Introduction
Angiogenic factors are expressed in human breast
cancer (1) and benign lesions
associated with high vascular density are correlated with an
increased risk of developing cancer (2,3).
Furthermore, breast cancer angiogenesis correlates directly with
the presence of bone marrow micro-metastases (4) and with survival (5). The quantification of angiogenesis
might also help to predict the possible occurrence of cancer
progression (6,7) and of tumor response to treatment
(reviewed in ref. 8). The
egfl7 gene is specifically expressed by blood vessel
endothelial cells during normal embryonic development and in the
adult (9,10). Three independent studies showed
that Egfl7 expression is deregulated in human cancer: high
expression levels of Egfl7 transcripts are correlated with a more
advanced stage of human colon cancer and with lymph node invasion,
with no correlation with overall survival or progression-free
survival (11). Egfl7 is expressed
by cancer cells of human hepatocarcinoma and high levels of
expression are correlated with poor survival (12). In a series of human gliomas, high
levels of Egfl7 in tumor tissue correlate with higher tumor grade,
Ki67 index and microvascular density (13). Here, we performed the first study
of the expression of Egfl7 transcripts levels and protein
localization in human breast cancer lesions. As reported in other
human cancers, Egfl7 is strongly expressed by breast cancer cells,
in addition to blood vessel endothelial cells. However, Egfl7
expression is associated with smaller lesions and with better
prognosis factors in human breast cancer, raising concerns about
the Egfl7-blocking therapies currently under development.
Materials and methods
Patients and samples
Paraffin-embedded or frozen samples were obtained
from 205 patients (203 women and 2 men) with 211 mammary carcinoma
and for whom the definitive diagnosis was made between January 1
and July 31, 2005 following surgical resection. Detailed clinical
data are provided in Table I. All
tissue samples were processed and stored at the Pathology
department of the Centre Oscar Lambret, Lille, France.
 | Table IClinical characteristics of the human
breast cancer samples. |
Table I
Clinical characteristics of the human
breast cancer samples.
| All | DCIS | Invasive
carcinoma |
|---|
| Tumors (n) | 211 | 37 | 174 |
| Patients (n) | 205 | 36 | 168 |
| Gender | | | |
| Female | 203 | 36 | 167 |
| Male | 2 | 0 | 2 |
| Age, years | | | |
| Mean (SD) | 58 (11) | 55 (10) | 59 (11) |
| Median
(min-max) | 57 (39–92) | 54 (39–75) | 57 (39–92) |
| Histological
type | | | |
| DCIS | 18% (37/211) | | 17% (30/174) |
| Ductal | 67% (141/211) | | 81% (141/174) |
| Lobular | 14% (30/211) | | 2% (3/174) |
| Other | 1% (3/211) | | |
| Tumor size, mm | | | |
| Mean (SD) | 18 (14) | 18 (17) | 18 (13) |
| Median
(min-max) | 15 (2–100) | 10 (2–60) | 15 (2–100) |
| T from TNM | | | |
| T1a | | | 8% (13/174) |
| T1b | | | 27% (47/174) |
| T1c | | | 40% (70/174) |
| T2 | | | 23% (40/174) |
| T3 | | | 2% (4/174) |
| SBR | | | |
| I | | | 27% (47/174) |
| II | | | 58% (101/174) |
| III | | | 15% (26/174) |
| ER | | | |
| Positive | 90% (182/203) | 88% (30/34) | 90% (152/169) |
| PR | | | |
| Positive | 65% (131/203) | 62% (21/34) | 66% (111/169) |
| HER2 (IHC and
FISH) | | | |
| 0+ | | | 42% (71/169) |
| 3+ | | | 6% (10/169) |
| Undetermined | | | 3% (5/169) |
| Positive | | | 6.5% (11/169) |
| Emboli | | | |
| Present | 21% (19/89) | 4 cases | 22% (19/85) |
| Necrosis | | | |
| Present | 73% (49/67) | 81% (21/26) | 68% (28/41) |
| DCIS grade | | | |
| 1 | 20% (29/142) | 16% (6/37) | 22% (23/105) |
| 2 | 36% (51/142) | 24% (9/37) | 40% (42/105) |
| 3 | 44% (62/142) | 60% (22/37) | 38% (40/105) |
| Lobular carcinoma
in situ | | | |
| Present | - | - | 16% (27/174) |
| pN | | | |
| pN0 | 57% (121/211) | 38% (14/37) | 61% (107/174) |
| pN0i+
and pmiN1 | 7% (15/211) | - | 9% (15/174) |
| pN+ | 25% (49/211) | 62% (23/37) | 29% (49/174) |
Antibodies
Primary goat anti-human Egfl7 antibody (AF3638
R&D Systems), mouse anti-human Egfl7 (sc-101349, Santa Cruz
Biotechnologies), goat anti-human actin (sc-1615, Santa Cruz),
biotinylated rabbit anti-goat (305-066-006, Jackson
Immunoreagents), peroxidase-mouse anti-goat (A9452-1VL, Sigma), and
mouse anti-mouse (NA-931, GE Healthcare) antibodies were
reconstituted and stored according to the manufacturer’s
instructions.
Immunohistochemistry protocol
Paraffin sections (4 μm) were processed using
a Discovery (Ventana) automat, including an internal reference
slide for quality control and a ‘no-antibody’ control slide in each
batch. Slides were heated 8 min at 75°C, incubated 8 min in EZ-PREP
(Ventana), rinsed with reaction buffer (Ventana), heated 2 min at
37°C and rinsed with reaction buffer. Slides were then incubated 8
min at 95°C in CC1 (Ventana), 40 min at 100°C and 8 min at room
temperature, rinsed with reaction buffer and heated 2 min at 37°C,
then rinsed again. Slides were then incubated 4 min in inhibitor D
solution at room temperature, rinsed and incubated with a
polyclonal anti-hEgfl7 antibody (1/30, AF3638 R&D) diluted in
Antibody Diluent (Cell Marque) for 5 h at room temperature. Slides
were then rinsed with reaction buffer, heated for 2 min at 37°C and
incubated 30 min at room temperature in biotinylated rabbit
anti-goat antibody (1/500, Jackson ImmunoResearch), rinsed and
processed for staining using the Discovery DAB Map Kit (Ventana).
Slides were then rinsed and dehydrated before mounting in
Vectamount Permanent Mounting Medium (Vector laboratories) and
dried. Following staining, slides were analyzed twice, each time by
two independent observers, including a breast pathologist
(Marie-Christine Baranzelli), using an Axioplan 2 microscope
(Zeiss) and compared to corresponding hematoxylin/phloxin
safran-stained slides used for identification of the tumor
sub-regions. Staining was analyzed on the entire tumor region of
the samples and a semi-quantitative Egfl7 intensity scale was
established by the observers. Immunostainings were analyzed in 5
different cell populations corresponding to infiltrating tumor
cells, DCIS or lobular carcinoma tumor cells, peritumoral blood
vessel endothelial cells, stromal cells and normal mammary cells
surrounding the tumors. In 11 cases, the staining was also analyzed
within in situ lobular carcinoma cells. The exact and
approximate concordance rates of both evaluations were respectively
77% (121/158) and 98% (155/158, p<0,001). The κ concordance
coefficient was 0.65 (SE 0005, p<0.0001) correcting for a 34%
chance agreement. In 35 cases, the double reading resulted in a
decreased estimation of the intensity of staining.
Cell culture and transfections
Primary human umbilical cord endothelial cells
(HUVEC) were from Lonza and cultured according to the supplier’s
recommendations between passage 2 and 5. 3T3 fibroblasts (ATCC
CRL-1658) were cultured in 78.5 cm2 dishes (Falcon) in
Dulbecco’s modified Eagle’s medium (Invitrogen), 10% calf or fetal
calf serum (Hyclone), 10 kU/l penicillin, 10 mg/l streptomycin. For
transient transfection, 3T3 cells were plated at 15,000
cells/cm2 in 10 cm2 plates, grown overnight
and transfected with 23 fmoles of pcDNA3-hEgfl7 expressing vector
in OptiMEM (Invitrogen) and in the presence of Exgen 500 reagent (3
μl/μg DNA, Euromedex) for 6 h at 37°C in a 5%
CO2/95% air atmosphere. For RNA interference, HUVEC were
plated in 4 cm2 well-plates (25,000
cells/cm2) and transfected the next day with 60 pmoles
siRNA (Dharmacon) in Primefect siRNA reagent (Lonza) mixed with
EGM-2. After 24 h, EGM-2 was added and cells cultured for 24 or 48
h. For western blot analysis, proteins were analyzed by 12%
SDS-PAGE, blotted onto Immobilon-P (Millipore) and probed using the
primary goat anti-human Egfl7 antibody (AF3638 R&D Systems,
1/1,000) or a goat anti-human actin antibody (c-11, sc-1615, Santa
Cruz Biotechnologies, 1/1,000) in PBS, 0.05% Tween-20, 5%
non-fat-dry milk overnight at 4°C under constant mixing and further
incubated with an horseradish peroxidase-coupled secondary antibody
(1/10,000, A9452-1VL Sigma) in PBS, 0.05% Tween-20, 5% non-fat dry
milk. Immunocomplexes were revealed using the Western Lightning-ECL
kit (Perkin-Elmer) after exposure to Hyperfilm ECL (GE
Healthcare).
Reverse-transcription quantitative
PCR
Total RNA was recovered from 10-μm paraffin
sections using the MasterPure RNA purification kit (Epicentre
Biotechnologies). Purified total RNA was treated with DNAse and
retro-transcribed using the High Capacity cDNA Reverse
Transcription (Life Technologies). Quantitative PCR (qPCR) was
performed using a StepOne system (Applied Biosystems) and the egfl7
(Hs_00211952_m1) and β2-microglobulin (B2M) TaqMan assays (Applied
Biosystems) using the ΔCT method with B2M levels as
reference. ΔΔCT was calculated using one arbitrarily
chosen sample set to 1.
Statistical analyses
Comparisons of percentages were performed using the
χ2 and Fisher’s exact tests. Mean comparisons were
performed using non-parametric rank Kruskall-Wallis test (for >2
categories) and Mann-Whitney U test (2 categories). Correlations
were analyzed using the non-parametric Spearman correlation
coefficient. The non-parametric Kaplan-Meier method was used for
analysis of survival data. The search for potential prognosis
factors was performed using log-rank test for categorical data and
Cox model for continuous variables. Variables were considered as
significant at the p=0.01 level due to the multiplicity of tests.
Reliability studies were performed using κ statistics.
Ethics
Sample storage, handling and analysis were done
according to the European regulations and the Helsinki Declaration.
Patient consent and legal authorizations were obtained for all the
analyses performed and for the processing of patients personal
data. The protocol was approved by the ‘Comité de Protection des
Personnes Nord-Ouest IV’ on January 12, 2010.
Results
Specificity of detection of Egfl7 in
breast cancer tissues
Egfl7 transcripts and protein expression levels were
evaluated in human breast cancer samples. Conditions for detecting
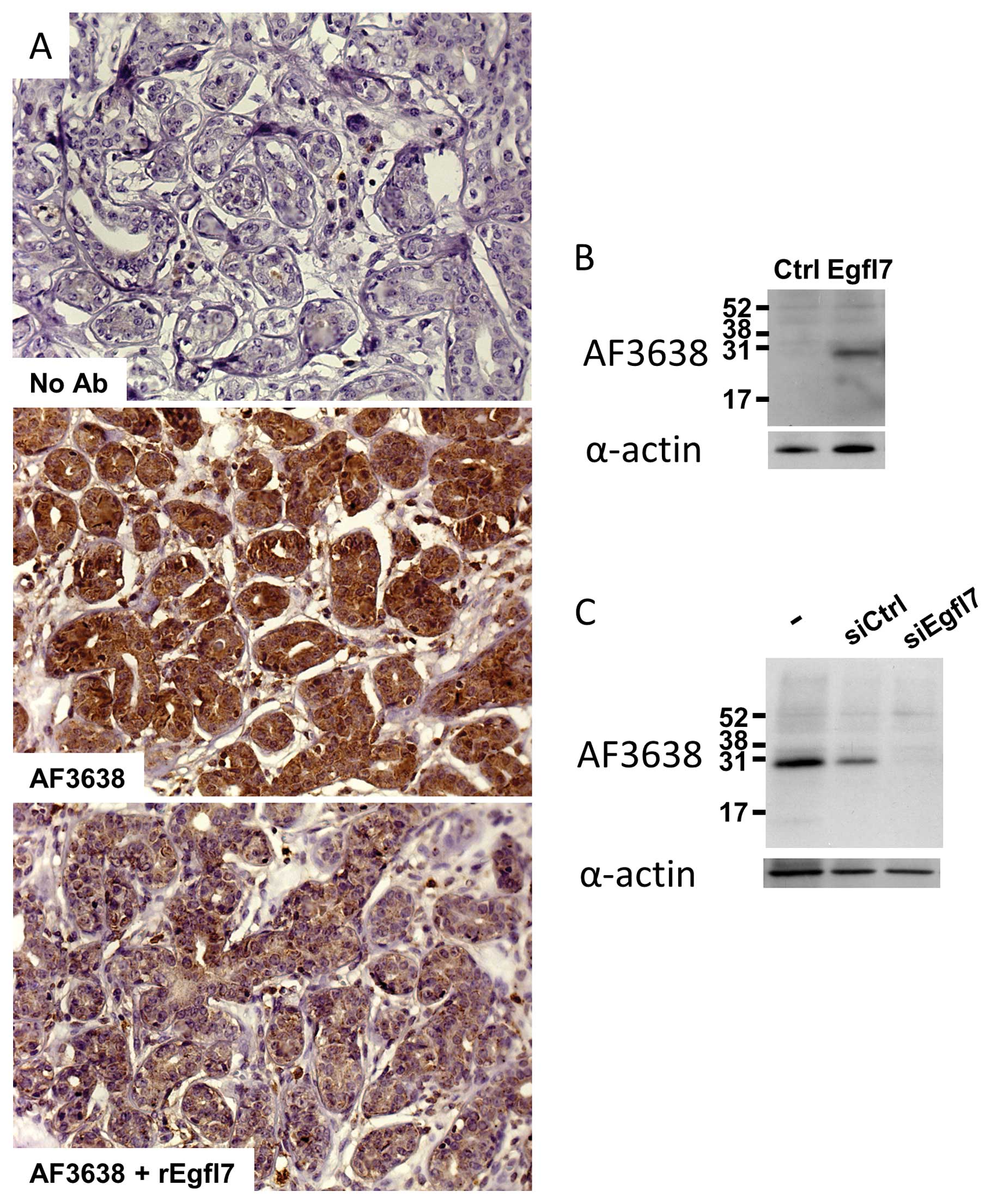
Egfl7 by IHC were setup and the specificity of detection was
assessed by staining a positive infiltrating ductal carcinoma with
the antibody in the presence of a 2-fold molar excess of
recombinant Egfl7 protein (14).
As expected from a specific interaction, the titration of the
antibody with the recombinant protein strongly reduced the signal
(Fig. 1A). Furthermore, when 3T3
cells, which do not normally express significant amounts of Egfl7
(9), were transfected with a
plasmid encoding the full-length human Egfl7, immunoblot analysis
of cell extracts using the antibody also used in IHC showed a major
band at the Egfl7 expected size (30 kDa), whereas no signal was
detected when using cells transfected with an empty vector
(Fig. 1B). Finally, when primary
human umbilical vein endothelial cells (HUVEC), which spontaneously
express high amounts of Egfl7, were treated with a siRNA
specifically targeting endogenous egfl7 transcripts
(15), a strong decrease in
protein levels was detected by immunobloting using the IHC
antibody, whereas no differences were noted when using a control
siRNA which does not target egfl7 (Fig. 1C). Altogether, these data indicate
that the chosen antibody recognizes both the endogenous endothelial
cell Egfl7 and the recombinant protein.
Expression of Egfl7 in invasive breast
carcinoma
Two-hundred and eleven (211) human samples were
analyzed for Egfl7 expression and localization by IHC,
corresponding to 174 invasive carcinoma and 37 DCIS (Table I). Since no references were
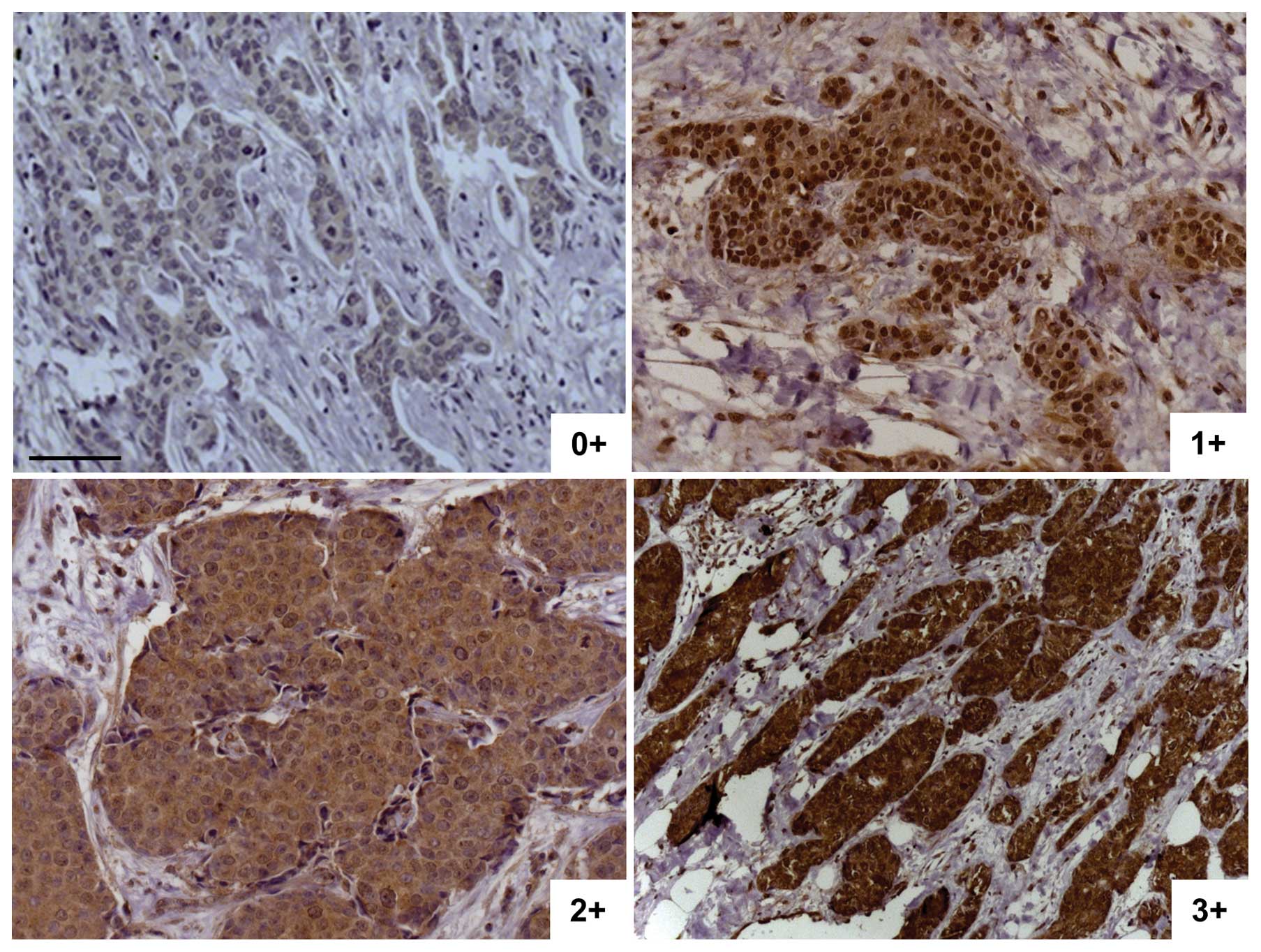
established prior to this work, a semi-quantitative scaling was
established within the sample set. Slides were scored from 0+ (no
cytoplasmic staining) to 3+ (intense cytoplasmic staining, Fig. 2). In most cases, the staining was
homogeneous within the same tumor. In case of heterogeneous
staining within a tumor, the sample was ranked with the highest
score. Invasive breast carcinoma samples were analyzed for Egfl7
protein localization by IHC. Expression of Egfl7 was significantly
higher in invasive tumor cells than in normal epithelial cells (71
and 41% of Egfl7-positive tumors added together, respectively,
p=0.003, Table II). Furthermore,
the levels of Egfl7 among the lesions analyzed were significantly
higher in invasive ductal when compared to invasive lobular
carcinoma (78 and 43%, respectively, p=0.001 χ2 test,
p=0.003 Fisher’s exact test, Table
III). On the other hand, no differences in protein staining
were noted between ER+ and ER− tumors and the
quantities of Egfl7 protein were not correlated with the tumor size
(p=0.56), the patient age (p=0.45), the HER2 status (p=0.25), the
presence of emboli (p=0.36) or with necrosis (p=0.93). It should
also be noted that there was no specific expression profile in the
triple-negative (ER= 0, PR= 0, HER2= 0) invasive lesions (n=8).
 | Table IIEgfl7 expression levels are higher in
tumor cells than in normal cells. |
Table II
Egfl7 expression levels are higher in
tumor cells than in normal cells.
| Egfl7 score | Invasive tumor
cells (n=158) | Normal glandular
cells (n=111) |
|---|
| 0 | 29% (45) | 59% (65) |
| 1+ | 49% (78) | 36% (40) |
| 2+ | 18% (29) | 5% (6) |
| 3+ | 4% (6) | 0% (0) |
 | Table IIIEgfl7 expression levels are higher in
ductal than in lobular invasive carcinoma. |
Table III
Egfl7 expression levels are higher in
ductal than in lobular invasive carcinoma.
| Egfl7 score | Invasive ductal
(n=127) | Invasive lobular
(n=28) |
|---|
| 0 | 22% (28) | 57% (16) |
| 1+ | 53% (67) | 36% (10) |
| 2+ | 20% (26) | 7% (2) |
| 3+ | 5% (6) | 0% (0) |
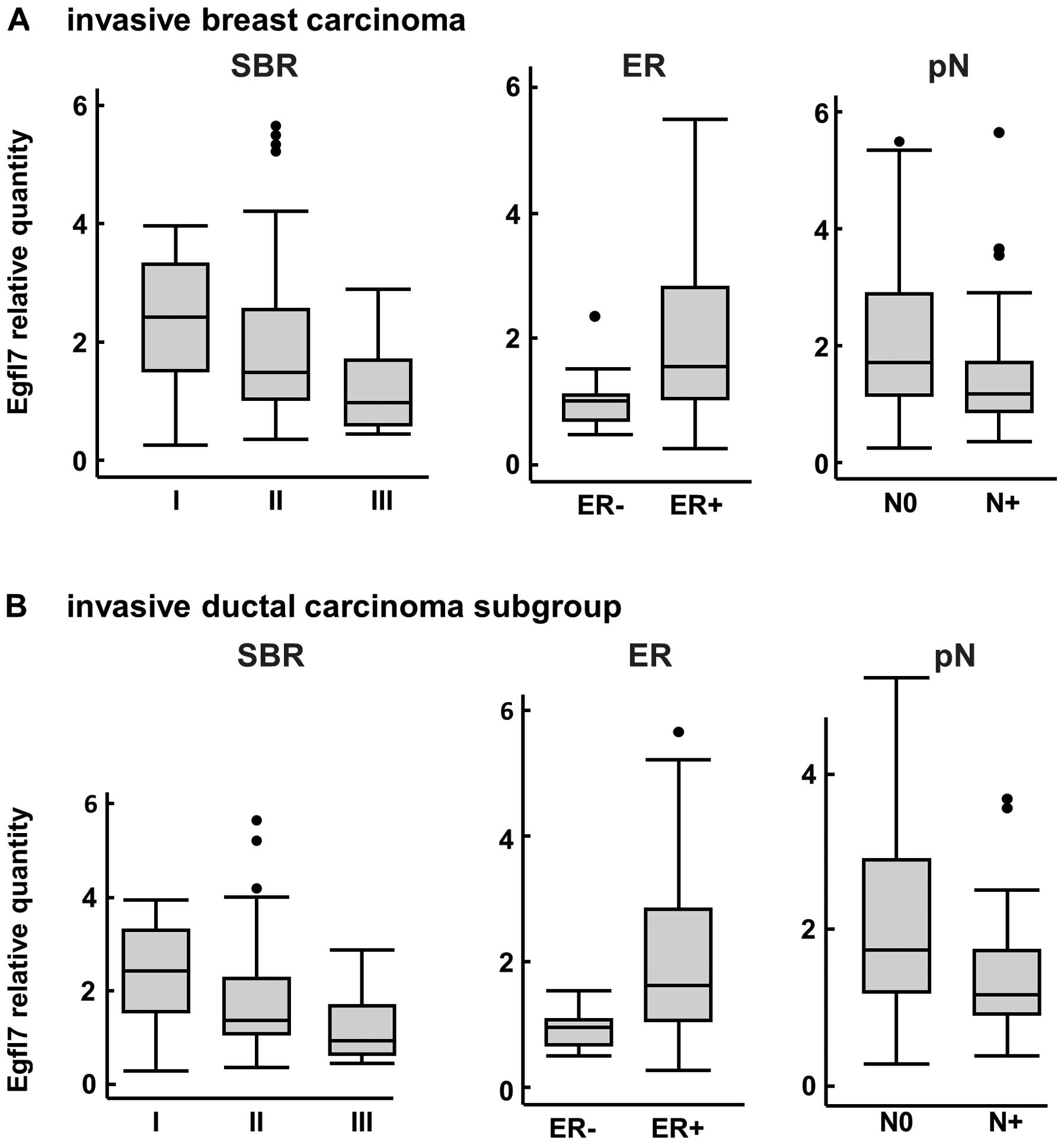
Regarding the expression levels of the Egfl7
transcripts within the same population, no differences between
invasive ductal and invasive lobular carcinoma were noted (data not
shown). However, the Egfl7 transcript levels were inversely
correlated with the Scarf-Bloom-Richardson (SBR) score (p=0.008,
Fig. 3A). Egfl7 transcript levels
were also significantly higher in ER+ tumors than in
ER− tumors (p= 0.002) whereas no correlations were
observed when comparing the progesterone receptor status to Egfl7
expression levels (data not shown). Within the primary invasive
lesions, the expression levels of Egfl7 transcripts were
significantly higher in tumors which were not associated with
axillary lymph node invasion (p=0.003, Fig. 3A) and the protein staining followed
a similar trend (p= 0.06, data not shown). There were no
correlations between the Egfl7 RNA levels and the size of the
tumor, the age, or with the HER2 status, the presence of emboli, or
with necrosis.
Interestingly, within the invasive ductal carcinoma
sub group, we confirmed that the SBR score significantly correlated
with the expression levels of Egfl7 transcripts (p=0.008, Fig. 3B). In both instances, a higher SBR
score corresponded to lower Egfl7 levels. The correlation between
the ER status and Egfl7 transcript levels was also confirmed in
this subpopulation, as ER+ tumors expressed higher
levels of Egfl7 than ER− (p=0.008, Fig. 3B). Furthermore, there was a
significant correlation between the expression levels of Egfl7
transcripts and the axillary lymph node invasion when analyzing
this subgroup (p=0.05 and p=0.01, Fig.
3B), confirming the trend initially observed within the
invasive carcinoma global population.
Overall, within the invasive breast carcinoma
population, Egfl7 expression was preferentially observed in the
invasive ductal breast carcinoma and was associated with better
prognosis factors and with no lymph node invasion.
Expression of Egfl7 in in situ
carcinoma
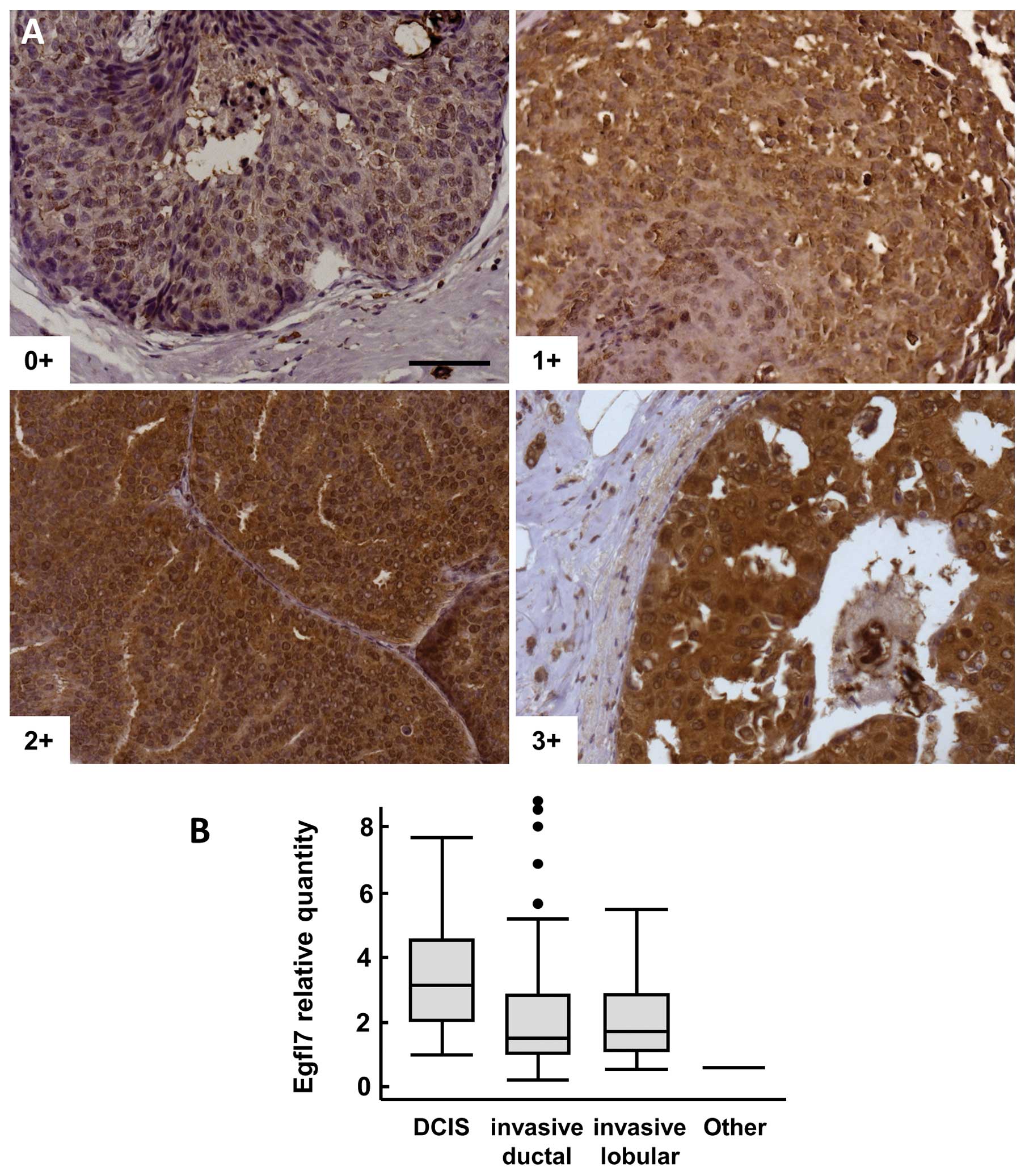
Ductal carcinoma in situ
Pure DCIS lesions (i.e., not associated with
invasive carcinoma) were also analyzed by IHC, 82% of the samples
were positive (score ≥1+) for Egfl7 (Fig. 4A). Among these samples, Egfl7
staining intensities were significantly higher in tumor cells than
in normal peritumoral glandular cells (p=0.008, Table IV). There was no significant
correlation with tumor grade, but a trend to a higher frequency of
samples ranked 2+ and 3+ when compared to 0+ and 1+ (p=0.07) was
noted in this population.
 | Table IVComparison of expression of Egfl7 in
pure DCIS and in DCIS associated with invasive lesions. |
Table IV
Comparison of expression of Egfl7 in
pure DCIS and in DCIS associated with invasive lesions.
| Pure DCIS
| DCIS associated
with invasive lesions
|
|---|
| Egfl7 score | Tumor cells
(n=34) | Normal glandular
cells (n=31) | Invasive tumor
cells (n=158) | DCIS tumor cells
(n=79) |
|---|
| 0+ | 18% (6) | 48% (15) | 29%
(45) | 10% (8) |
| 1+ | 26% (9) | 52% (16) | 49%
(78) | 49% (39) |
| 2+ | 32% (11) | 0% (0) | 18%
(29) | 34% (27) |
| 3+ | 24% (8) | 0% (0) | 4% (6) | 6% (5) |
In DCIS lesions associated with invasive carcinomas,
the frequency of Egfl7-positive samples (score ≥1+) was
significantly higher in adjacent DCIS lesions (90%) than in
invasive tumor cells (71%, p=0.001, Table IV). Interestingly, the levels of
staining were also significantly higher in smaller lesions (T1a, b
and c) than in T2 and T3 lesions altogether (p=0.02, Table V), showing again that Egfl7
expression is associated with smaller lesions in breast cancer
patients.
 | Table VPercentage of Egfl7-positive cases in
DCIS lesions associated with invasive carcinoma, comparison of the
Tn score to that of Egfl7. |
Table V
Percentage of Egfl7-positive cases in
DCIS lesions associated with invasive carcinoma, comparison of the
Tn score to that of Egfl7.
| Egfl7 score
|
|---|
| 0 | 1+ | 2+ | 3+ |
|---|
| T1a+T1b+T1c
(n=64) | 5% (3) | 52% (33) | 36% (23) | 8% (5) |
| T2+T3 (n=15) | 33% (5) | 40% (6) | 27% (4) | 0% (0) |
When comparing pure DCIS with DCIS lesions
associated with invasive carcinoma, the number of 3+ lesions was
significantly higher in pure DCIS lesions than within DCIS lesions
associated with infiltrating carcinoma (23 and 6%, respectively, p=
0.009 χ2 test, p= 0.02 Fisher’s exact test). Similarly,
the expression levels of Egfl7 transcripts were significantly
higher in pure DCIS lesions than all other lesions taken together
(p=0.0008, Fig. 4B).
Lobular carcinoma in situ
Lobular carcinoma in situ were associated
with lobular invasive carcinoma in 11 cases. Staining for Egfl7 was
essentially cytoplasmic, noted as 0+ in 27% of the cases, 1+ in 64%
of the cases, 2+ in 9% of the cases (data not shown). No 3+ lesions
were observed and no statistical analyses were performed on this
small sample population.
Expression of Egfl7 in axillary lymph
node metastasis
The lymph nodes of 171 patients diagnosed with
invasive lesions were available, among which no node invasion was
noted in 61% of the cases (n=107, pN0). In 9% of the cases,
isolated cancer cells or micrometastasis were too small to be
analyzed. Egfl7 protein expression and localization was thus
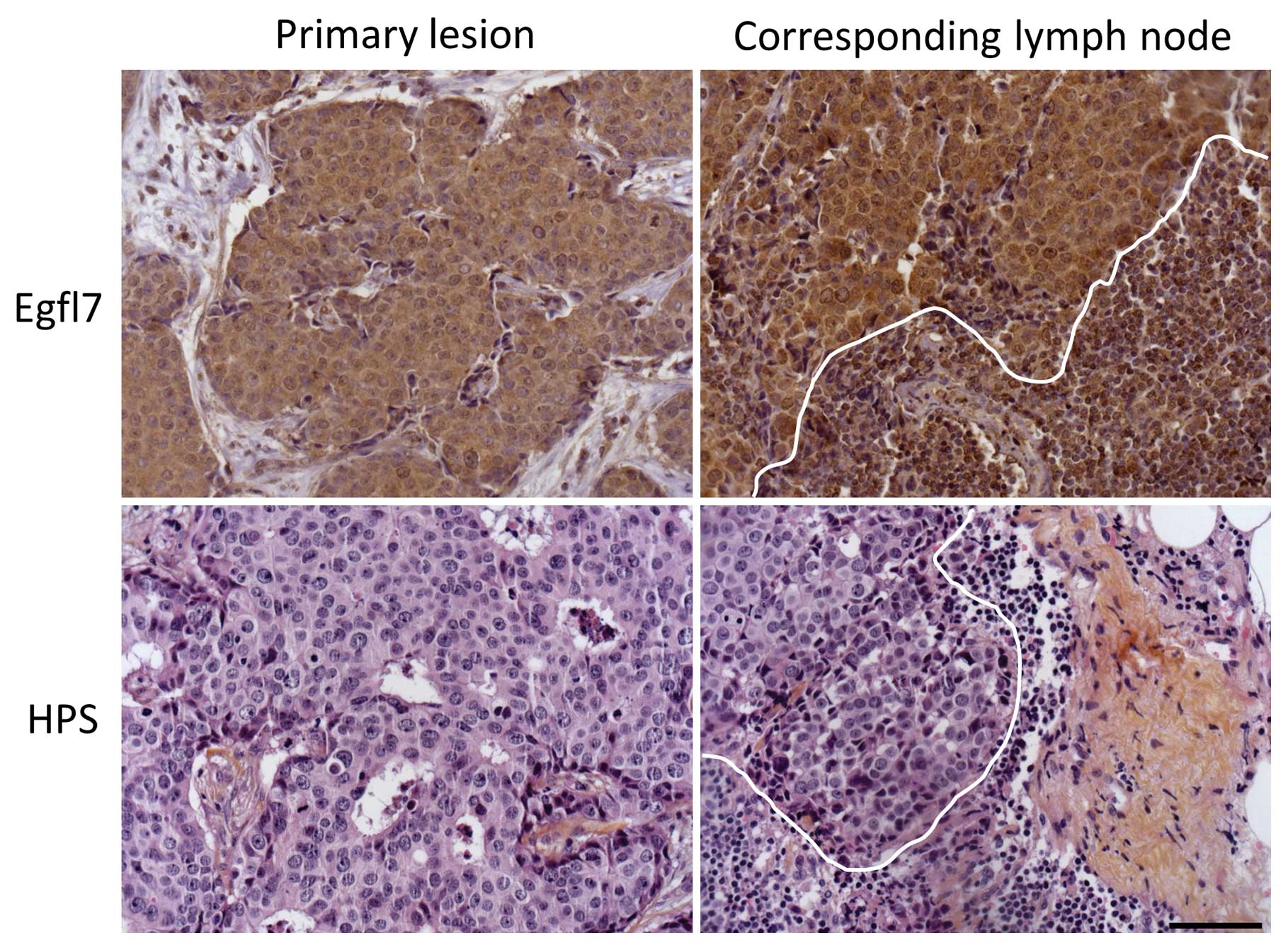
evaluated by IHC in 33 invaded lymph nodes (Fig. 5 and Table VI). Again, and using univaried
analysis, the quantities of Egfl7 were correlated with the
histological type of the lesions, i.e., they were higher in ductal
vs lobular carcinoma (p=0.01 χ2 test, p=0.02 Fisher’s
exact test). On the other hand, there were no correlations with the
histological SBR grade, size of the primary lesion, age, hormone
receptors status, HER2, or the presence of emboli.
 | Table VIPercentage of Egfl7-positive cases in
invaded lymph nodes scored according to Egfl7 expression. |
Table VI
Percentage of Egfl7-positive cases in
invaded lymph nodes scored according to Egfl7 expression.
| Egfl7 score
(n=33) | |
|---|
| 0+ | 39% (13) |
| 1+ | 36% (12) |
| 2+ | 24% (8) |
| 3+ | 0% (0) |
Expression levels of the Egfl7 transcripts were
analyzed by RT-qPCR in 24 pN1 samples. The transcript levels
correlated with the size of the associated primary invasive tumor
(p= 0.02, data not shown), including in the invasive ductal
carcinoma sub-population (p=0.02). On the other hand, there were no
correlations between the levels of expression of the Egfl7
transcripts and the age, the histological type of the primary
tumor, tumor size, pN rank, or SBR grade, nor with progesterone
receptors status, HER2 status, the presence of emboli or
necrosis.
Relapse-free and overall survival
The median follow-up of the sampled population was
4.75 years (0–6 years). The 2-year relapse-free and overall
survival values reached 94% (88–97%) and 99% (94–100%),
respectively, and the 5-year relapse-free and global survival
values were 83% (75–89%) and 95% (89–98%), respectively. The 2- and
5-year values of relapse-free and of overall survival in the DCIS
group were 100%. As may be expected from such low degrees of
variation, no correlation could be measured between the levels of
expression of Egfl7 and relapse-free or overall survival in the
non-DCIS population when assessed by immunostaining and by
RT-qPCR.
Discussion
Expression of Egfl7 in human cancer has not been
extensively studied and this is the first study which addresses the
expression of the protein and of its transcripts in human breast
cancer. The specificity of the chosen antibody was verified using
several independent approaches. Furthermore, most of the results
obtained by IHC were confirmed by the analysis of the levels of
Egfl7 transcripts in the same lesions using RT-qPCR, adding further
strength to the IHC results.
Overall, the clinical characteristics of the sampled
population corresponded to the usual epidemiological
characteristics of a breast cancer population (median age 57, 80%
ductal vs. 17% lobular invasive carcinoma). However, it should be
noted that a large number of small lesions was collected (18 mm
mean diameter, 15 mm median diameter for the invasive carcinoma),
and that most (90%) of the tumors were ER+ while only
6.5% were HER2-positive. This is mainly due to the specificity of
our institute which recruits patients at early stages of the
disease and it does not affect the statistical significance of our
observations.
The main finding of this study was that Egfl7 is
expressed by tumor cells and is associated with lower grade and
better prognosis lesions. The fact that Egfl7 expression is
deregulated in breast cancer lesions and that cancer cells show a
much stronger and frequent cytoplasmic signal for Egfl7 than the
surrounding tissues is consistent with previous reports in other
cancers. Egfl7 protein was indeed detected in the cytoplasm of
cancer cells from human hepatocarcinoma (12). On the other hand, here, higher
levels of Egfl7 expression are associated with smaller and of
better prognosis primary lesions as, in invasive carcinoma, Egfl7
was more highly expressed in ductal lesions, in low scored SBR
lesions, and in ER+ than in their respective
counterparts. Similarly, in in situ carcinoma, Egfl7 was
more highly expressed in smaller lesions, and in pure DCIS than in
DCIS associated with infiltrating lesions. This seems apparently
contradictory with the fact that Egfl7 expression was previously
associated with the progression of other human cancers: in
hepatocarcinoma, Egfl7 expression was correlated with the presence
of multiple nodules, venous invasion and the absence of capsule. It
should be however noted that Egfl7 expression was not correlated
with the tumor size nor with the Edmonson-Steiner grade in these
hepatic lesions (12). In colon
carcinoma and in glioma, high expression levels of Egfl7
transcripts were associated with higher tumor grades (11,13)
and with higher microvascular density in the case of glioma
(13). However, since there was no
separate analysis of expression of Egfl7 in endothelial, stromal
and tumor cells in these studies and since Egfl7 is highly
expressed in endothelial cells, it is most probable that these
correlations were at least partly due to the high vascularization
of advanced tumors, as seen in gliomas (13). These analyses need to be
complemented with a histologic identification, such as that
performed here.
We observed that Egfl7 is more highly expressed in
ductal carcinoma than in lobular carcinoma. Ductal and lobular
carcinoma have distinct biological characteristics, such as
E-cadherin expression (16),
cellular cohesion, different metastasis targets, and blood vessel
formation (17) which may account
for these differences. It should also be noted that Egfl7
expression is associated with lower grade SBR tumors, which are
themselves associated with ER+ lesions, whereas the
incidence of ER+ lesions is higher in lobular invasive
carcinoma than ductal carcinoma. This suggests that Egfl7
expression is regulated by mechanisms independent from that
regulating estrogen receptor expression in breast cancer.
Although Egfl7 expression was associated with lower
grade lesions and better prognosis, it showed no correlation with
survival rates. This is most certainly due to the size of the
population tested which, when associated with high survival rates,
could not allow a meaningful statistical analysis. It should be
noted that a similar absence of correlation between Egfl7
transcript expression and survival was also observed in colon
carcinoma (11), whereas a
positive correlation was found in a study performed in
hepatocarcinoma, although this study was performed on a very
limited number of cases (12).
Egfl7 expression in human cancer needs to be
carefully analyzed as Egfl7 may play complex roles in cancer
biology. Indeed, Egfl7 has different effects on tumor cells
depending on their origin: Egfl7 increases tumor cell migration of
hepatocarcinoma cells (12) while
it has no effects on breast cancer cell migration (18). Here, we found that Egfl7 is not
associated with a more advanced disease when expressed by human
breast cancer epithelial cells. Thus, it probably does not provide
per se a growth advantage to cancer cells which express it. This is
consistent with the observations that Egfl7 is not an oncogene as
it does not increase breast tumor cell proliferation, migration,
nor clone formation (18). The
molecular targets of Egfl7 and its functions are not well
characterized. We have previously shown that Egfl7 inhibits the
activity of the extracellular lysyl oxidases, thus preventing
elastogenesis within the vascular walls (15). Egfl7 was also shown to inhibit the
Notch signaling by a direct interaction with the Notch family of
receptors (19). Interestingly,
the Notch receptors seem to play opposite roles in human breast
cancer as Notch-1 seems to promote cancer while Notch-2 correlates
with a better outcome (20).
Expression of the Notch ligand Jag1 is also associated with poor
outcome in human breast cancer (21). Whether Egfl7 interacts with Notch
receptors during the development of breast cancer and whether this
interaction might be important for the progression of the disease
is not known at present.
Based on this study, it seems that Egfl7 is not
expressed similarly in different human cancers, raising specific
concerns regarding the planned therapies which target Egfl7 in
order to treat cancer.
Acknowledgements
We would like to acknowledge the help
of Ms. Delphine Bertin, Dr Xavier Leroy and Dr Yves Marie-Robin. We
are grateful to the Cancéropole Nord-Ouest Tumorothèque. This study
was supported by Ligue Nationale contre le Cancer (Equipe
Labellisée La Ligue to F.S.), Institut National du Cancer (no.
2008-053 to F.S.) and Association pour la Recherche sur le Cancer
(to F.S.). G.L.P. was supported by a fellowship ‘Année Recherche’
from Ministère de l’Enseignement Supérieur et de la Recherche. F.S.
is Directeur de Recherche of the Institut National de la Santé et
de la Recherche Médicale.
References
|
1
|
Relf M, LeJeune S, Scott PA, Fox S, Smith
K, Leek R, Moghaddam A, Whitehouse R, Bicknell R and Harris AL:
Expression of the angiogenic factors vascular endothelial cell
growth factor, acidic and basic fibroblast growth factor, tumor
growth factor beta-1, platelet-derived endothelial cell growth
factor, placenta growth factor, and pleiotrophin in human primary
breast cancer and its relation to angiogenesis. Cancer Res.
57:963–969. 1997.
|
|
2
|
Weidner N, Semple JP, Welch WR and Folkman
J: Tumor angiogenesis and metastasis--correlation in invasive
breast carcinoma. N Engl J Med. 324:1–8. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weidner N, Folkman J, Pozza F, Bevilacqua
P, Allred EN, Moore DH, Meli S and Gasparini G: Tumor angiogenesis:
a new significant and independent prognostic indicator in
early-stage breast carcinoma. J Natl Cancer Inst. 84:1875–1887.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fox SB, Leek RD, Bliss J, Mansi JL,
Gusterson B, Gatter KC and Harris AL: Association of tumor
angiogenesis with bone marrow micrometastases in breast cancer
patients. J Natl Cancer Inst. 89:1044–1049. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Uzzan B, Nicolas P, Cucherat M and Perret
GY: Microvessel density as a prognostic factor in women with breast
cancer: a systematic review of the literature and meta-analysis.
Cancer Res. 64:2941–2955. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guidi AJ, Fischer L, Harris JR and Schnitt
SJ: Microvessel density and distribution in ductal carcinoma in
situ of the breast. J Natl Cancer Inst. 86:614–619. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Engels K, Fox SB, Whitehouse RM, Gatter KC
and Harris AL: Distinct angiogenic patterns are associated with
high-grade in situ ductal carcinomas of the breast. J Pathol.
181:207–212. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fox SB, Generali DG and Harris AL: Breast
tumour angiogenesis. Breast Cancer Res. 9:2162007. View Article : Google Scholar
|
|
9
|
Soncin F, Mattot V, Lionneton F, Spruyt N,
Lepretre F, Begue A and Stehelin D: VE-statin, an endothelial
repressor of smooth muscle cell migration. EMBO J. 22:5700–5711.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parker LH, Schmidt M, Jin SW, Gray AM,
Beis D, Pham T, Frantz G, Palmieri S, Hillan K, Stainier DY, De
Sauvage FJ and Ye W: The endothelial-cell-derived secreted factor
Egfl7 regulates vascular tube formation. Nature. 428:754–758. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diaz R, Silva J, Garcia JM, Lorenzo Y,
Garcia V, Pena C, Rodriguez R, Munoz C, Garcia F, Bonilla F and
Dominguez G: Deregulated expression of miR-106a predicts survival
in human colon cancer patients. Genes Chromosomes Cancer.
47:794–802. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu F, Yang LY, Li YF, Ou DP, Chen DP and
Fan C: Novel role for epidermal growth factor-like domain 7 in
metastasis of human hepatocellular carcinoma. Hepatology.
50:1839–1850. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang CH, Li XJ, Zhou YZ, Luo Y, Li C and
Yuan XR: Expression and clinical significance of EGFL7 in malignant
glioma. J Cancer Res Clin Oncol. 136:1737–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Caetano B, Drobecq H and Soncin F:
Expression and purification of recombinant vascular
endothelial-statin. Protein Expr Purif. 46:136–142. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lelièvre E, Hinek A, Lupu F, Buquet C,
Soncin F and Mattot V: VE-statin/egfl7 regulates vascular
elastogenesis by interacting with lysyl oxidases. EMBO J.
27:1658–1670. 2008.PubMed/NCBI
|
|
16
|
Moll R, Mitze M, Frixen UH and Birchmeier
W: Differential loss of E-cadherin expression in infiltrating
ductal and lobular breast carcinomas. Am J Pathol. 143:1731–1742.
1993.PubMed/NCBI
|
|
17
|
Lee AH, Dublin EA, Bobrow LG and Poulsom
R: Invasive lobular and invasive ductal carcinoma of the breast
show distinct patterns of vascular endothelial growth factor
expression and angiogenesis. J Pathol. 185:394–401. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Delfortrie S, Pinte S, Mattot V, Samson C,
Villain G, Caetano B, Lauridant-Philippin G, Baranzelli M-C,
Bonneterre J, Trottein F, Faveeuw C and Soncin F: Egfl7 promotes
tumor escape from immunity by repressing endothelial cell
activation. Cancer Res. 71:7176–7186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schmidt MH, Bicker F, Nikolic I, Meister
J, Babuke T, Picuric S, Muller-Esterl W, Plate KH and Dikic I:
Epidermal growth factor-like domain 7 (EGFL7) modulates Notch
signalling and affects neural stem cell renewal. Nat Cell Biol.
11:873–880. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Parr C, Watkins G and Jiang WG: The
possible correlation of Notch-1 and Notch-2 with clinical outcome
and tumour clinicopathological parameters in human breast cancer.
Int J Mol Med. 14:779–786. 2004.PubMed/NCBI
|
|
21
|
Dickson BC, Mulligan AM, Zhang H, Lockwood
G, O’Malley FP, Egan SE and Reedijk M: High-level JAG1 mRNA and
protein predict poor outcome in breast cancer. Mod Pathol.
20:685–693. 2007. View Article : Google Scholar : PubMed/NCBI
|