Introduction
Cholangiocarcinoma (CCA) is a malignant tumor which
arises from bile duct epithelium. The northeastern region of
Thailand, where liver fluke (Opisthorchis viverrini)
infection is highly endemic, is reported to have the highest
incidence rate of CCA worldwide. Chronic inflammation caused by
liver fluke infestation leads to oxidative DNA damage and malignant
transformation of the infected bile ducts (1). CCA is categorized according to its
anatomic location as either intrahepatic (ICC) or extrahepatic
(ECC) (2). The majority of CCA
patients have a poor prognosis with a rather short mean overall
survival (<30 weeks) due to the delayed diagnosis and different
chemo-therapeutic responses, even at the same stages of the
disease. To date, the availability of effective prognostic markers
for predicting CCA progression and therapeutic outcome is
limited.
p53 is a tumor suppressor gene that regulates
cell cycle arrest and apoptosis. Two p53 protein family members,
p63 and p73, have structures similar to the p53 protein and their
transactivation, DNA binding and oligomerization domains enable
them to promote cell cycle arrest and apoptosis (3–5).
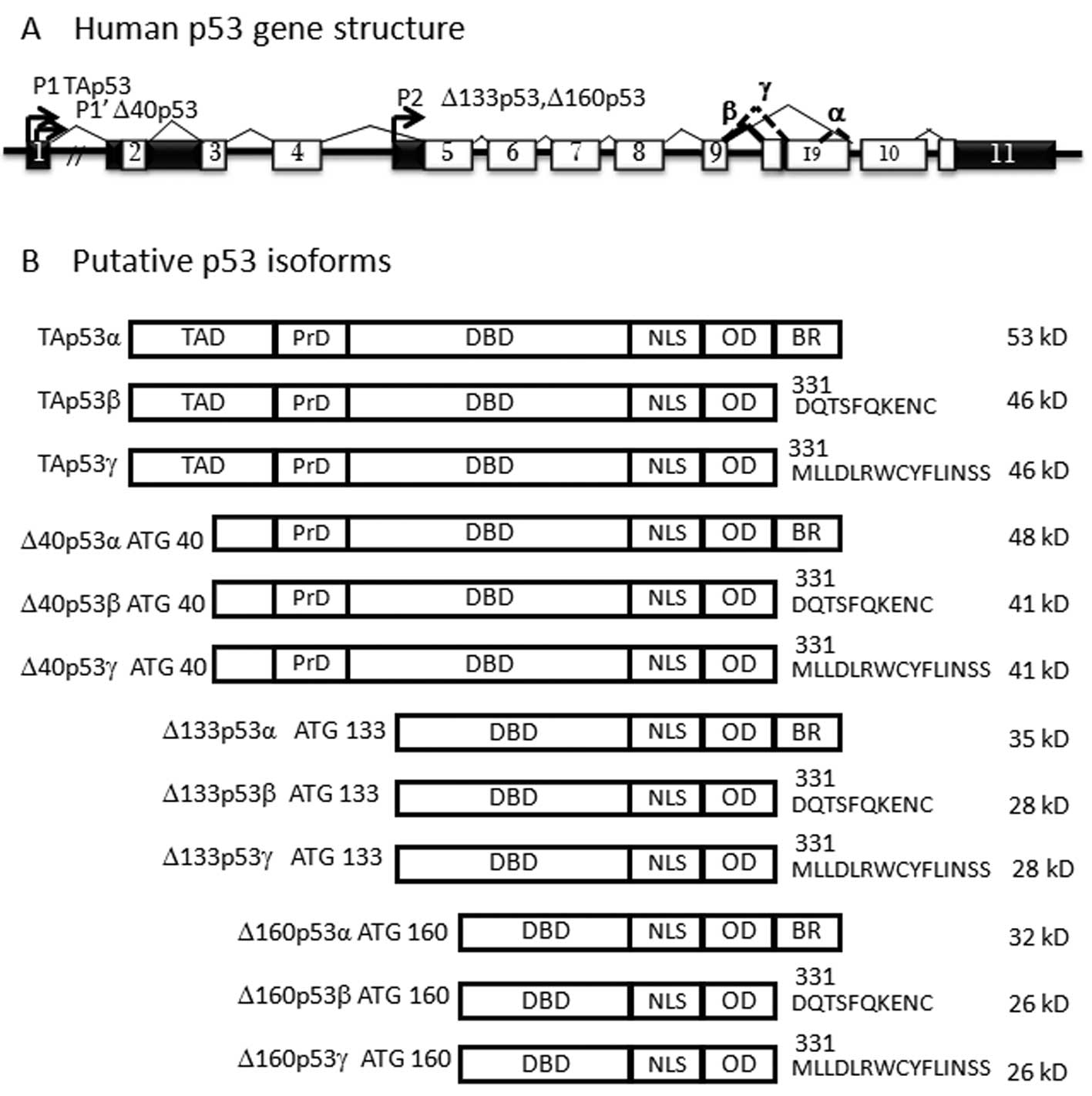
Several protein isoforms of p53, p63 and p73, generated by
alternative splicing and promoter use (6), have been identified as the truncated
proteins at the amino (N-; ΔNp53, ΔNp63 and ΔNp73 isoforms) and the
carboxy (α, β and γ isoforms) termini. p53 is known to contain a
second intronic promoter that generates the N-terminally truncated
ΔN proteins, Δ133p53 (7) and
Δ160p53 (8). Δ40p53 isoforms can
also be generated by alternative splicing and alternative
initiation of translation at intron 2 (7). The N-terminal domain is essential for
the transactivation of target genes and the transactivating
full-length isoforms or TAp53 are functionally distinguished from
the transactivation-compromised ΔN isoforms that exhibit
anti-apoptotic properties. Moreover, intron 9 can be spliced in 3
different ways, leading to the formation of α, β and γ isoforms. As
a whole, the human p53 gene can express 12 different isoforms of
the p53 protein (TAp53, TAp53β, TAp53γ, Δ133p53, Δ133p53β,
Δ133p53γ, Δ40p53, Δ40p53β, Δ40p53γ, Δ160p53, Δ160p53β and
Δ160p53γ), containing different domains of the protein, due to
alternative splicing, alternative promoter use and alternative
initiation of translation (Fig.
1). Deletion of their N-terminal domains not only contributes
to the loss of transactivation but also interferes with the
transactivation of their full-length isoforms (TAp53, TAp63 and
TAp73), via tetramerization of the deleted isoform and the
full-length protein (9).
Therefore, overexpression of the ΔN isoform proteins can inactivate
the full-length p53 family proteins (10–13).
A significant correlation has been reported between
overexpression of ΔN isoforms and a poor prognosis in cervical,
colon and ovarian cancer (10–12,14),
but not in CCA. The incidence of p53 gene mutations in ICC
is approximately 41.6% (15),
while there has been no report of mutation in p73. However,
promoter hypermethylation has been previously reported (16). Taken together, these data suggest a
different mechanism underlying p53 inactivation. Thus, in
this study, we aimed to examine the expression pattern of the ΔN
and TA isoforms of the p53 family at the mRNA and protein
levels. The correlation between the ΔN/TA p53 ratio and
clinical outcome was investigated for its potential use as a
prognostic marker in CCA.
Materials and methods
CCA samples and mRNA extraction
The CCA-derived cell lines, KKU-M055, KKU-M156,
KKU-100, KKU-M139 and KKU-M213, established from CCA patients used
in this study, were obtained from the Liver Fluke and
Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen
University, Khon Kaen, Thailand. The HeLa cell line was used as a
positive control of p53 protein expression. All cell lines were
maintained at 37°C in an atmosphere of 5% CO2 in DMEM
high-glucose medium supplemented with 10% fetal bovine serum (FBS)
and 1% penicillin-streptomycin. Cells were harvested when they
reached 90% confluence and mRNA was extracted using the RNeasy Mini
kit (Qiagen, Hilden, Germany). cDNA was prepared using the
ImProm-II™ Reverse Transcription system (Promega, Madison, WI, USA)
according to the manufacturer’s instructions and maintained at
−20°C until use.
Resected ICC samples were collected from 48 patients
who were admitted to Srinagarind Hospital, Faculty of Medicine,
Khon Kaen University. This study was approved by the Ethics
Committee of Khon Kaen University (HE52202) and written informed
consent was obtained from each patient. Tissue samples were used
for mRNA extraction, as mentioned above.
Primers designed for detection of ΔN and
TA isoform transcripts using RT-PCR
All primers used to detect the mRNA expression of
p53, p63 and p73 isoforms are summarized in Table I. Δ133p53 and TAp53 primers were
designed in this study using free Primer3 software (available at:
http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi).
Each specific isoform product obtained from CCA cell lines was
cloned into the pGEM®-T vector and verified by direct
sequencing. The plasmid construct containing each isoform was used
for setting a standard curve for the quantification of each isoform
level using real-time RT-PCR.
 | Table IOligonucleotide sequences for
quantitation of p53, p63 and p73. |
Table I
Oligonucleotide sequences for
quantitation of p53, p63 and p73.
| Primer name | Sequences
(5′→3′) | Nucleotide
residues | Product size
(bp) | Authors/(Ref.) |
|---|
| TAp53 | F:
CGCAGTCAGATCCTAGCGTC | 262 | 171 | Designed in this
study |
| R:
CTGGACCTGGGTCTTCAGTG | 432 |
| Δ133p53 | F:
GGTTGCAGGAGGTGCTTACAC | 144 | 128 | Designed in this
study |
| R:
GTTGAGGGCAGGGGAGTACTG | 271 |
| TAp63 | F:
GTCCCAGAGCACACAGACAA | 210 | 266 | Lin, et
al(17) |
| R:
GAGGAGCCGTTCTGAATCTG | 475 |
| ΔNp63 | F:
CTGGAAAACAATGCCCAGAC | 151 | 197 | Lin, et
al(17) |
| R:
GGGTGATGGAGAGAGAGCAT | 348 |
| TAp73 | F:
GGCTGCGACGGCTGCAGAGC | 61 | 257 | Stiewe (3) |
| R:
GCTCAGCAGATTGAACTGGGCCATG | 317 |
| ΔNp73 | F:
CAAACGGCCCGCATGTTCCC | 53 | 256 | Stiewe (3) |
| R:
TGGTCCATGGTGCTGCTCAGC | 308 |
| GAPDH | F:
TCATCAGCAATGCCTCCTGCA | 635 | 118 | Stiewe (3) |
| R:
TGGGTGGCAGTGATGGCA | 752 |
Quantification of each isoform using
real-time RT-PCR
The final volume of 25 μl of RT-PCR reaction
contained 20 ng cDNA, 5 pmol of each primer and ABsolute™ QPCR
SYBR®-Green Mix (Thermo Fisher Scientific, Loughborough,
UK). The reaction was conducted on a Rotor-Gene 6000 thermal cycler
(Qiagen) using PCR cycling conditions as follows: 94°C for 1 min,
57°C for 1 min and 72°C for 1 min for 40 cycles, with a final
extension at 72°C for 10 min. All experiments were performed in
triplicate. The absolute copy numbers were estimated from standard
curves generated from a serial dilution of plasmid construct,
ranging from 30 to 3×106 copies. The relative copy
numbers were normalized to those of GAPDH. Coefficient of variation
<15% and PCR efficiency >0.85 were considered acceptable.
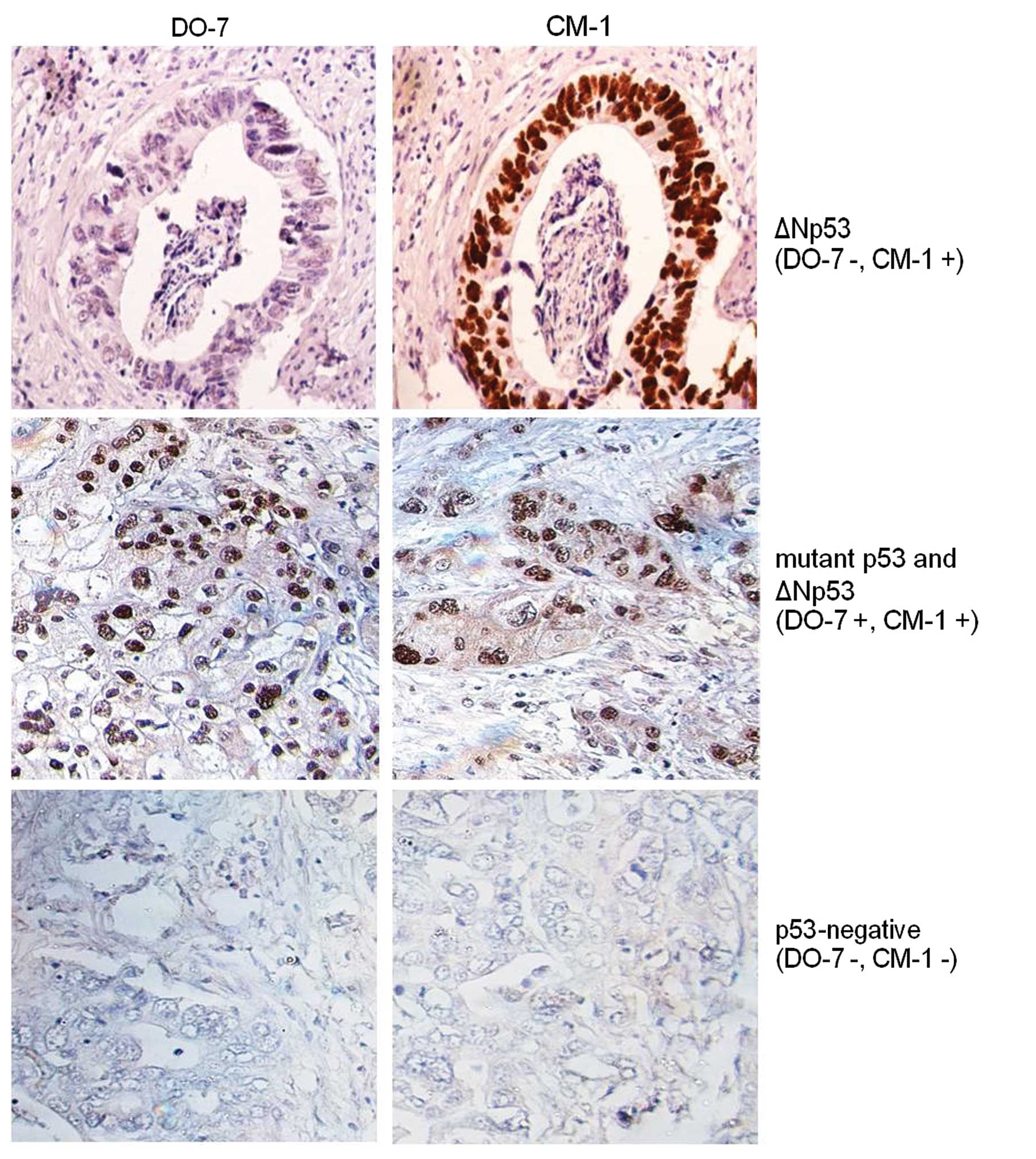
Immunostaining of ΔN isoforms
CCA cell lines were pelleted and embedded in
paraffin. The paraffin-embedded section (5 μm) of either
tissue or cell pellet was deparaffinized and was used for antigen
retrieval in boiled 0.01 M citrate buffer (pH 6.0). Endogenous
peroxidase was inactivated with 100 μl of 3%
H2O2. Non-specific binding was further
blocked with blocking buffer containing phosphate-buffered saline
with Tween-20 (PBST), 30% casein and 5% FBS. Each isoform was
detected with primary antibodies: p53: clone DO-7, epitope 1–45 aa
(Dako, Glostrup, Denmark) and clone CM-1, epitope located in
DNA-binding domain (Signet, Emeryville, CA, USA); ΔNp63: clone 4A4,
epitope 1–205 aa (Dako); and ΔNp73: clone 38c674.2, epitope 2–13 aa
(Imgenex, San Diego, CA, USA). Proteins were detected using the
EnVision system (Dako). The slides were counterstained with
hematoxylin. Positive staining was observed as brown color in the
nuclei and graded as positive when the percentage of positive cells
was >10%, according to a previous study (18). The mutant p53 was defined when
staining was positive for DO-7 and CM-1, while Δ133p53 was positive
only for CM-1
Western blot analysis
Protein was prepared from CCA tissues and cell lines
using TRIzol (Invitrogen, Paisley, UK) and fractionated on 15%
SDS-polyacrylamide gels. The transferred proteins were detected
with 1:100 of CM-1 (Signet) as the primary antibody and
peroxidase-labeled anti-rabbit (Abcam, Cambridge, UK) as the
secondary antibody. Chemiluminescence was detected with the ECL
Plus system (GE Healthcare, Chalfont St. Giles, UK).
Statistical analysis
The significance of isoform expression was analyzed
using the Wilcoxon test. Survival was determined with the
univariate and multivariate Cox regression models, Kaplan-Meier
analysis and the log-rank test. Statistical analyses utilized SPSS
for Windows, version 15.0 (SPSS Inc., Chicago, IL, USA). A p-value
<0.05 was considered to indicate a statistically significant
difference.
Results
Significant increase of ΔN and TAp53
isoforms in CCA cell lines
We examined p53 family isoform transcripts in
CCA cell lines using real-time PCR. The mRNA levels of ΔN and TA
isoforms of p53, p63 and p73 genes were
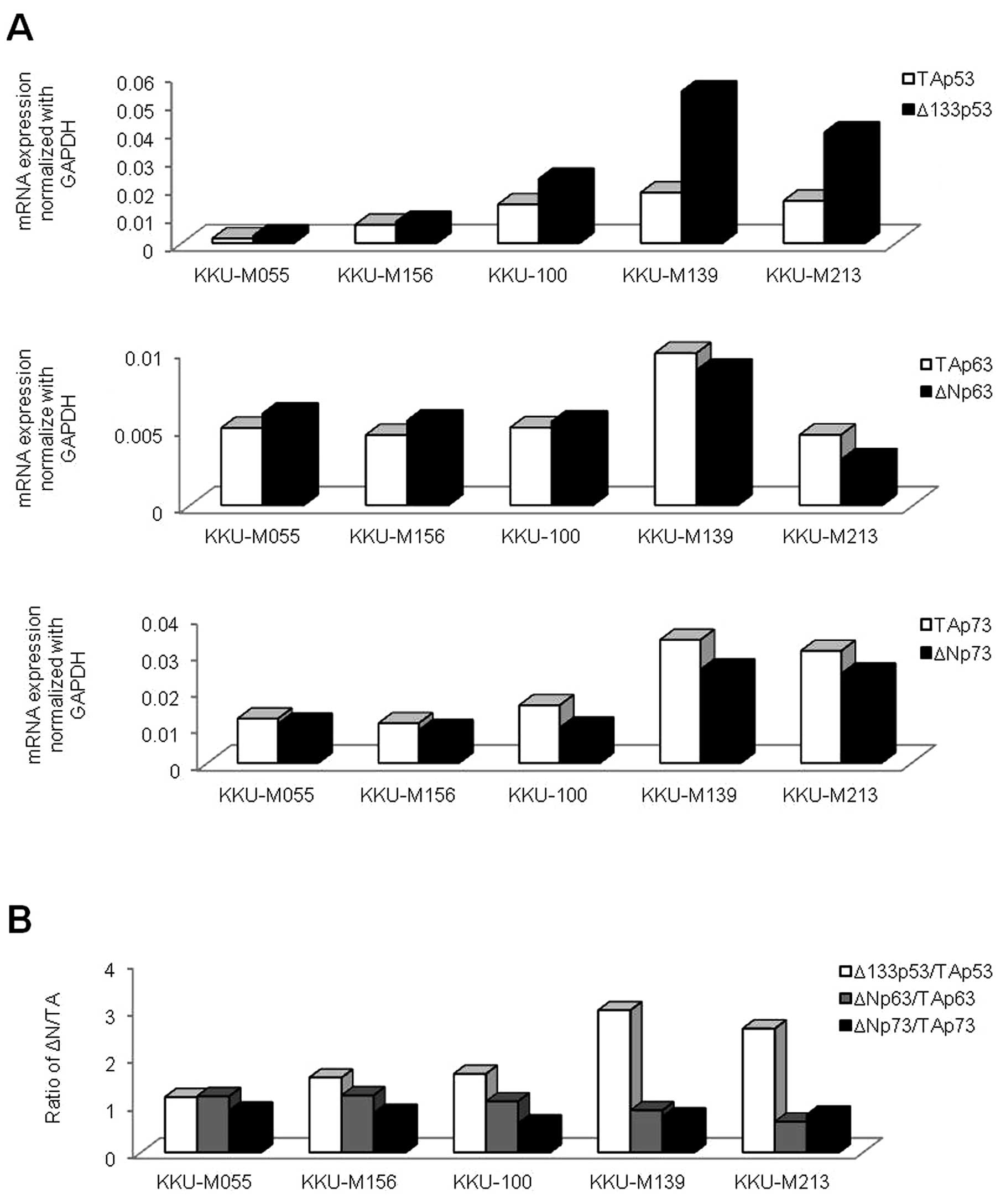
plotted as a relative number to GAPDH (Fig. 2A). The expression of the p53
family was observed in all CCA cell lines, although to a different
extent. Of note, only the Δ133p53/TAp53 expression ratio was
markedly increased (>1.0) in all the CCA cell lines, compared to
ΔNp63/TAp63 and ΔNp73/TAp73 (Fig. 2B). Therefore, we were particularly
interested in the Δ133p53 isoform, since it harbors no TA
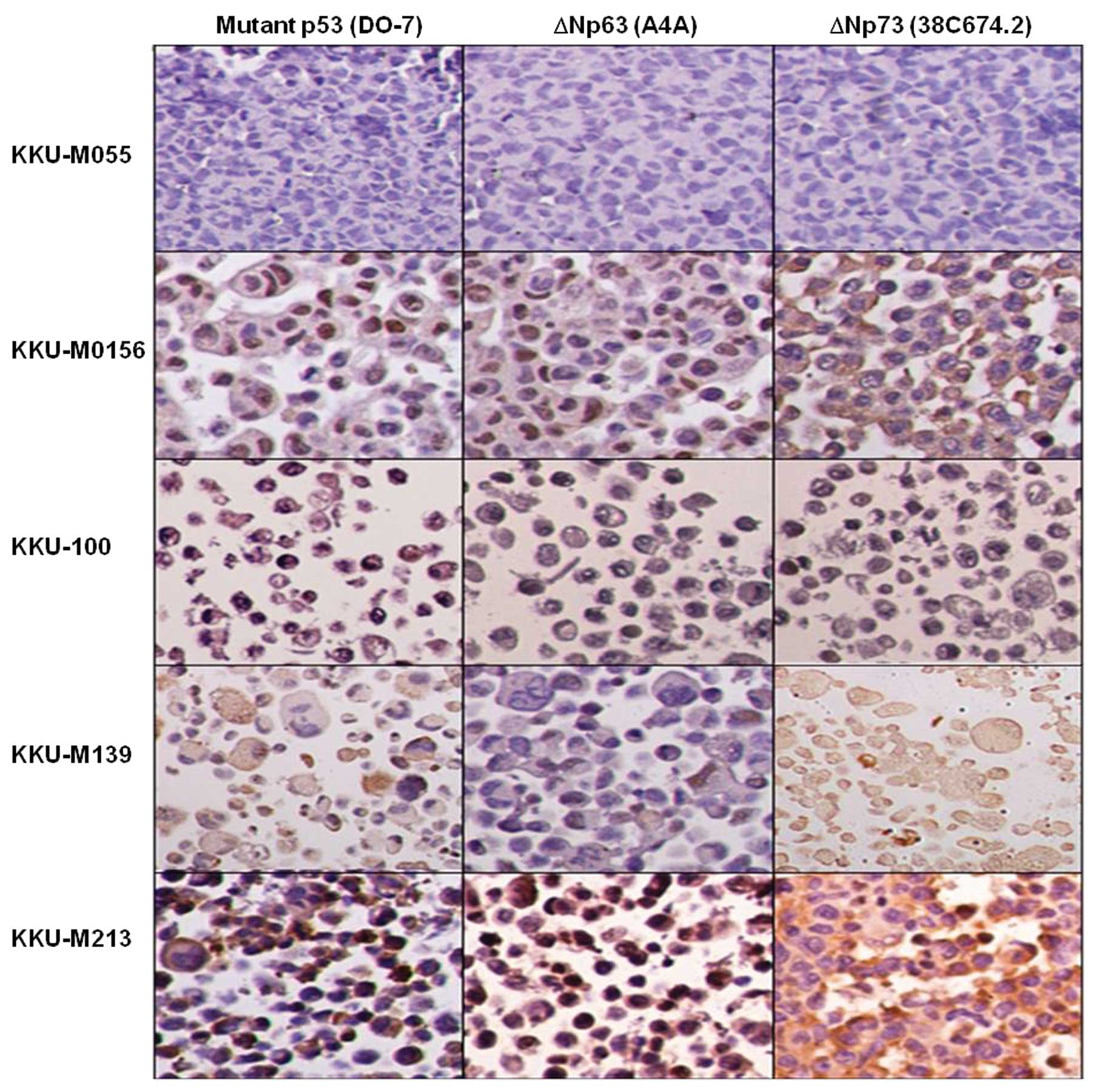
domain. ΔN isoforms of p63 and p73 were also detected by
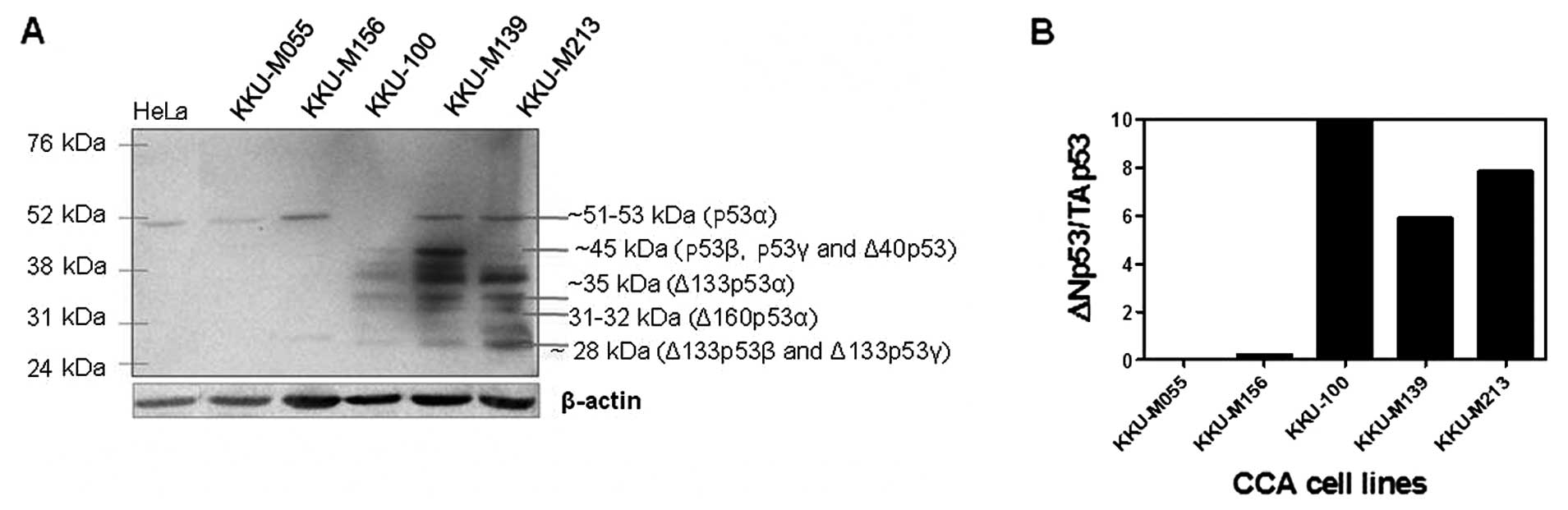
immunostaining (Fig. 3). p53
isoform variants and the full-length (51–53 kD) protein were
detected by western blot analysis, in which the Δ133p53 protein was
highly expressed in the KKU-100, KKU-M139 and KKU-M213 cell lines
(Fig. 4A). Moreover, the
full-length p53 was observed in all the CCA cell lines, with the
exception of KKU-100. The high relative ratio of ΔN/TA p53 protein
was found in the KKU-100, KKU-M139 and KKU-M213 cell lines
(Fig. 4B), suggesting the
disruption of the expression between ΔN and TAp53.
Overexpression of Δ133p53 isoform at the
mRNA level in CCA tissues
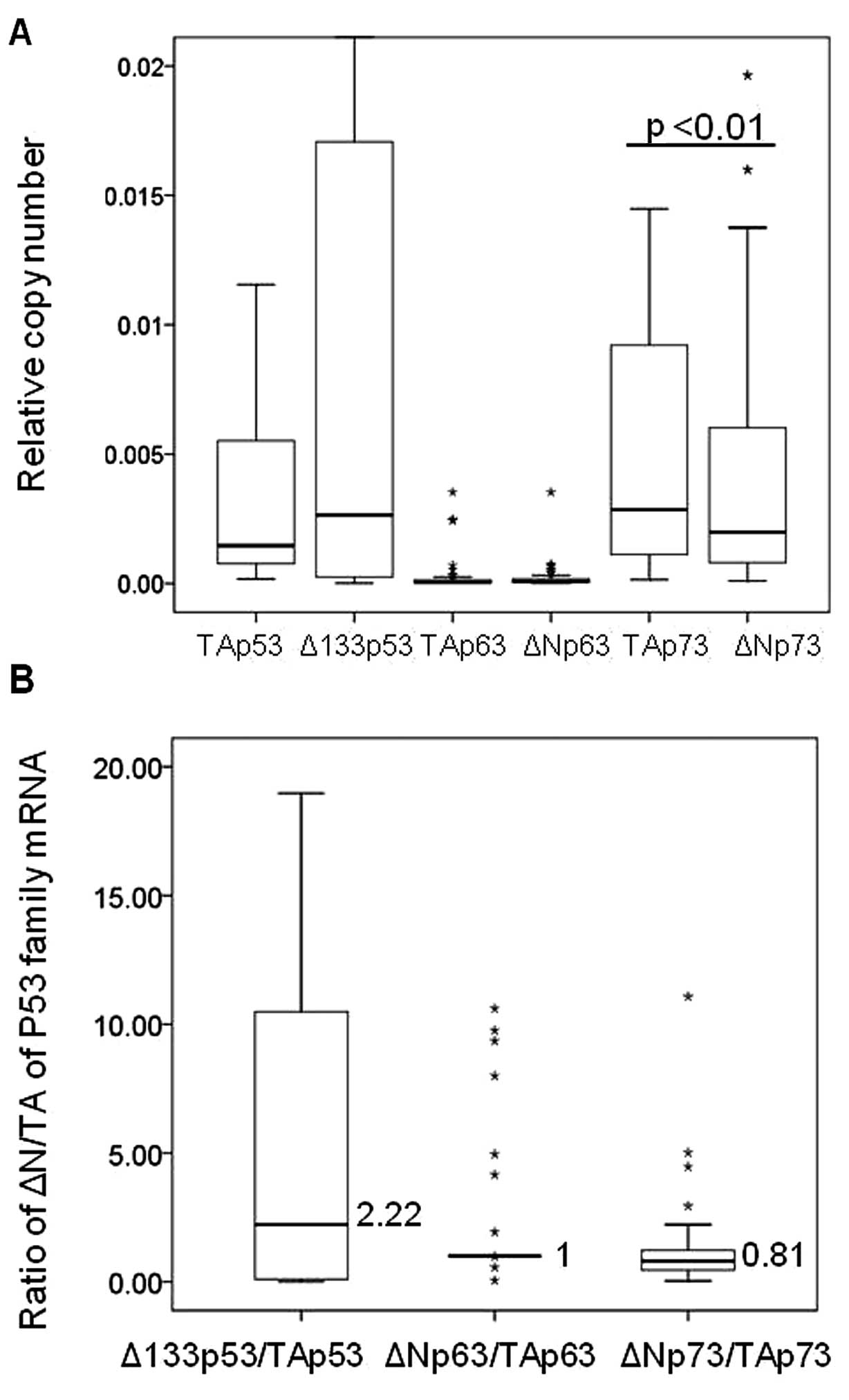
The distribution of mRNA levels for the p53,
p63 and p73 isoforms among the 48 CCA tumor tissues
is shown in Fig. 5A. The median
expression level of Δ133p53 tended to increase compared to
its full-length isoform, whereas TAp73 was significantly
increased compared to ΔNp73 (p<0.01). In addition, the
highest relative ratio of ΔN over the full-length isoform was
clearly obtained in p53 (2.2-fold) (Fig. 5B). These results demonstrate the
overexpression of Δ133p53 in CCA tissues.
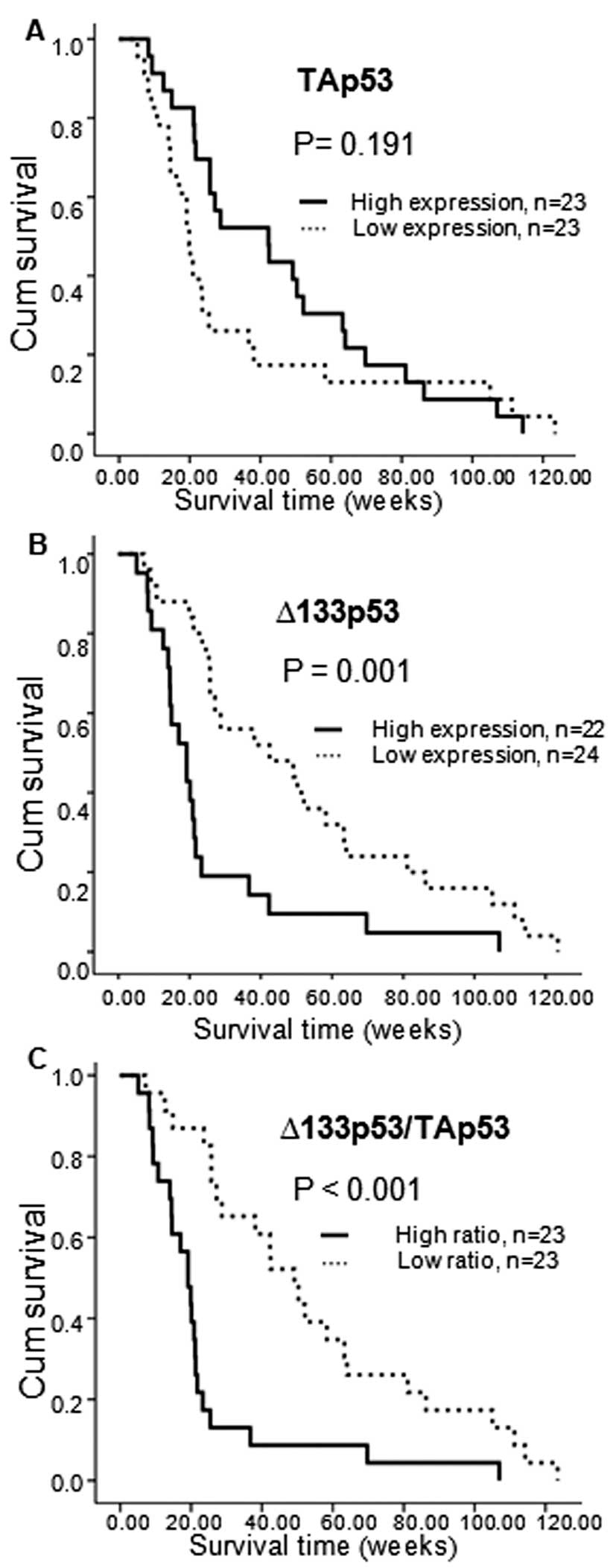
The association of the Δ133p53 transcript
with patient survival was demonstrated using the Kaplan-Meier
analysis. The 48 CCA patients were divided into 2 groups: those
with high and low mRNA expression, according to the individual
median values. Patients with high Δ133p53 and Δ133p53/
TAp53 expression demonstrated a poor overall survival (p=0.001
and p<0.001, respectively) (Fig. 6B
and C).
Overexpression of defective p53
correlates with poor survival
Immunostaining was performed to determine the
predominant p53 isoform expressed in the 48 CCA samples. Out of the
46 CM-1-positive samples, 26 (54.2%) were classified as ΔN isoform
(DO-7-negative) and 20 (41.6%) as mutant p53 (DO-7-positive)
(Fig. 7), suggesting that the
mutant and ΔNp53 isoforms were predominantly expressed in CCA.
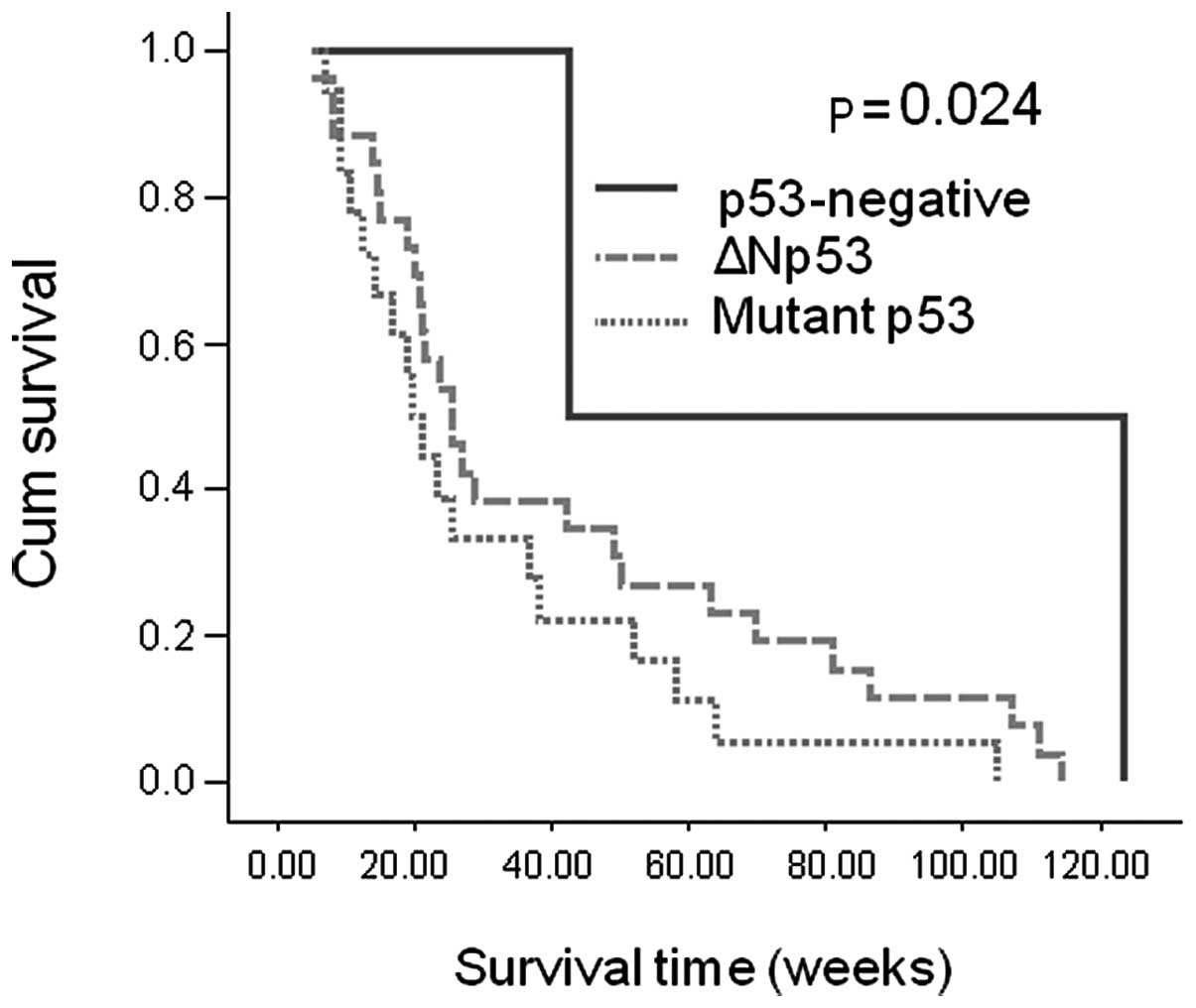
Patients with wild-type p53 exhibited a longer overall survival
than those with defective p53 (p=0.024) (Fig. 8). In addition, multivariate
analysis demonstrated that Δ133p53/TAp53 and mutant p53
protein may be used as independent prognostic factors for CCA
(Table II).
 | Table IICox regression analysis of p53
isoform expression and clinicopathological parameters. |
Table II
Cox regression analysis of p53
isoform expression and clinicopathological parameters.
| Parameters (n) | Univariate
|
Multivariateb
|
|---|
| HR (95% CI) | p-valuea | HR (95% CI) | p-valuec |
|---|
| Age | | | | |
| ≤57 years
(26) | Reference | | | |
| >57 years
(20) | 1.77
(0.96–3.26) | 0.067 | - | - |
| Gender | | | | |
| Male (30) | Reference | | | |
| Female (16) | 0.77
(0.41–1.42) | 0.396 | - | - |
| Histopathology | | | | |
| Invasive
papillary carcinoma (21) | Reference | | | |
| Well
differentiated (17) | 1.65
(0.85–3.22) | 0.042 | NS | NS |
| Moderately
differentiated (4) | 1.08
(0.36–3.26) | 0.896 | NS | NS |
| Poorly
differentiated (4) | 3.99
(1.13–14.05) | 0.031 | NS | NS |
| Staging | | | | |
| I–II (7) | Reference | | | |
| III–IV (39) | 5.67
(1.70–18.69) | 0.011 | 2.43
(1.39–4.24) | 0.002 |
| Chemotherapy | | | | |
| Treatment
(18) | Reference | | | |
| No treatment
(28) | 3.72
(1.80–7.66) | <0.001 | 1.93
(1.17–3.34) | 0.015 |
| Mutant p53
protein | | | | |
| Negative
(26) | Reference | | | |
| Positive
(20) | 2.59
(1.35–4.99) | 0.003 | 1.71
(1.21–2.64) | 0.005 |
| TAp53 | | | | |
| Low expression
(23) | Reference | 0.195 | - | - |
| High expression
(23) | 0.67
(0.37–1.22) | | | |
| Δ133p53 | | | | |
| Low expression
(24) | Reference | 0.002 | NS | NS |
| High expression
(22) | 2.67
(1.44–4.97) | | | |
| Δ133p53/
TAp53 | | | | |
| Low ratio
(23) | Reference | <0.001 | 3.73
(1.81–7.66) | 0.007 |
| High ratio
(23) | 3.25
(1.73–6.11) | | | |
Discussion
In this study, we demonstrated the expression of ΔN
isoforms of all p53 family members at the mRNA and protein
levels. A significant correlation between the mRNA expression of
Δ133p53/TAp53 and mutant p53 protein with poor overall
survival was observed, demonstrating its value as a prognostic
marker in CCA. In normal cells, the P1 promoter encodes the
TAp53 and Δ40p53 isoforms, while P2 encodes
Δ133p53. The autoregulation of any p53 isoform level depends
on switching between promoters (7). Therefore, the upregulation of
Δ133p53 expression, leading to the increased ratio of
Δ133p53/TAp53 in CCA, may reflect the preferential use of
the P2 promoter. The increase of Δ133p53 expression in CCA
may negatively regulate p53 transcriptional activity in the control
of cell cycle arrest and apoptosis, resulting in the pathogenesis
of CCA. An increase of Δ133p53 expression has been reported
in renal cell (19), breast
(7) and colon carcinomas (20). The overexpression of Δ133p53
has been shown to correlate with the progression of premalignant
lesions to colon cancer, by signaling an escape from the senescence
barrier (20). Our findings, as
well those from other studies, suggest the value of Δ133p53
as a prognostic biomarker. Moreover, the present study also
demonstrates the significance of the correlation between the
equilibrium ratio Δ133p53/TAp53 and poor clinical outcome in
CCA. The Δ133p53/TAp53 ratio is a more sensitive marker than
either TAp53 or Δ133p53 alone. Thus, several studies
have used the ΔN/TA isoform ratio as a biomarker. The
ΔNp73/TAp73 ratio has been associated with clinical response
to chemotherapy in hepatocellular carcinoma and various cancer cell
lines (13,21).
In this study, we detected mutant p53 and ΔNp53
simultaneously in CCA tissues, suggesting that mutation and the
ΔNp53 isoform play a critical role in p53 inactivation. The
incidence rate of p53 mutation in 20 out of the 48 CCA smples (42%)
in our study, is in agreement with data from a previous study
(41.6%) (15). Patients with
mutant p53 tended to have poorer overall survival compared to those
with ΔNp53 (p>0.05), suggesting that mutant p53 was completely
non-functional, while ΔNp53 enabled the mediation of p53
transcriptional activity. Further studies are required to elucidate
the role of ΔNp53 and its effect on TAp53 in CCA. The specific p53
isoforms could not be accurately detected by western blot analysis,
due to the limitation of the commercial availability of p53
antibodies. In addition, DO-7 detected mutant p53, while CM-1
detected all p53 isoforms. Therefore, the combination of these two
antibodies enables the discrimination between mutant p53 and ΔNp53.
We recommend immunohistochemistry rather than western blot analysis
for the detection of p53 isoforms in clinical specimens, since this
procedure is easier and less time-consuming. In conclusion, to our
knowledge, this study is the first to demonstrate the value of
Δ133p53/TAp53 as a prognostic biomarker in CCA.
Acknowledgements
The present study was supported by the
Higher Education Research Promotion and National Research
University Project of Thailand, Office of the Higher Education
Commission, through the Health Cluster (SHeP-GMS),
Khon Kaen University; the Centre for Research and Development of
Medical Diagnostic Laboratories, Faculty of Associated Medical
Sciences; and the Graduate School, Khon Kaen University, Khon Kaen,
Thailand.
References
|
1
|
Sripa B and Pairojkul C:
Cholangiocarcinoma: lessons from Thailand. Curr Opin Gastroenterol.
24:349–356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blechacz B and Gores GJ:
Cholangiocarcinoma: advances in pathogenesis, diagnosis, and
treatment. Hepatology. 48:308–321. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stiewe T: The p53 family in
differentiation and tumorigenesis. Nat Rev Cancer. 7:165–168. 2007.
View Article : Google Scholar
|
|
4
|
Murray-Zmijewski F, Lane DP and Bourdon
JC: p53/p63/p73 isoforms: an orchestra of isoforms to harmonise
cell differentiation and response to stress. Cell Death Differ.
13:962–972. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mills AA, Zheng B, Wang XJ, Vogel H, Roop
DR and Bradley A: p63 is a p53 homologue required for limb and
epidermal morpho-genesis. Nature. 398:708–713. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bourdon JC: p53 and its isoforms in
cancer. Br J Cancer. 97:277–282. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bourdon JC, Fernandes K, Murray-Zmijewski
F, et al: p53 isoforms can regulate p53 transcriptional activity.
Genes Dev. 19:2122–2137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marcel V, Perrier S, Aoubala M, et al:
Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53
transcript. FEBS Lett. 584:4463–4468. 2010.
|
|
9
|
Helton ES, Zhu J and Chen X: The unique
NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the
PPXY motif are required for the transcriptional activity of the
DeltaN variant of p63. J Biol Chem. 281:2533–2542. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marchini S, Marabese M, Marrazzo E, et al:
DeltaNp63 expression is associated with poor survival in ovarian
cancer. Ann Oncol. 19:501–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu SS, Chan KY, Cheung AN, Liao XY, Leung
TW and Ngan HY: Expression of deltaNp73 and TAp73alpha
independently associated with radiosensitivities and prognoses in
cervical squamous cell carcinoma. Clin Cancer Res. 12:3922–3927.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marabese M, Marchini S, Marrazzo E, et al:
Expression levels of p53 and p73 isoforms in stage I and stage III
ovarian cancer. Eur J Cancer. 44:131–141. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Müller M, Schilling T, Sayan AE, et al:
TAp73/Delta Np73 influences apoptotic response, chemosensitivity
and prognosis in hepatocellular carcinoma. Cell Death Differ.
12:1564–1577. 2005.PubMed/NCBI
|
|
14
|
Soldevilla B, Díaz R, Silva J, et al:
Prognostic impact of ΔTAp73 isoform levels and their target genes
in colon cancer patients. Clin Cancer Res. 17:6029–6039. 2011.
|
|
15
|
Limpaiboon T, Sripa B, Wongkham S,
Bhudhisawasdi V, Chau-in S and Teerajetgul Y: Anti-p53 antibodies
and p53 protein expression in cholangiocarcinoma.
Hepatogastroenterology. 51:25–28. 2004.PubMed/NCBI
|
|
16
|
Yang B, House MG, Guo M, Herman JG and
Clark DP: Promoter methylation profiles of tumor suppressor genes
in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol.
18:412–420. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin Z, Nan Y, Zhang X, Zhao Y, Kim C and
Kim I: Reverse transcription-polymerase chain reaction and western
blotting analysis for detection of p63 isoforms in uterine cervical
cancers. Int J Gynecol Cancer. 16:1643–1647. 2006. View Article : Google Scholar
|
|
18
|
Furubo S, Harada K, Shimonishi T,
Katayanagi K, Tsui W and Nakanuma Y: Protein expression and genetic
alterations of p53 and ras in intrahepatic cholangiocarcinoma.
Histopathology. 35:230–240. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song W, Huo SW, Lü JJ, et al: Expression
of p53 isoforms in renal cell carcinoma. Chin Med J (Engl).
122:921–926. 2009.PubMed/NCBI
|
|
20
|
Fujita K, Mondal AM, Horikawa I, et al:
p53 isoforms Delta133p53 and p53beta are endogenous regulators of
replicative cellular senescence. Nat Cell Biol. 11:1135–1142. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Conforti F, Yang AL, Agostini M, et al:
Relative expression of TAp73 and ΔNp73 isoforms. Aging (Albany,
NY). 4:202–205. 2012.
|