to the principle of design
purchased from GenePharma Co., Ltd. (pGPU6/GFP/Neo; Shanghai,
China). The targeting sequences of three different shRNAs and one
negative control shRNA were designed using RNAi algorithm available
online, one sequence with the most effective silencing effect
targeting FOXQ1 was chosen for knockdown expression of FOXQ1 and
subsequent experiment. The RNAi vector can be distinguished by
neomycin resistance screening marker and green fluorescent protein
(GFP) expression driven by the U6 promoter. T24 cells were seeded
at a density of 6×105 cells per 24-well plate in
RPMI-1640 with 10% FBS and grown overnight. Then preserved with
RPMI-1640 free of serum or antibiotics and prepared for
transfection assays. The cells were divided into two groups:
shRNA-FOXQ1 and shRNA-NC (non-specificity sequence for a negative
control). When the cells were 90–95% confluent after washing twice
with 1 ml phosphate buffer saline (PBS), the shRNA plasmid was
transfected into T24 cells using Lipofectamine 2000 transfection
reagent (Invitrogen, Carlsbad, CA) at the ratio of 1:2. After 24 h,
GFP activity of the cells was detected by IX71 fluorescent
microscope (Olympus, Tokyo, Japan).
RT-PCR
To measure the relative expression of mRNAs of each
sample, total RNA was isolated from the indicated tissues,
transfected and control bladder cancer cells, using TRIzol reagent
(Invitrogen) according to the manufacturer’s protocol. After being
washed with 75% ethanol, the final RNA extracts were eluted in a 20
μl volume of distilled water treated with diethyl
pyrocarbonate. The concentration and purity of RNA were measured
with a spectrophotometer. All the RNA preparations had an optical
density OD 260:OD 280 ratio of 1.8–2.0. Then, total RNA from each
sample was reverse transcripted (RT) in a first-strand cDNA
synthesis reaction with PrimeScript RT-PCR kit as recommended by
the supplier (Takara Biotechnology Dalian, China), by reverse
transcription at 37°C for 25 min, followed by incubation at 85°C
for 5 sec in 20 μl of reaction volume. Resultant cDNA (1
μg) was used for semiquantitative polymerase chain reaction
(PCR) amplification, which was performed with primers (Invitrogen)
for: FOXQ1 5′-ATTTCTTGCTATTGACCGATGC-3′ (sense) and
5′-CCCAAGGAGACCACAGTTAGAG-3′ (antisense), TGF-β1
5′-AACCCACAACGAAATCTA-3′ (sense) and 5′-TGAGGTAT CGCCAGGAAT-3′
(antisense), E-cadherin 5′-AACGCATTGC CACATACAC-3′ (sense) and
5′-GAGCACCTTCCATGACA GAC-3′ (antisense), Vimentin
5′-ACAGGCTTTAGCGAGT TATT-3′ (sense) and 5′-GGGCTCCTAGCGGTTTAG-3′
(anti-sense), GAPDH 5′-GGTGAAGGTCGGAGTCAACGG-3′ (sense) and
5′-CCTGGAAGATGGTGATGGGATT-3′ (anti-sense) served as an internal
control to determine the relative amount of RNA of the samples in
the process of RT-PCR. The PCR (32 cycles) was conducted in a
Mastercycler thermal cycler (Eppendorf, Hamburg, Germany). Each
cycle included denaturation (94°C, 30 sec), annealing (58°C, 30
sec) and extension (72°C, 30 sec) performed in 25 μl of
reaction mixture. The initial denaturation period was 4 min and the
final extension was 7 min. Amplified products were analyzed by DNA
gel electrophoresis in 2.0% agarose and were visualized by ethidium
bromide staining under ultraviolet illumination. The result of gel
electrophoresis was analyzed by Quantity One 4.5.0 software for the
optical density.
qRT-PCR
Quantification of mRNA expression was performed with
1 μl of cDNA in a final volume of 20 μl containing 10
μl of SYBR Green master mixture from Takara Clontech (Kyoto,
Japan) and 10 pmol of sense and antisense primers of FOXQ1
5′-ATTTCTTGCTATTGACCGATGC-3′ (sense) and 5′-CCCA
AGGAGACCACAGTTAGAG-3′ (antisense), E-cadherin 5′-CTG
GACGCTCGGCCTGAAGT-3′ (sense) and 5′-GGGTCAGTAT CAGCCGCTTT-3′
(antisense), Vimentin 5′-ACAGGCTTTAG CGAGTTATT-3′ (sense) and
5′-GGGCTCCTAGCGGTTTAG-3′ (antisense) GAPDH
5′-GGTGAAGGTCGGAGTCAACGG-3′ (sense) and
5′-CCTGGAAGATGGTGATGGGATT-3′ (anti-sense) respectively. qPCR was
carried out in a DNA engine, Option TM2 Real-Time Detector
(Bio-Rad, Hercules, CA) using the following thermal cycling
profile: 95°C for 30 sec, followed by 40 cycles of amplification
(95°C for 5 sec, 58°C for 30 sec). All samples were run in
triplicate, the relative levels of individual mRNA in each sample
transcript to control GAPDH were calculated using the
2−ΔΔCt method.
Western blotting
Similarly to shRNA transfected bladder cancer cells,
T24 and bladder tissue samples and protein extracts were prepared
by washing cells with PBS and lysing in RIPA buffer containing
protease inhibitor. The concentrations of protein were detected by
BCA protein assay. Proteins of the samples in each group were
resolved on 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE) (Bio-Rad) and transferred to a NC
membrane (Bio-Rad), blocked with TBST buffer composed of 50 mM Tris
(pH 7.6), 150 mM NaCl and 0.05% Tween supplemented with 5% fat-free
milk at 4°C for 1 h. The membrane was incubated overnight at 4°C in
TBST containing 5% bovine serum albumin (BSA) with rabbit
anti-human monoclonal antibody FOXQ1 (Abcam) (1:500), TGF-β1,
E-cadherin and Vimentin (Cell Signaling Technology, Beverly, MA)
(1:500). Rabbit anti-human GAPDH polyclonal antibody (Santa Cruz
Biotechnology Inc., Santa Cruz, CA) (1:500) was used for detecting
the internal control protein. Then, blots were washed 3 times using
TBST for 1 h at room temperature. Antibody binding was detected
using peroxidase-conjugated goat anti-rabbit IgG and visualized
with ECL chemiluminescent reagents (Pierce Biotechnology, Rockford,
IL) according to the manufacturer’s instructions.
Immunohistochemical staining
analysis
Immunohistochemical staining was done on
conventional paraffin-embedded tissue sections obtained from normal
bladder and different stages of bladder cancers. Tissue sections
were incubated with monoclonal antibody at 1:60 dilution, followed
by treatment with a streptavidin-peroxidase immunohistochemical
staining kit (Zymed Laboratories Inc., South San Francisco, CA),
according to the manufacturer’s protocol. Finally, tissue sections
were counterstained with hematoxylin to discriminate nucleus from
cytoplasm, upgraded alcohols, mounted and analyzed by standard
light microscopy. Immunohistochemical staining of FOXQ1, TGF-β1,
Vimentin and E-cadherin was defined as detectable immunoreactions
in perinuclear and/or cytoplasm. Expression of FOXQ1, TGF-β1 and
Vimentin was considered negative when no or <49% of the tumor
cells were stained (23). Cancer
cells that were immunostained with <10% staining were defined as
having a reduced E-cadherin expression (24).
Cell wound healing assay
Wound healing assay was performed to examine the
capacity of cell migration and invasion. Briefly, T24 cells were
transfected in a 24-well plate with Lipofectamine 2000 after the
cells grew to 90–95% confluence. At 48 h post-transfection, the
wound was generated by scratching the surface of the plates with a
0–100-μl pipette tip. Cell migration into the wounded empty
space was photographed using a microscope after 24 h, well images
were evaluated and the relative migration distance was
analyzed.
Matrigel invasion assay
Forty-eight hours after shRNA plasmid transfection,
cells were harvested, counted and resuspended in serum-free
RPMI-1640 culture medium. A total 200 μl of 5,000 cells of
T24 cells were added to the upper compartment of the matrigel
invasion chamber (BD Biosciences, Bedford, MA) in 24-well tissue
culture plates and 500 μl of RPMI-1640 containing 10% FBS
was added to the lower compartment. After a 24-h incubation period
at 37°C in 5% CO2, membranes were fixed in methanol and
stained with crystal violet. Cells on the upper surface of the
filter were removed carefully with a cotton swab and experiments
were performed in triplicate. Invasion power of the cells was
determined by counting the number of cells that have migrated to
the lower side of the membrane under a light microscope. Five
visual fields (×400) were determined in each chamber. Finally, the
mean value of fields was calculated.
MTT assay
Bladder cancer cells described above were seeded at
4×103 cells per well in flat-bottomed 96-well plates.
After 24 h, cells were transduced with shRNA plasmid for 1, 2, 3
and 4 days. At the end of culture, 10 μl of MTT (5 mg/ml)
was added to each well and plates were placed at 37°C for 4 h. The
medium was then removed and 100 μl of dimethylsulfoxide
(DMSO) solution was added to each well to lyse the cells.
Absorbance was measured at 570 using a microplate reader to
determine cell viability. Four replicate wells were tested per
assay and each experiment was repeated 3 times.
Apoptosis assay
The effect of FOXQ1 silencing on cell apoptosis was
determined by flow cytometry. Briefly, T24 cells at
1×106 cells per well were cultured in 6-well plates
overnight and transfected with 50 nM of negative control shRNA-NC
and FOXQ1-shRNA respectively. The cells were harvested and stained
with Annexin-V and PI, using the Annexin V-FITC apoptosis detection
kit (Keygentec, China). The apoptotic cells were assessed using a
FACSCalibur instrument.
Statistical analysis
Data were expressed as the means ± standard
deviation and were analyzed using independent samples t-test by the
statistical software program SPSS 17.0 (SPSS Inc., Chicago, IL).
Values of P<0.05 were considered statistically significant.
Results
High expression levels of TGF-β1, FOXQ1,
Vimentin, or low level of E-cadherin in bladder cancer tissues
RT-PCR and western blot analysis were performed to
determine whether a high-level of expression of FOXQ1 was
accompanied by an alteration in the expression of epithelial
markers, E-cadherin and Vimentin. Although there were significantly
higher levels of FOXQ1, TGF-β1 and Vimentin were also found in
bladder cancer tissues of differential stages, paracancer tissues,
in which E-cadherin expression levels were markedly decreased
(Fig. 1A and B). FOXQ1 expression
was inversely correlated to E-cadherin, but positively to Vimentin.
Similar results could be obtained by IHC staining for detecting
TGF-β1, FOXQ1, Vimentin and E-cadherin in clinical tissue samples
(Fig. 1C). The results from IHC
staining indicated that FOXQ1 localized mainly in the cytoplasm in
bladder cancer cells (Fig.
1C).
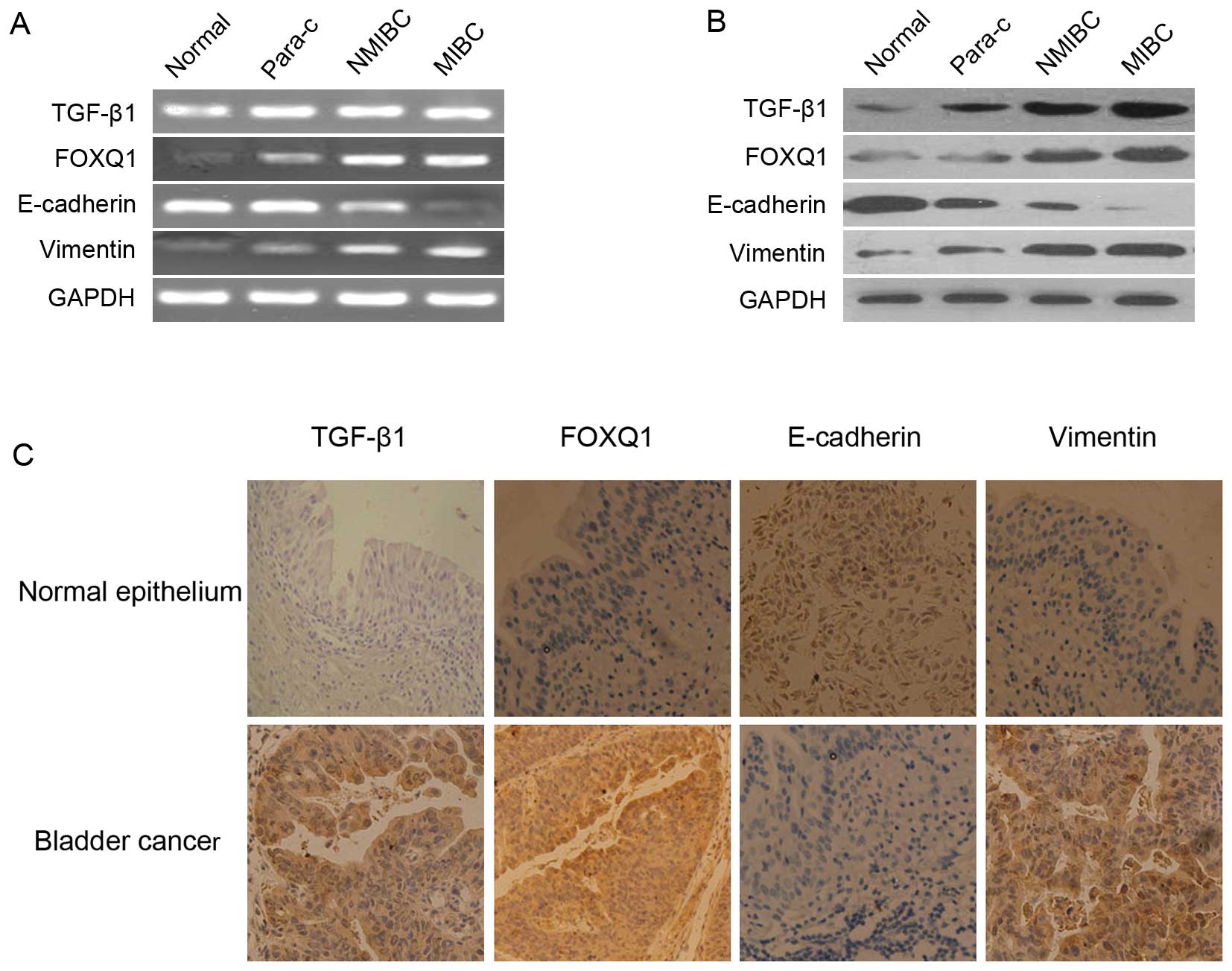 | Figure 1Expression levels of TGF-β1, FOXQ1,
E-cadherin and Vimentin in the normal bladder and tumor tissues.
(A) RT-PCR (left panel) was performed using total RNA isolated from
various samples derived from normal bladder tissues (normal),
bladder paracancer tissues (para-c), non-muscle-invasive bladder
cancer (NMIBC), muscle-invasive bladder cancer (MIBC), The
experiment was repeated 3 times with reproducible results. (B)
Protein levels of TGF-β1, FOXQ1, E-cadherin and Vimentin in the
bladder tissue samples, bladder cancer tissues were examined by
western blot analysis (right panel) using corresponding monoclonal
antibodies. (C) IHC staining of the normal bladder tissues, tumor
tissues resected from bladder cancer patients with anti-TGF-β1,
anti-FOXQ1, anti-E-cadherin and anti-Vimentin antibodies was
performed as described in Materials and methods. Original
magnification, ×200. |
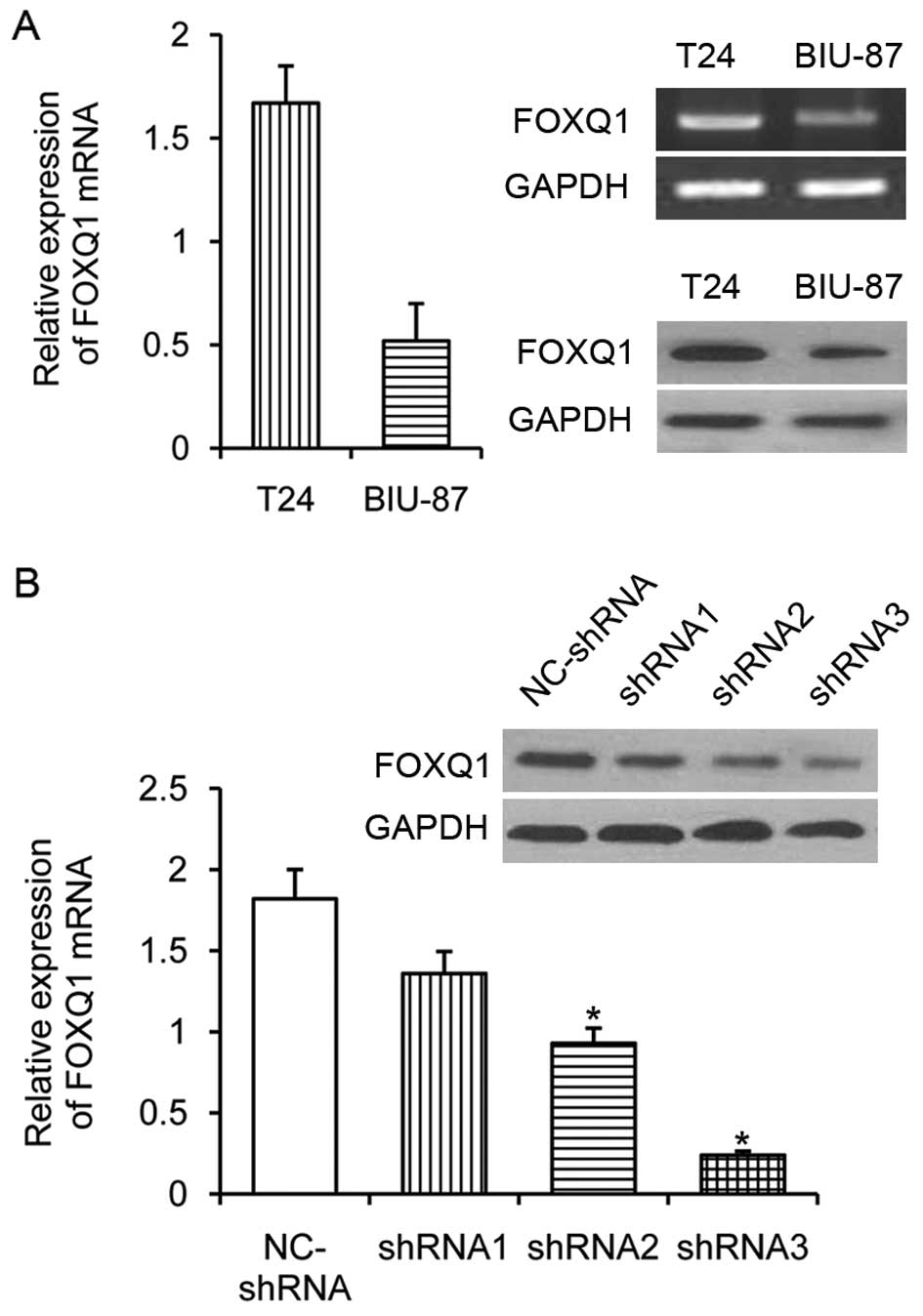
Higher expression of FOXQ1 in T24 cells
compared with BIU-87 cells and the knockdown effect of FOXQ1-shRNA
in T24 cells
To investigate the expression of FOXQ1, we analyzed
the mRNA expression levels of FOXQ1 in human bladder cancer cell
lines T24 and BIU-87 using qRT-PCR. High levels of FOXQ1 expression
were observed in the highly invasive and mesenchymal-like bladder
cancer T24 cells, relatively weak expression levels were detected
in the none-invasive epithelial bladder cancer BIU-87 cells.
Similar results could be obtained by western blot analysis for
detecting FOXQ1 (Fig. 2A). To
unveil the possible role of FOXQ1 in the progression of bladder
cancer, a more invasive cell line model T24 was chosen for
knockdown expression of FOXQ1 by stable transfection of FOXQ1
shRNA. The three positive plasmids were named shRNA1, shRNA2,
shRNA3; one negative plasmid was named NC-shRNA. To compare the
effect of the shRNA-mediated silence of the target FOXQ1 gene
expression, control shRNA, the FOXQ1-shRNAs and 50 nM of each was
transfected into T24 cells for 24 h. The efficacy of individual
inhibitory RNAs in downregulating the expression of FOXQ1 gene was
measured by qRT-PCR. As shown in Fig.
2B, both the FOXQ1-shRNAs effectively inhibited the FOXQ1 gene
transcription. However, the shRNA3 displayed the strongest
inhibitory activity among these inhibitory shRNAs tested. The
relative levels of FOXQ1 mRNA transcripts significantly decreased
by nearly 90%, as compared with the negative control group
(NC-shRNA) and were significantly lower than those of other
inhibitory RNAs (P<0.05). Expression of the internal standard
GAPDH did not differ significantly between the groups (P>0.05).
These data show that shRNA3 specifically inhibited the expression
of FOXQ1 in T24 cells. Therefore, the FOXQ1-shRNA3 with the most
effective silencing effect was chosen for knockdown expression of
FOXQ1 and subsequent experiment.
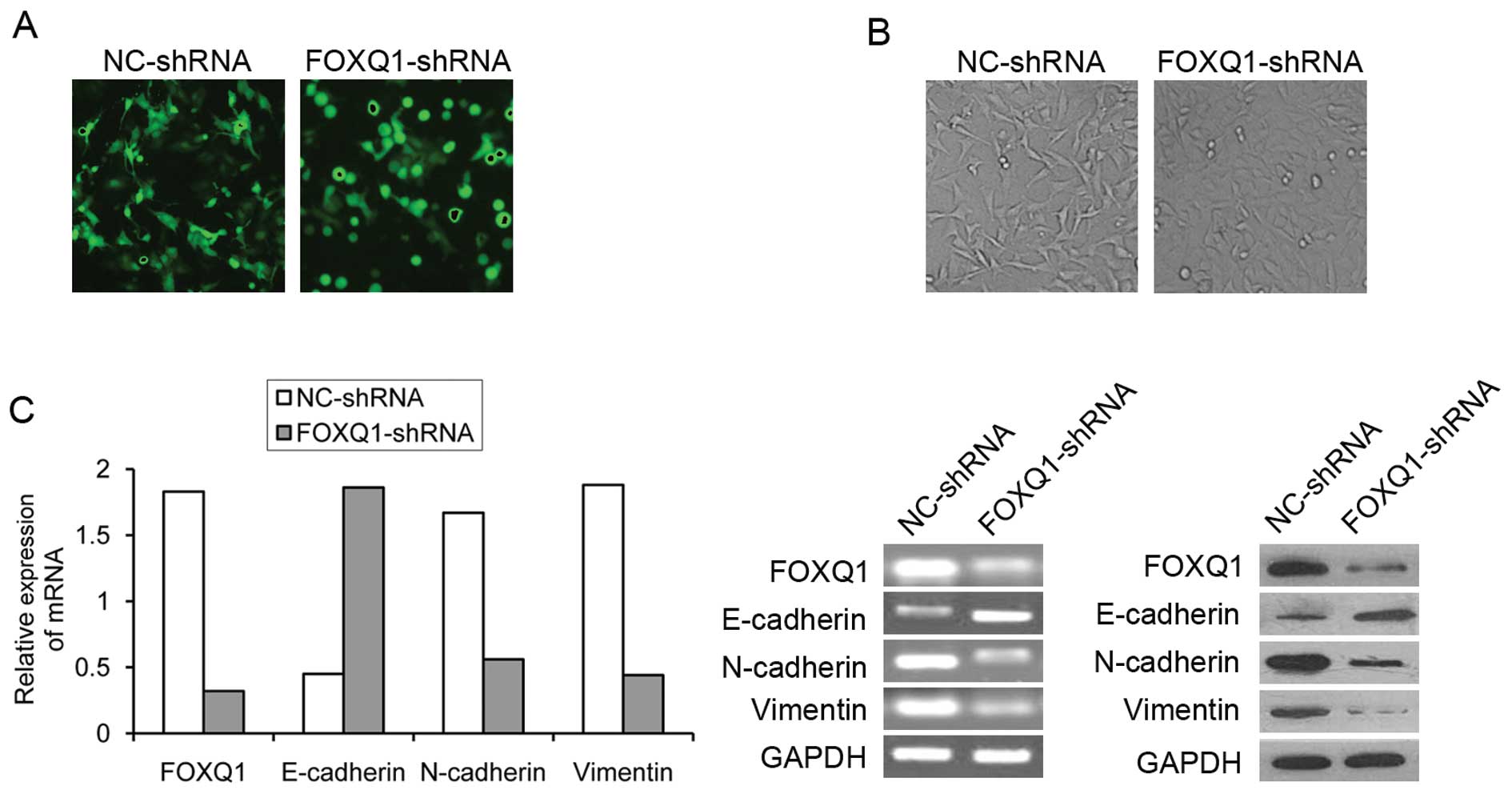
FOXQ1 gene silencing induced changes in
the morphology of T24 cells
Changes in transfection efficiency after 12–48 h
were determined through the observation of green fluorescence; the
strongest green fluorescence occurred at 48 h. After transfection
of recombinant plasmid through Lipofectamine 2000, the cells that
had clear structure and form took on full green fluorescent protein
in cytoplasm and nucleus (Fig.
3A). Interestingly, we found that NC-shRNA transfected T24
cells displayed irregular fibroblastoid morphology (Fig. 3B, left panel). In contrast, T24
cells treated with a reduced FOXQ1 expression appeared enlarged in
cell size, while the integrity of the epithelial sheet structure
remained intact. Compared to NC-shRNA group cells, FOXQ1-shRNA
group T24 cells had a rounded shape, typical of an epithelial
cobblestone appearance (Fig. 3B,
right panel). These changes in phenotype suggested that the
inhibition of FOXQ1 expression could potentially reverse EMT of T24
cells.
Changes in markers of epithelial and
mesenchymal phenotypes
To further validate whether FOXQ1 gene silencing was
able to reverse EMT phenotype in T24 cells, we determined the
expression of epithelial marker E-cadherin (CDH1) and mesenchymal
markers N-cadherin and Vimentin using qRT-PCR and western blot
analysis in FOXQ1-shRNA group and NC-shRNA group. Notably, FOXQ1
ablation induced a change from spindle-like mesenchymal morphology
of T24 cells into epithelial morphology by manifesting an increased
cell-to-cell adhesion (Fig. 2B).
Consistent with the phenotypic change associated with FOXQ1
knockdown was an upregulation of epithelial marker E-cadherin
concomitant with decreased expression of mesenchymal markers
Vimentin and N-cadherin, as determined at both mRNA and protein
levels (Fig. 3C). Thus, FOXQ1
depletion resulted in a reversal of EMT in T24 cells, close to a
normal epithelial phenotype and displayed a mesenchymal-epithelial
transition (MET).
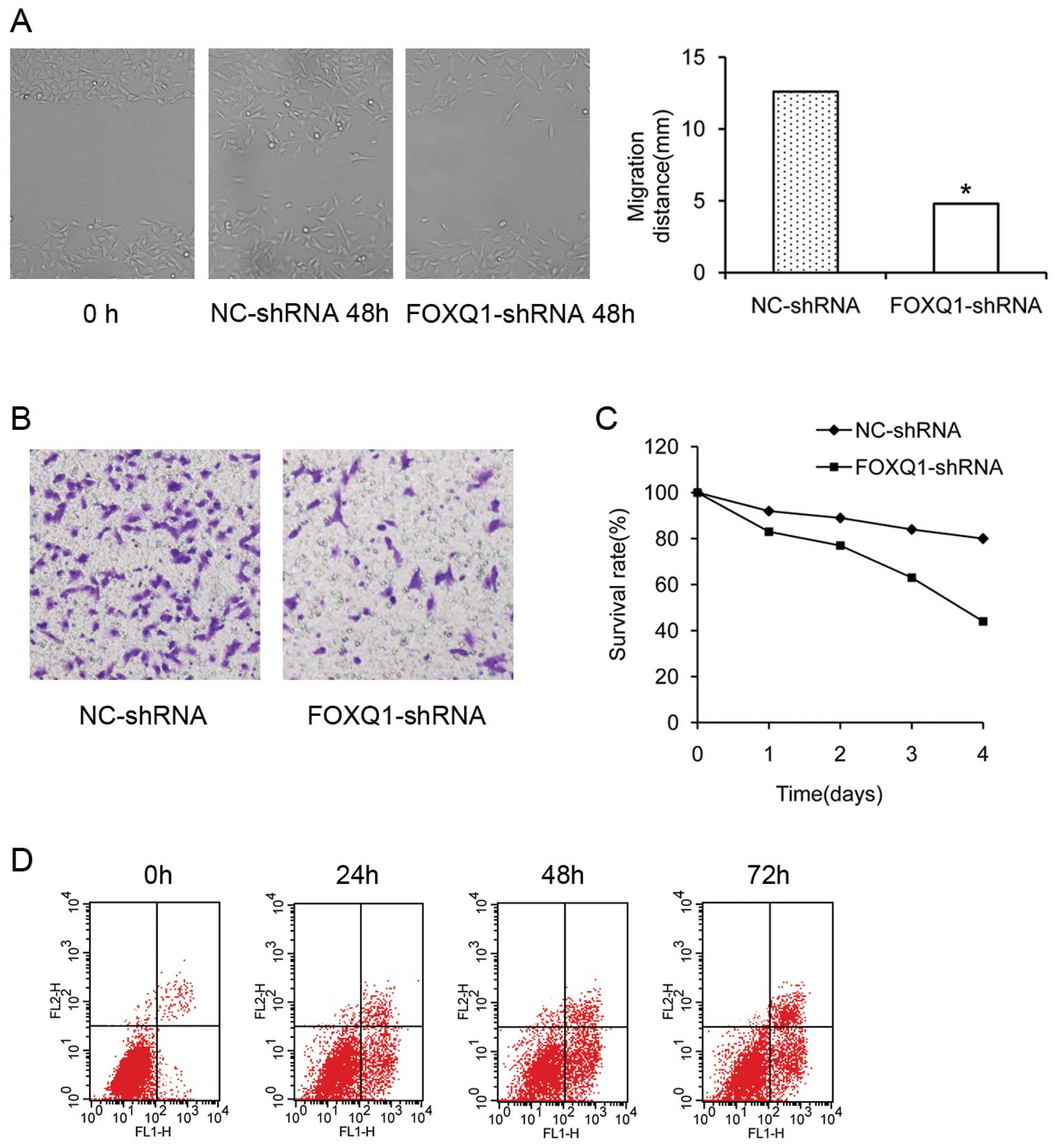
Effect of FOXQ1 gene silencing on the
migration and invasion of T24 cells
FOXQ1 activity has been consistently correlated with
the migratory and invasive potential of tumor cells (25). To functionally confirm the role of
FOXQ1 in aggressive bladder cancers, we used a cell wound healing
assay to examine the effect of FOXQ1 inhibition on cell migration
in T24 cells. As shown in Fig. 4A,
cells showed wounds that were healed nearly 48 h after scraping
with pipette tips. Cells in the FOXQ1-shRNA group showed a reduced
distance of migration compared with the NC-shRNA group.
Furthermore, T24 cells freely invaded the matrigel and passed into
the lower chamber in the NC-shRNA group. In contrast, treatment of
the cells with FOXQ1-shRNA significantly reduced the number of
cells that invaded (Fig. 4B), the
difference was significant (P<0.05). Collectively, these
findings indicated that the knockdown of FOXQ1 inhibited the
migration and invasion of bladder cancer cells in vitro.
The knockdown effect of FOXQ1 on cell
viability and apoptosis
As shown in Fig.
4C, knockdown of FOXQ1 expression with the shRNA plasmid
decreased cell viability of T24 cells by 24 h post-transfection,
which was measured by MTT assay. FOXQ1 gene silencing promotes
apoptosis in T24 cells. Flow cytometry analysis of T24 cells was
performed 24, 48 and 72 h after transfection, the apoptosis rates
were (8.63±1.47, 19.65±1.24 and 26.82±1.35%) respectively, and the
cells at 72 h showed obvious apoptosis compared with cells of the
24-h group (Fig. 4D). These
results suggest that our process of FOXQ1-shRNA plasmid
transduction and FOXQ1 gene silencing in bladder cancer cells did
suppress their viability.
Discussion
A great deal of literature indicates that EMT plays
a pivotal role during embryonic development and in the formation of
fibroblasts during inflammation and wound healing (26–28).
Emerging evidence suggests that EMT also participates in the course
of carcinogenesis. Accompanying with EMT process, cancer cells
often acquire the capability to disseminate from the primary tumor
site and to localise and group in distant organs, a process called
metastasis (29). Recent studies
have identified additional EMT regulators within the Forkhead
transcription factor family and shown that of all 43 Forkhead
family members presented in the array FOXQ1 was the most highly
overexpressed in the highly invasive and mesenchymal-like breast
cancer MDA-MB-231 cells as compared to non-invasive epithelial-like
breast cancer MCF7 cells (19).
Consistently, we have shown that FOXQ1 was expressed in higher
levels in aggressive bladder cancer T24 cells than that in
non-aggressive bladder cancer BIU-87 cells.
In the present study, we have shown that the
expression of TGF-β1, FOXQ1 and Vimentin was significantly
increased in human bladder cancer tissue compared to that of in
background tissue. Patients with high TGF-β1 (50/65) expression
displayed strong FOXQ1 expression (58/65). Moreover, the patients
with strong E-cadherin expression showed no or less staining of
FOXQ1. A significant correlation between expression levels of FOXQ1
and E-cadherin was obvious in these human specimens (P<0.05). We
have also shown that more patients with high Vimentin (54/65)
expression displayed low E-cadherin expression (18/65), which
confirmed a previous study (30).
These results showed that there was an inverse relationship between
FOXQ1 overexpression and loss of E-cadherin expression, a positive
relationship between TGF-β1 upregulated expression and FOXQ1
overexpression. FOXQ1 is one positive regulation factor for EMT
phenotype, suggesting a potential impact of FOXQ1 expression
regulated by TGF-β1 signal pathway during EMT process of bladder
tumor tissue.
Several reports have shown that EMT-inducing
transcription factors such as SNAIL (4,31),
SIP1 (32), SLUG (33,34),
TWIST1 (35) repress expression of
epithelial marker E-cadherin, and could act as important molecular
regulators of EMT. Recent studies have identified that FOXQ1
repressed E-cadherin expression by targeting the E-box in its
promoter region similarly to the E-cadherin repressors (17). E-cadherin plays a critical role in
cell adhesion, development of epithelial organs and the maintenance
of epithelial polarity (36). The
progression of benign tumors to invasive metastatic cancer involves
partial or complete loss of E-cadherin expression, or an impairment
of its adhesive function (37).
Direct transcriptional repression of E-cadherin was considered for
transcription factors to trigger EMT (4), suggesting that FOXQ1, like other
transcription factors, is also a critical mediator of EMT by
repressing transcriptional activity of E-cadherin. FOXQ1 has become
one of the few transcription factors involved in both EMT process
and cancer metastasis.
To address the precise mechanism for FOXQ1 induced
EMT-like phenotypic changes in bladder cancer cells, in this
experiment, we designed independently and constructed shRNA
directed to FOXQ1 and imported recombinant plasmid into bladder
cancer T24 cells. The shRNA was dependent on the double-strand
structure of the RNA and had target site specificity. We
demonstrated initially that shRNA specific for FOXQ1 could be
successfully transfected into T24 cells, resulting in significantly
reduced gene transcription or expression level of protein.
Interestingly, downregulation of FOXQ1 expression was accompanied
by upregulation of E-cadherin and a lower level of N-cadherin were
observed in T24 cells treated with shRNA plasmid (Fig. 3C). This change in cadherin
expression is referred to as ‘cadherin-switch’ (38). The observation that bladder cancer
cells may gain mesenchymal characteristics, associated with the
expression of mesenchymal markers (e.g., N-cadherin and Vimentin),
has been proposed to contribute to the acquisition of the malignant
phenotype (39,40). In knockdown expression of FOXQ1,
cancer cells expressing mesenchymal markers show cadherin-switch
and reversal of EMT phenotype. However, the inhibition of Slug,
Snail or Twist action through interfering RNA (siRNA or shRNA)
resulted in tumor metastasis or growth inhibition (41–43),
whether there is a possibility that FOXQ1 represses E-cadherin
through regulating other EMT promoting genes is unclear, because
mutual regulation is very common between the EMT promoting genes.
These results strongly suggest that knockdown expression of
transcription factors of EMT induction attain more effective
multifunctional result. Therefore, transcription factors as
therapeutic targets, compared with the inhibition of a single
effector molecule or signal pathway is more reasonable and
feasible.
In addition, the shRNA against FOXQ1 induced changes
in the morphology of T24 cells, which were similar to a normal
epithelial phenotype (Fig. 3B,
right panel). Furthermore, knockdown of FOXQ1 expression caused a
significant increase of multinuclear cells and significantly
reduced the migration and invasion of bladder cancer cells in
vitro assays (Fig. 4A and B),
subsequently inhibiting proliferation and leading to cancer cell
apoptosis (Fig. 4C and D). The
result of our research suggested that RNA interference of FOXQ1 may
inhibit metastasis of T24 cells through reversal of EMT.
In this study we found that knockdown expression of
FOXQ1 leads to the reversal of EMT phenotype by upregulation in the
protein expression of epithelial cell marker, E-cadherin and by
downregulation of mesenchymal cell markers, N-cadherin and Vimentin
in T24 cells. These results strongly suggested the importance of
FOXQ1 signaling in tumor cell aggressiveness through the
acquisition of EMT phenotype in urothelium cancer cells. Therefore,
targeting FOXQ1 signaling by novel approaches would be useful for
reversing the EMT phenotype, which would likely result in the
reversal of neoplasm recurrence and elimination of cancer
cells.
Acknowledgements
This study was supported by grants
from the Chinese Natural Science Foundation (NSFC) (no. 30872561)
to Zhaohui Zhu and (no. 30973008) to Yifei Xing.
References
|
1
|
Scheel C, Onder T, Karnoub A and Weinberg
RA: Adaptation versus selection: the origins of metastatic
behavior. Cancer Res. 67:11476–11480. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelialmesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Batlle E, Sancho E, Franci C, et al: The
transcription factor snail is a repressor of E-cadherin gene
expression in epithelial tumour cells. Nat Cell Biol. 2:84–89.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bolos V, Peinado H, Perez-Moreno MA, et
al: The transcription factor slug represses E-cadherin expression
and induces epithelial to mesenchymal transitions: a comparison
with Snail and E47 repressors. J Cell Sci. 116:499–511. 2003.
View Article : Google Scholar
|
|
6
|
Eger A, Aigner K, Sonderegger S, et al:
DeltaEF1 is a transcriptional repressor of E-cadherin and regulates
epithelial plasticity in breast cancer cells. Oncogene.
24:2375–2385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perez-Moreno MA, Locascio A, Rodrigo I, et
al: A new role for E12/E47 in the repression of E-cadherin
expression and epithelialmesenchymal transitions. J Biol Chem.
276:27424–27431. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Battula VL, Evans KW, Hollier BG, et al:
Epithelial-mesenchymal transition-derived cells exhibit
multilineage differentiation potential similar to mesenchymal stem
cells. Stem Cells. 28:1435–1445. 2010. View
Article : Google Scholar
|
|
9
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Katoh M: Human FOX gene family. Int J
Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
11
|
Hader C, Marlier A and Cantley L:
Mesenchymal-epithelial transition in epithelial response to injury:
the role of Foxc2. Oncogene. 29:1031–1040. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bao B, Wang Z, Ali S, Kong D, Banerjee S,
et al: Over-expression of FoxM1 leads to epithelial-mesenchymal
transition and cancer stem cell phenotype in pancreatic cancer
cells. J Cell Biochem. 112:2296–2306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bieller A, Pasche B, Frank S, Gläser B,
Kunz J, et al: Isolation and characterization of the human forkhead
gene FOXQ1. DNA Cell Biol. 20:555–561. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feuerborn A, Srivastava PK, Küffer S,
Grandy WA, Sijmonsma TP, et al: The Forkhead factor FoxQ1
influences epithelial differentiation. J Cell Physiol. 226:710–719.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao D, Hustinx SR, Sui G, Bala P, Sato N,
Martin S, et al: Identification of novel highly expressed genes in
pancreatic ductal adenocarcinomas through a bioinformatics analysis
of expressed sequence tags. Cancer Biol Ther. 3:1081–1091. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Paoni NF, Feldman MW, Gutierrez LS,
Ploplis VA and Castellino FJ: Transcriptional profiling of the
transition from normal intestinal epithelia to adenomas and
carcinomas in the APCMin/t mouse. Physiol Genom. 15:228–235. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, et al: FOXQ1 is overexpressed in colorectal cancer and
enhances tumorigenicity and tumor growth. Cancer Res. 70:2053–2063.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi
SC, et al: FOXQ1 regulates epithelial-mesenchymal transition in
human cancers. Cancer Res. 71:3076–3086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang H, Ethier SP, Miller F, Wu G, et al:
Forkhead transcription factor Foxq1 promotes epithelial-mesenchymal
transition and breast cancer metastasis. Cancer Res. 71:1292–1301.
2011. View Article : Google Scholar
|
|
20
|
Bubenick J, Baresora M, Viklicky V, et al:
Established cell line of urinary bladder carcinoma (T24) containing
tumor-specific antigen. Int J Cancer. 11:765–773. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sobin DH and Witteking C: TNM
Classification of Malignant Tumors. 6th edition. Wiley-Liss; New
York, NY: 2003, View Article : Google Scholar
|
|
22
|
Miyamoto H, Miller JS, Fajardo DA, et al:
Non-invasive papillary urothelial neoplasms: the 2004 WHO/ISUP
classification system. Pathol Int. 60:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu Q, Zhang K, Wang X, Liu X and Zhang Z:
Expression of transcription factors snail, slug, and twist in human
bladder carcinoma. J Exp Clin Cancer Res. 29:1192010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shiozaki H, Tahara H, Oka H, Miyata M, et
al: Expression of immunoreactive E-cadherin adhesion molecules in
human cancers. Am J Pathol. 139:17–23. 1991.PubMed/NCBI
|
|
25
|
Feng J, Zhang X, Zhu H, Wang X, Ni S and
Huang J: FoxQ1 overexpression influences poor prognosis in
non-small cell lung cancer, associates with the phenomenon of EMT.
PLoS One. 7:e399372012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peter ME: Let-7 and miR-200 microRNAs:
guardians against pluripotency and cancer progression. Cell Cycle.
8:843–852. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: at the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Desmouliere A: Factors influencing
myofibroblast differentiation during wound healing and fibrosis.
Cell Biol Int. 19:471–476. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Onder TT, Gupta PB, Mani SA, Yang J,
Lander ES and Weinberg RA: Loss of E-cadherin promotes metastasis
via multiple downstream transcriptional pathways. Cancer Res.
68:3645–3654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cano A, Perez-Moreno MA, Rodrigo I, et al:
The transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Comijn J, Berx G, Vermassen P, et al: The
two-handed E box binding zinc finger protein SIP1 downregulates
E-cadherin and induces invasion. Mol Cell. 7:1267–1278. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hajra KM, Chen DY and Fearon ER: The SLUG
zinc-finger protein represses E-cadherin in breast cancer. Cancer
Res. 62:1613–1618. 2002.PubMed/NCBI
|
|
34
|
De Craene B, van Roy F and Berx G:
Unraveling signalling cascades for the Snail family of
transcription factors. Cell Signal. 17:535–547. 2005.PubMed/NCBI
|
|
35
|
Liu AN, Zhu ZH, Chang SJ, Hang XS, et al:
Twist expression associated with the epithelial-mesenchymal
transition in gastric cancer. Mol Cell Biochem. 367:195–203. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gumbiner BM: Cell adhesion: the molecular
basis of tissue architecture and morphogenesis. Cell. 84:345–357.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Christofori G and Semb H: The role of the
cell-adhesion molecule E-cadherin as a tumour-suppressor gene.
Trends Biochem Sci. 24:73–76. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
McConkey DJ, Lee S, Choi W, et al:
Molecular genetics of bladder cancer: emerging mechanisms of tumor
initiation and progression. Urol Oncol. 28:429–440. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Iwatsuki M, Mimori K, Yokobori T, et al:
Epithelial-mesenchymal transition in cancer development and its
clinical significance. Cancer Sci. 101:293–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang A, Chen G, Meng L, Wang Q, et al:
Antisense-Snail transfer inhibits tumor metastasis by inducing
E-cadherin expression. Anticancer Res. 28:621–628. 2008.PubMed/NCBI
|
|
42
|
Cheng GZ, Chan J, Wang Q, et al: Twist
transcriptionally upregulates AKT2 in breast cancer cells leading
to increased migration, invasion, and resistance to paclitaxel.
Cancer Res. 67:1979–1987. 2007. View Article : Google Scholar
|
|
43
|
Vannini I, Bonafe M, Tesei A, et al: Short
interfering RNA directed against the SLUG gene increases cell death
induction in human melanoma cell lines exposed to cisplatin and
fotemustine. Cell Oncol. 29:279–287. 2007.PubMed/NCBI
|