Introduction
Despite extensive investigations of therapeutic
improvements in surgical techniques, chemotherapy and
chemo-radiotherapy, esophageal squamous cell carcinoma (ESCC)
remains one of the most aggressive and fatal malignancies and the
prognosis of patients with ESCC remains poor (1). Although curative surgical resection
can be performed, half of all patients develop recurrences within a
few years after surgery and the 5-year survival rate is only
approximately 50% (2). Therefore,
more effective therapies are urgently needed to improve the
prognosis of patients with ESCC.
The overexpression of epidermal growth factor
receptor (EGFR) and HER2 can be observed in a variety of human
malignancies and the roles of such overexpressions in cancer
development, progression and aggressiveness have been widely
recognized (3,4). Approximately 50–70% of ESCC tumors
express EGFR protein when examined using
immunohisto-chemistry (IHC), while 15–28% of specimens exhibit
EGFR gene amplification when examined using fluorescence
in situ hybridization (FISH) (5,6).
Similarly, HER2 protein overexpression has been observed in 30–41%
of specimens examined using IHC, while HER2 gene
amplification has been observed in 11–19% of specimens using FISH
(7–9). These results indicate that EGFR and
HER2 overexpression and gene amplification are frequently observed
in ESCC, strongly suggesting that signaling involving these factors
may play important biological roles and may be useful molecular
targets in ESCC. Somatic mutations of EGFR tyrosine kinase in
non-small cell lung cancer (NSCLC) have been shown to increase
kinase activity and to be associated with hypersensitivity to
gefitinib, a selective EGFR tyrosine kinase inhibitor (EGFR-TKI)
(10,11). A recent phase III study
demonstrated that first-line gefitinib for patients with advanced
NSCLC with EGFR mutations improved progression-free
survival, compared with standard chemotherapy (12). Therapeutics targeting EGFR and
HER2, such as small-molecule inhibitors or specific monoclonal
antibodies, are now under intensive investigation in clinical
settings and some of them have achieved clinical success in the
treatment of diverse solid cancers (4,13).
Fibroblast growth factor receptor (FGFR) signaling
is deregulated in a wide variety of cancers (14). We previously reported that
FGFR2 amplification was observed in 4.1% of gastric cancers
and that FGFR2 amplification confers hypersensitivity to
FGFR inhibitor in gastric cancer cell lines both in vitro
and in vivo(15,16), strongly suggesting that
FGFR2 amplification may be a promising molecular target for
gastric cancer treatment. In ESCC, information regarding
FGFR2 amplification remains unclear. Additionally,
hepatocyte growth factor (HGF)-MET receptor signaling provides
important signals for cell survival and migration in cancer cells;
thus, these molecules have also emerged as promising molecular
targets for cancer therapy (17).
Very limited information is available regarding the
gene amplification of EGFR, HER2, FGFR2 and MET and
the EGFR mutation status in relation to the prognostic
impact for post-curative surgery in ESCC. In an attempt to advance
molecular-targeted therapy for ESCC, we retrospectively studied
these issues using formalin-fixed, paraffin-embedded (FFPE) samples
from patients with ESCC who had undergone surgery.
Materials and methods
Cell culture
KYSE170, KYSE180 and KYSE270 were maintained in a
1:1 mixture of Ham’s F12 medium and RPMI-1640 medium (Sigma, St.
Louis, MO) supplemented with 2% heat-inactivated fetal bovine serum
(FBS; Gibco BRL, Grand Island, NY). T.T. was maintained in a 1:1
mixture of Dulbecco’s modified Eagle’s medium (DMEM; Nissui
Pharmaceutical, Tokyo, Japan) and Ham’s F12 medium with 10% FBS.
KYSE30 and KYSE50 were maintained in DMEM with 10% FBS. KYSE70 was
maintained in DMEM with 2% FBS. KYSE150 was maintained in Ham’s F12
with 2% FBS.
Patients
This study was performed retrospectively. The
criteria for eligibility were histologically confirmed ESCC,
surgery for stage I–III disease, absence of prior radiotherapy or
chemo-therapy before surgery and the availability of a FFPE sample.
Tumor specimens were collected from 246 patients with ESCC who were
treated at the Kinki University Faculty of Medicine between 2001
and 2011. One sample was excluded because of poor DNA quality and
245 ESCC samples were finally evaluated. The World Health
Organization Classification of Tumors was used for histologically
grading. The tumors were staged according to the
tumor-node-metastasis (TNM) classification of the American Joint
Committee on Cancer (AJCC)/Union for International Cancer Control
(UICC). The present study was approved by the institutional review
board of the Kinki University Faculty of Medicine.
Isolation of genomic DNA
Macro-dissection of the surgical specimens preserved
as FFPE tissues was performed after deparaffinization to select a
region of cancer tissue. Genomic DNA samples were extracted using a
QIAamp DNA Micro Kit (Qiagen, Hilden, Germany) according to the
manufacturer’s instructions. The DNA concentration was determined
using the NanoDrop2000 (Thermo Scientific, Waltham, MA).
Copy number assay
The DNA copy numbers of EGFR, HER2, FGFR2 and
MET were determined using commercially available and
pre-designed TaqMan Copy Number Assays (Applied Biosystems, Foster
City, CA), as described previously (15). The primer IDs used in this study
were as follows: EGFR, Hs00997424_cn; HER2,
Hs05475431_cn; FGFR2, HS05182482_cn (introns 14 and 15); and
MET, Hs05005660_cn (introns 16 and 17). The TERT locus was
used for the internal reference copy number. Human genomic DNA
(Takara, Otsu, Japan) and DNA from non-cancer FFPE tissue were used
as normal controls. The PCR analysis was performed using the ABI
PRISM 7900HT Sequence Detection System (Applied Biosystems) and the
results were analyzed using SDS 2.2 and CopyCaller software
(Applied Biosystems).
FISH analysis
FISH analysis of EGFR and HER2
amplification was performed using the Vysis EGFR/CEP7 FISH Probe
Kit (Abbott Laboratories, Abbott Park, IL) or the PathVysions HER2
DNA Probe Kit (Abbott Laboratories), according to the
manufacturer’s instructions. Amplification was determined based on
a HER2/CEP17 signal ratio of >2.2. A two or more increase
in the EGFR gene signal relative to the CEP7 signal
was considered to indicate gene amplification. The
FGFR2-FISH method has been previously described (15), as has the MET-FISH method
(18).
Detection of EGFR mutations
EGFR mutations (exons 18–21) were detected using the
Therascreen RGQ PCR kit (Qiagen), which combines Scorpions
technology and the amplified refractory mutation system (ARMS) to
detect mutations using real-time PCR. This sensitive method can
detect 29 types of active mutations in the EGFR gene. All
the reactions were performed according to the manufacturer’s
instructions, as previously described (19).
Cell growth inhibitory assay
To evaluate growth inhibition in the presence of
various concentrations of EGFR-TKI AG1478 (Sigma), we used an MTT
assay and a previously described method (20). Briefly, the cells were seeded at a
density of 2×103 cells/well in 96-well plates.
Twenty-four hours later, AG1478 was added and the incubation was
further continued for 72 h at 37°C. The assay was conducted in
triplicate.
Immunoblotting
A western blot analysis was performed as described
previously (21). The following
antibodies were used: polyclonal EGFR antibody, polyclonal
phospho-EGFR antibody, polyclonal HER2 antibody, monoclonal HER4
antibody, polyclonal Akt antibody, monoclonal phosphor-Akt
antibody, polyclonal p44/42 MAPK antibody, polyclonal
phosphop-44/42 MAPK antibody, β-actin antibody and HRP-conjugated
secondary antibody (Cell Signaling Technology, Beverly, MA); and
monoclonal HER3 antibody (Upstate Biotechnology, Lake Placid, NY).
The cells were cultured overnight in serum-starved medium and
exposed to 0.1–10 μmol/l of AG1478 for 3 h before the
addition of EGF (10 ng/ml) for 15 min.
Comparative genomic hybridization (CGH)
analysis
The CGH analysis was performed using a SurePrint G3
Human CGH Microarray (Agilent Technologies, Santa Clara, CA)
according to the manufacturer’s instructions. For the analysis, 0.2
μg of DNA was extracted from each FFPE sample of ESCC and an
FGFR2-amplified tumor or a non-cancer tissue were used as a
control. The copy number changes were analyzed using Partek Genomic
Suite 6.4 software (Partek Inc., St. Louis, MO).
Statistical analysis
The prognostic analyses of the clinicopathological
features and molecular factors were performed using a Cox
regression. In the multivariate Cox models, the variable selection
was based on the presence of significance (P<0.10) in a
univariate analysis; variables that were not significant in the
final model were removed using the stepwise method. The
disease-free survival (DFS) and overall survival (OS) curves were
constructed using the Kaplan-Meier method and were compared using
the log-rank test. Statistical analyses were performed using PAWS
Statistics 18 (SPSS Japan Inc., Tokyo, Japan).
Results
Patient results
Of the 245 patients evaluated in this study, all the
patients had undergone surgery for histologically confirmed stage
I–III ESCC. The patient characteristics are shown in Table I. The percentages of the
pathological stages were as follows: stage I, 24%; stage II, 27%;
and stage III, 49%. Fourteen (6%) patients had residual cancer at
the time of surgery and tumor recurrence occurred in 98 (42%)
patients. The median follow-up period was 24 months (range 0–126
months).
 | Table IPatient characteristics. |
Table I
Patient characteristics.
|
Characteristics | No. |
|---|
| Age | |
| Range
(years) | 34 – 83 |
| Median
(years) | 65 |
| <60/≥60 | 55/190 |
| Sex | |
| Male/female | 208/37 |
| Location | |
| Ut/Mt/Lt/Ae | 20/149/68/8 |
| pT | |
| T1/T2/T3/T4 | 69/45/124/7 |
| pN | |
| N0/N1/N2/N3 | 89/77/50/29 |
| pM | |
| M0/M1 | 245/0 |
| pStage | |
| I/II/III/IV | 59/65/121/0 |
| Diff. | |
| Well/mod/por | 48/141/56 |
| Ly | |
| 0/1 | 93/152 |
| V | |
| 0/1/2 | 205/40/0 |
| Residual | |
| 0/1/2 | 231/7/7 |
| Recurrence | |
| (−)/(+) | 133/98 |
| Total | 245 |
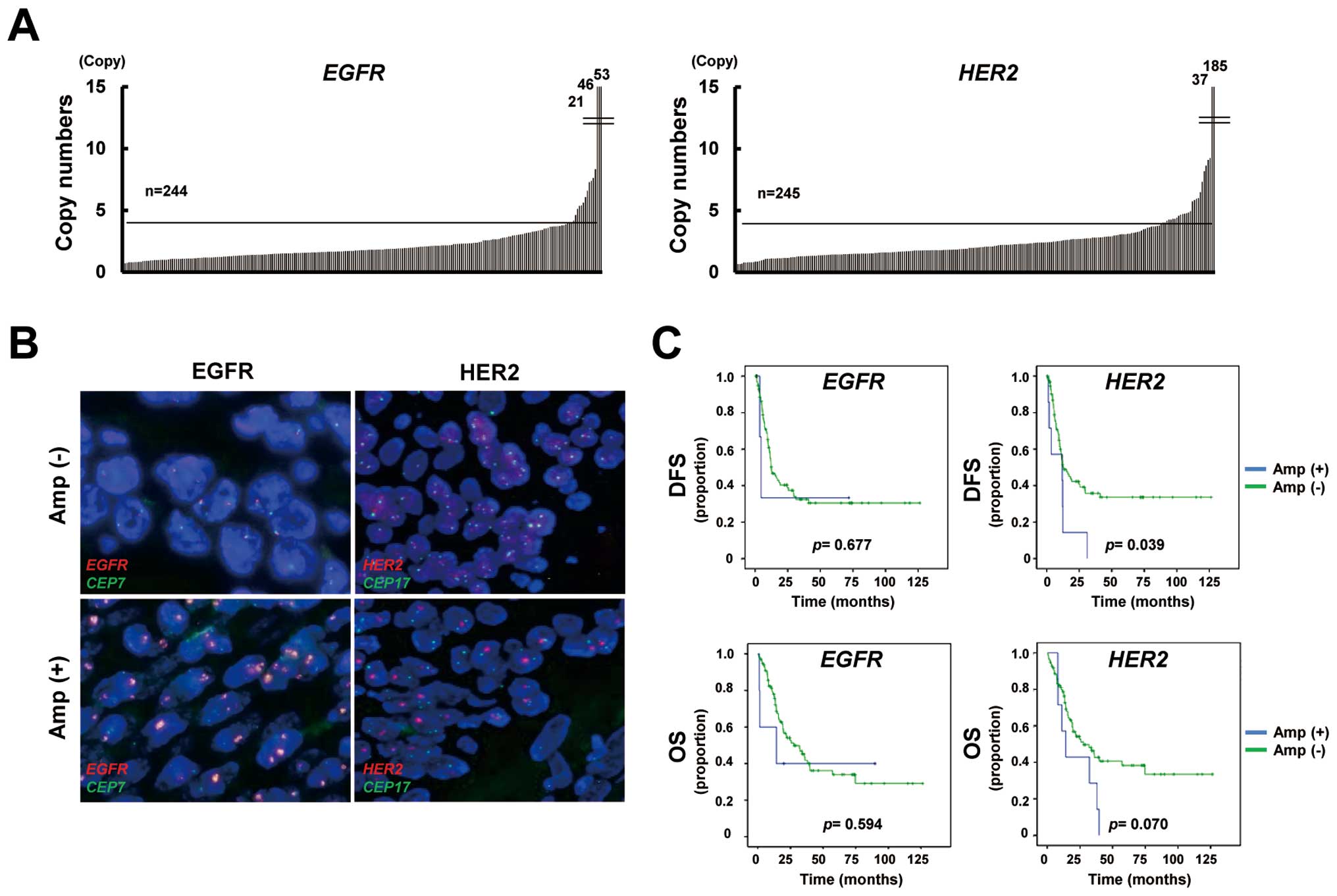
Gene amplification of EGFR and HER2 in
ESCC
To determine the gene amplification of EGFR
and HER2 in FFPE samples, we used a high-throughput and
real-time PCR-based copy number assay, as previously reported
(15). Gene amplification was
defined as more than four copies. The copy number assay showed that
EGFR and HER2 were amplified in 7% (16/244, one not
determined; range 0.6–52.8 copies) and 11% (27/245; range 0.4–185.0
copies) of the ESCC specimens, respectively (Fig. 1A). FISH analysis demonstrated that
the EGFR/CEP7 signal ratio was increased in
EGFR-amplified samples, while the ratio was not increased in
a non-amplified sample (Fig. 1B).
Similarly, the HER2/CEP17 signal ratio was consistent with
the results of a copy number assay for HER2. FISH analysis
verified the results of the copy number assays for EGFR and
HER2.
Prognostic impact of clinicopathological
and gene amplification in ESCC
Of the 121 patients with stage III ESCC, 14 were
excluded because of residual cancer and three were excluded because
of the lack of copy number results; finally, 104 patients with
stage III ESCC were evaluated to determine the prognostic impact of
post-operative ESCC findings. The correlations between
clinicopathological features, including age, sex, pathological
tumor stage, pathological lymph node stage, tumor differentiation,
lymphatic vessel invasion (Ly), vascular invasion (V) and the gene
amplification statuses of EGFR and HER2 and the DFS
or OS were evaluated. A univariate analysis showed that the
pathological lymph node stage, Ly grade, V grade and HER2
amplification status were significant predictors of a poor DFS
(Table II). A multivariate
analysis revealed that the pathological lymph node stage
(P=0.00003) and HER2 amplification (P=0.021) were
significant predictors of a poor DFS. Meanwhile, the pathological
lymph node stage, tumor differentiation, Ly grade and V grade were
significant predictors of a poor OS. A multivariate analysis
demonstrated that the pathological lymph node stage (P=0.004) was a
significant predictor of a poor OS. The Kaplan-Meier curves for DFS
and OS plotted according to the gene amplification status are shown
in Fig. 1C. These results
indicated that HER2 amplification, but not EGFR
amplification, was a predictor of a poor outcome among
postoperative patients with stage III ESCC in the present
study.
 | Table IIUnivariate and multivariate analysis
of clinical and molecular factors for disease-free and overall
survival in stage III ESCC. |
Table II
Univariate and multivariate analysis
of clinical and molecular factors for disease-free and overall
survival in stage III ESCC.
| Disease-free
survival | Overall
survival |
|---|
|
|
|
|---|
| Univariate
analysis | Multivariate
analysis | Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Factor | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (≥60
vs.<60) | 1.31 | (0.75–2.30) | 0.340 | | | | 1.15 | (0.66–2.01) | 0.625 | | | |
| Gender (male vs.
female) | 1.19 | (0.56–2.49) | 0.653 | | | | 1.85 | (0.80–4.33) | 0.153 | | | |
| pT (T3,T4 vs. T1,
T2) | 0.81 | (0.45–1.47) | 0.492 | | | | 0.65 | (0.36–1.17) | 0.151 | | | |
| pN (N3 vs. N1,
N2) | 3.44 | (2.02–5.84) | 0.000005 | 3.20 | (1.85–5.53) | 0.00003 | 2.81 | (1.60–4.93) | 0.0003 | 2.35 | (1.32–4.20) | 0.004 |
| Diff. (por vs.
well, mod) | 1.49 | (0.85–2.64) | 0.168 | | | | 2.10 | (1.22–3.63) | 0.008 | 1.70 | (0.95–3.04) | 0.092 |
| Ly (1 vs. 0) | 2.20 | (1.16–4.18) | 0.016 | 1.75 | (0.81–3.82) | 0.157 | 2.31 | (1.15–4.64) | 0.018 | 2.05 | (0.89–4.74) | 0.095 |
| V (1, 2 vs. 0) | 2.07 | (1.21–3.53) | 0.008 | 1.64 | (0.95–2.82) | 0.076 | 2.05 | (1.20–3.53) | 0.009 | 1.62 | (0.92–2.85) | 0.074 |
| EGFR amp (+ vs.
−) | 1.39 | (0.34–5.70) | 0.650 | | | | 1.40 | (0.44–4.48) | 0.575 | | | |
| HER2 amp (+ vs.
−) | 2.31 | (1.05–5.09) | 0.038 | 2.59 | (1.16–5.81) | 0.021 | 2.15 | (0.97–4.75) | 0.060 | 2.05 | (0.92–4.60) | 0.081 |
Active EGFR mutation in ESCC cell lines
and clinical samples
We next examined the growth inhibitory effect of the
EGFR-TKI AG1478 against eight ESCC cell lines to evaluate the
effect of EGFR-TKI treatment on ESCC. Notably, the KYSE270 cells
were hypersensitive to AG1478 at a sub-micro molar level of
IC50 (0.45 μM), which is similar to the
hypersensitivity of lung cancer cells harboring an EGFR
mutation (Fig. 2A). The possible
presence of 29 types of EGFR mutations in the eight ESCC
cell lines was examined using the Scorpion-ARMS method. The KYSE270
cells, which exhibited hypersensitivity to AG1478, harbored the
L861Q type of EGFR mutation, whereas the other cell lines
carried no mutations (Fig. 2B). A
copy number assay revealed that EGFR was amplified in KYSE30
cells, while no significant amplifications of HER2 were
observed (Fig. 2C). The western
blot analysis showed no significant overexpression of HER2,
HER3 or HER4 (Fig.
2D), compared with a positive control (data not shown). The
phosphorylation and protein expression levels of EGFR were
increased in KYSE30, KYSE50, KYSE180 and KYSE270 cells. In the
KYSE270 cells (L861Q), AG1478 completely inhibited the
phosphorylation levels of MAPK, AKT and EGFR at a concentration of
100 nM, while phosphorylation was not inhibited in the KYSE170
cells (wild-type) at this concentration (Fig. 2E). These results indicate that an
active EGFR mutation conferring hypersensitivity to EGFR-TKI
was found in an ESCC cell line. Finally, we examined the presence
of EGFR mutations in 107 clinical samples of ESCC. One ESCC
tumor exhibited a del745-750 type of EGFR mutation (Fig. 2F). Thus, although the frequency of
EGFR mutation was not high compared with NSCLC, we did find
a mutation in a cell line and in a clinical sample of ESCC.
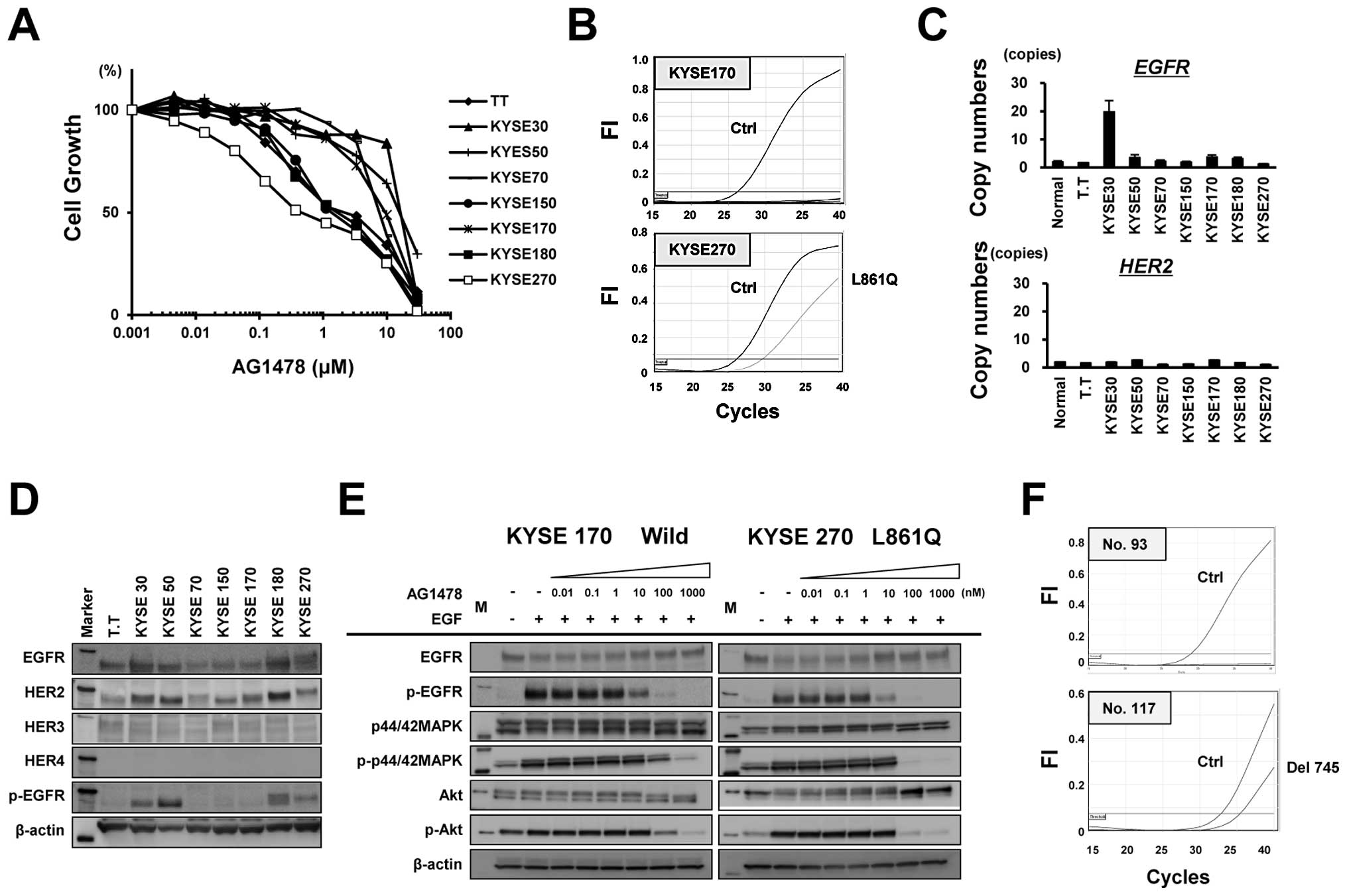 | Figure 2EGFR mutation in esophageal
squamous cell carcinoma (ESCC) cell lines and clinical samples. (A)
Growth inhibition in response to the EGFR tyrosine kinase inhibitor
AG1478 was evaluated at the indicated concentrations using an MTT
assay. (B) The status of the 29 types of EGFR mutation
determined using the Scorpion-ARMS method in eight ESCC cell lines.
Notably, KYSE270 cells, which were hypersensitive to AG1478,
harbored the L861Q type of EGFR mutation, whereas the other
cell lines did not exhibit any EGFR mutations. (C) The
TaqMan copy number assay was used to determine the copy numbers of
EGFR and HER2 in ESCC cell lines. (D and E) Western
blot analysis for EGFR, HER2, HER3, HER4 and phospho-EGFR
expression in ESCC cell lines. β-actin was used as an internal
control. Marker, molecular marker. Western blot analysis for
expression levels of EGFR, phospho-EGFR, MAPK, phospho-MAPK, AKT
and phospho-AKT in KYSE270 cells (L861Q) and KYSE170 cells
(EGFR wild-type). The cells were exposed to AG1478 at the
indicated concentrations for 3 h and then were stimulated with 10
ng/ml of EGF. β-actin was used as an internal control. M, molecular
marker. (F) Among the 107 clinical ESCC samples that were
evaluated, one (no. 117) carried a del745-750 type EGFR
mutation. |
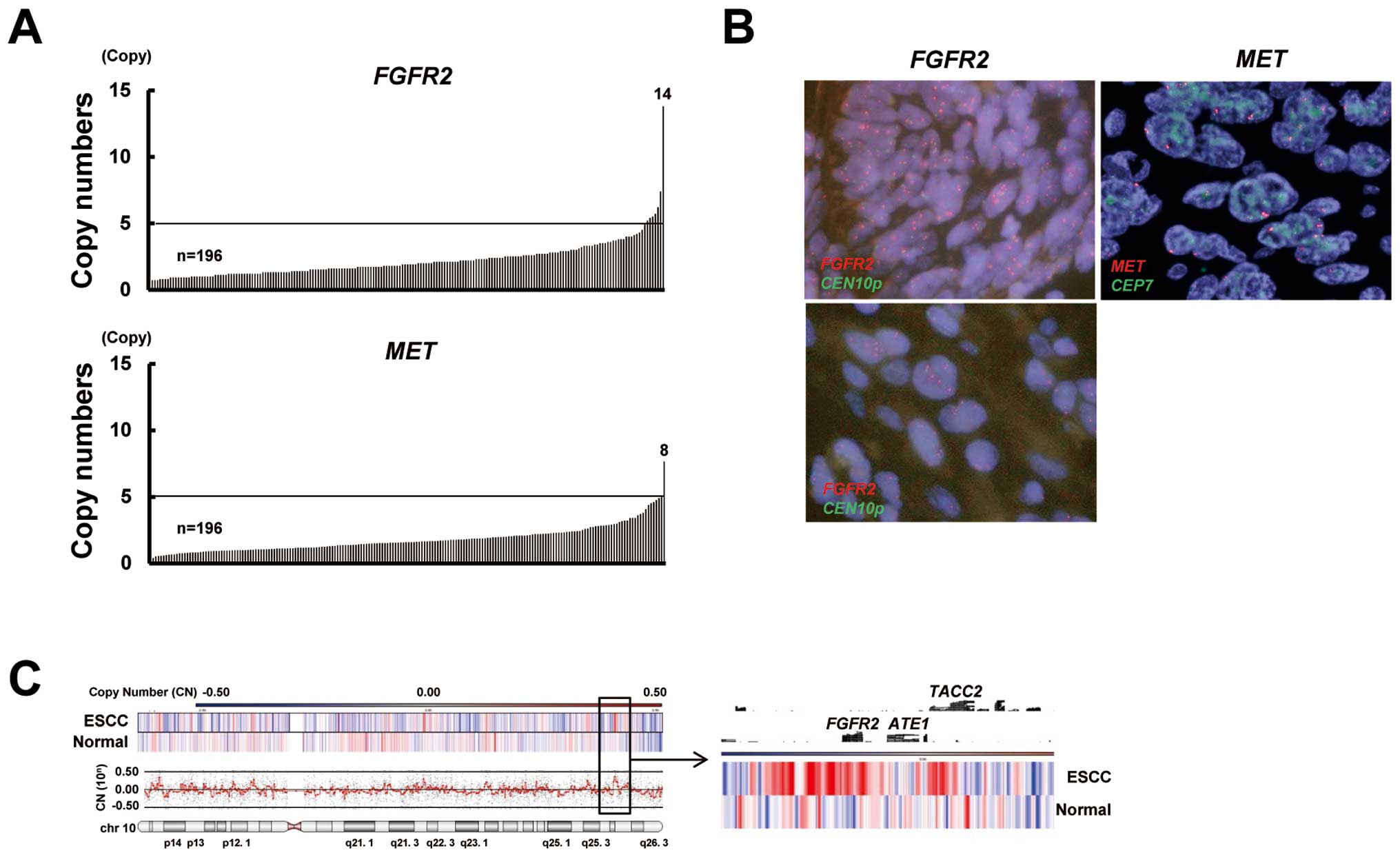
Gene amplification of FGFR2 and MET in
ESCC
To gain insight into molecular therapy targeting
FGFR2 or MET amplification in ESCC, we evaluated the
amplifications of these genes. FGFR2 and MET were
amplified in 4% (8/196; range 0.4–13.8 copies) and 1% (2/196; range
0.4–7.7 copies) of the ESCC specimens, respectively (Fig. 3A). A FISH analysis confirmed the
FGFR2 and MET amplification (Fig. 3B). A CGH analysis showed that the
FGFR2 locus was amplified in FGFR2-amplified ESCC and
that the amplicon seemed to consist of a relatively narrow region
(Fig. 3C). The clinical features
of FGFR2-amplified or MET-amplified ESCC are shown in
Table III. Although the numbers of
amplified cases were relatively small and, accordingly, definitive
evidence could not be obtained, patients who had tumors with
FGFR2 or MET amplification seemed to have no
significant trends regarding clinicopathological factors, including
patient outcome. Collectively, these findings indicate that
FGFR2 amplification is present but that MET
amplification is rare in ESCC.
 | Table IIIClinical features of
FGFR2-amplified or MET-amplified ESCC. |
Table III
Clinical features of
FGFR2-amplified or MET-amplified ESCC.
| No. | Age | Sex | Location | Macroscopic
type | pT | pN | pM | pStage | Ly | V | Histology | Rec. | Rec. sites | FGFR2
copies | MET
copies | FGFR2 | MET |
|---|
| 1 | 79 | M | Mt | 3 | T1b | N0 | M0 | I A | 0 | 0 | Mode | (−) | | 13.8 | 3.2 | Amp | n |
| 2 | 68 | M | Mt-Lt | 0-IIa | T1b | N0 | M0 | I A | 0 | 0 | Por | (−) | | 7.4 | 4.9 | Amp | n |
| 3 | 79 | M | Mt | 0-IIc | T1b | N0 | M0 | I A | 0 | 0 | Well | (−) | | 5.4 | 2.8 | Amp | n |
| 4 | 69 | M | Mt | 0-IIc+IIa | T2 | N0 | M0 | IB | 0 | 0 | Mod | (−) | | 5.5 | 1.9 | Amp | n |
| 5 | 67 | M | Lt | 1 | T2 | N1 | M0 | II B | 0 | 0 | Por | (−) | | 5.7 | 0.7 | Amp | n |
| 6 | 71 | M | Mt | 0-I | T1b | N2 | M0 | III A | 0 | 0 | Mod | (−) | | 5.2 | 1.2 | Amp | n |
| 7 | 64 | M | Lt | 3 | T3 | N1 | M0 | III A | 0 | 0 | Well | (+) | Liver | 5.0 | 3.8 | Amp | n |
| 8 | 63 | M | Mt | 2 | T3 | N2 | M0 | III B | 0 | 1 | Mod | (+) | Adrenal | 6.2 | 2.4 | Amp | n |
| 9 | 63 | F | Mt | 1 | T3 | N0 | M0 | II A | 0 | 0 | Mod | (−) | | 3.0 | 7.7 | n | Amp |
| 10 | 75 | M | Mt | 0-IIc+IIa | T1b | N2 | M0 | III A | 0 | 0 | Mod | (+) | Lung | 2.4 | 5.0 | n | Amp |
Discussion
FGFR2 is frequently amplified in gastric
cancer cell lines, especially in poorly differentiated type cells
and amplification confers hypersensitivity to FGFR inhibitors
(16,22). Regarding the mutation of
FGFR2, somatic mutations of FGFR2 have been found in
12% (15/122) of endometrial carcinomas and these FGFR2
mutations have an oncogenic property that confers hypersensitivity
to FGFR inhibitors (23). We
demonstrated, for the first time, that FGFR2 amplification
was observed in ESCC. Our findings provide novel insight into
FGFR-targeting therapy and further prospective studies evaluating
FGFR2 amplification in ESCC are needed. Meanwhile, recent
study has demonstrated that 2% of patients (10/489) with
esophagogastric adenocarcinoma harbored MET amplification
and two of four patients with MET-amplified tumors treated
with a MET inhibitor experienced tumor shrinkage (24). Although MET amplification is
rare in ESCC, MET-targeted therapy may be a useful therapeutic
approach in some cases.
The presence of active EGFR mutations or drug
sensitivity to EGFR-TKI in ESCC cells remains unknown (25). We found that one out of eight ESCC
cell lines harbored an L861Q mutation with hypersensitivity to
EGFR-TKI and 1% (1/107) of clinical ESCC samples had a del745-750
type of mutation when examined using a highly sensitive detection
method. In an EGFR-vIII-based method of overexpression,
L861Q mutation reportedly enhances EGFR kinase activity and
transforms activity without an increase in sensitivity to EGFR-TKI
but with an increase in sensitivity to irreversible
second-generation EGFR-TKI (26).
Our results indicate that L861Q is an active mutation to EGFR-TKIs
in ESCC cell lines; however, the 1% frequency of EGFR
mutation in ESCC makes it difficult to stratify patients who may
benefit from EGFR-TKI treatment, compared with NSCLC. For
HER2-positive advanced gastric or gastroesophageal junction
cancer, recent advances in the clinical development of molecular
targeted therapy have enabled the use of trastuzumab as a standard
therapy (27); however, similar
regimens for the treatment of ESCC remain elusive. EGFR
family-targeted therapy is considered to be the most promising
approach to date, because EGFR and HER2 overexpression and
amplification are frequently observed in ESCC. The anti-EGFR
antibody cetuximab used in combination with radiotherapy or
chemotherapy exhibited a significant clinical benefit when used
against head and neck squamous cell carcinoma (28,29),
leading to an ongoing intensive clinical trial using cetuximab for
the treatment of ESCC. We detected EGFR and HER2
amplification in 7 and 11% of ESCC specimens and our results
support a rationale for introducing anti-EGFR and anti-HER2
antibody therapies to the treatment of patients with ESCC.
In conclusion, we determined the frequency of
EGFR, HER2, FGFR2 and MET amplification in ESCC and
the presence of EGFR mutations among ESCC cell lines and
clinical samples. Our results warrant serious consideration of the
development of EGFR family inhibitors and FGFR-targeted therapies
for ESCC exhibiting gene amplification.
Acknowledgements
We thank Ms. Tomoko Kitayama and Ms.
Fusako Kamada for their technical assistance. This study was
supported by the Third-Term Comprehensive 10-Year Strategy for
Cancer Control and a Grant-in-Aid for Cancer Research from the
Ministry of Health, Labour and Welfare.
References
|
1
|
Ando N, Ozawa S, Kitagawa Y, Shinozawa Y
and Kitajima M: Improvement in the results of surgical treatment of
advanced squamous esophageal carcinoma during 15 consecutive years.
Ann Surg. 232:225–232. 2002.PubMed/NCBI
|
|
2
|
Nakagawa S, Kanda T, Kosugi S, Ohashi M,
Suzuki T and Hatakeyama K: Recurrence pattern of squamous cell
carcinoma of the thoracic esophagus after extended radical
esophagectomy with three-field lymphadenectomy. J Am Coll Surg.
198:205–211. 2004. View Article : Google Scholar
|
|
3
|
Normanno N, De Luca A, Bianco C, et al:
Epidermal growth factor receptor (EGFR) signaling in cancer. Gene.
366:2–16. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shepard HM, Brdlik CM and Schreiber H:
Signal integration: a framework for understanding the efficacy of
therapeutics targeting the human EGFR family. J Clin Invest.
118:3574–3581. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanawa M, Suzuki S, Dobashi Y, et al: EGFR
protein overexpression and gene amplification in squamous cell
carcinomas of the esophagus. Int J Cancer. 118:1173–1180. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sunpaweravong P, Sunpaweravong S,
Puttawibul P, et al: Epidermal growth factor receptor and cyclin D1
are independently amplified and overexpressed in esophageal
squamous cell carcinoma. J Cancer Res Clin Oncol. 131:111–119.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhan N, Dong WG, Tang YF, Wang ZS and
Xiong CL: Analysis of HER2 gene amplification and protein
expression in esophageal squamous cell carcinoma. Med Oncol.
29:933–940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sato-Kuwabara Y, Neves JI, Fregnani JHTG,
Sallum RA and Soares FA: Evaluation of gene amplification and
protein expression of HER-2/neu in esophageal squamous cell
carcinoma using fluorescence in situ hybridization (FISH) and
immunohistochemistry. BMC Cancer. 9:1207–1471. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mimura K, Kono K, Hanawa M, et al:
Frequencies of HER-2/neu expression and gene amplification in
patients with oesophageal squamous cell carcinoma. Br J Cancer.
92:1253–1260. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paez JG, Jänne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maemondo M, Inoue A, Kobayashi K, et al:
Gefitinib or chemo-therapy for non-small-cell lung cancer with
mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peták I, Schwab R, Orfi L, Kopper L and
Kéri G: Integrating molecular diagnostics into anticancer drug
discovery. Nat Rev Drug Discov. 9:523–535. 2010.PubMed/NCBI
|
|
14
|
Turner N and Grose R: Fibroblast growth
factor signalling: from development to cancer. Nat Rev Cancer.
10:116–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matsumoto K, Arao T, Hamaguchi T, et al:
FGFR2 gene amplification and clinicopathological features in
gastric cancer. Br J Cancer. 106:727–732. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takeda M, Arao T, Yokote H, et al: AZD2171
shows potent antitumor activity against gastric cancer
over-expressing fibroblast growth factor receptor 2/keratinocyte
growth factor receptor. Clin Cancer Res. 13:3051–3057. 2007.
View Article : Google Scholar
|
|
17
|
Gherardi E, Birchmeier W, Birchmeier C and
Vande Woude G: Targeting MET in cancer: rationale and progress. Nat
Rev Cancer. 12:89–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cappuzzo F, Marchetti A, Skokan M, et al:
Increased MET gene copy number negatively affects survival of
surgically resected non-small-cell lung cancer patients. J Clin
Oncol. 27:1667–1674. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kimura H, Kasahara K, Kawaishi M, et al:
Detection of epidermal growth factor receptor mutations in serum as
a predictor of the response to gefitinib in patients with
non-small-cell lung cancer. Clin Cancer Res. 12:3915–3921. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kaneda H, Arao T, Tanaka K, et al: FOXQ1
is overexpressed in colorectal cancer and enhances tumorigenicity
and tumor growth. Cancer Res. 70:2053–2063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Matsumoto K, Arao T, Tanaka K, et al: mTOR
signal and hypoxia-inducible factor-1 alpha regulate CD133
expression in cancer cells. Cancer Res. 69:7160–7164. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kunii K, Davis L, Gorenstein J, et al:
FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3
signaling for growth and survival. Cancer Res. 68:2340–2348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dutt A, Salvesen HB, Chen TH, et al:
Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl
Acad Sci USA. 105:8713–8717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lennerz JK, Kwak EL, Ackerman A, et al:
MET amplification identifies a small and aggressive subgroup of
esophagogastric adenocarcinoma with evidence of responsiveness to
crizotinib. J Clin Oncol. 29:4803–4810. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Okines A, Cunningham D and Chau I:
Targeting the human EGFR family in esophagogastric cancer. Nat Rev
Clin Oncol. 8:492–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kancha RK, Peschel C and Duyster J: The
epidermal growth factor receptor-L861Q mutation increases kinase
activity without leading to enhanced sensitivity toward epidermal
growth factor receptor kinase inhibitors. J Thorac Oncol.
6:387–392. 2011. View Article : Google Scholar
|
|
27
|
Bang YJ, Van Cutsem E, Feyereislova A, et
al: ToGA Trial Investigators. Trastuzumab in combination with
chemotherapy versus chemotherapy alone for treatment of
HER2-positive advanced gastric or gastro-oesophageal junction
cancer (ToGA): a phase 3, open-label, randomised controlled trial.
Lancet. 376:687–697. 2010. View Article : Google Scholar
|
|
28
|
Bonner JA, Harari PM, Giralt J, et al:
Radiotherapy plus cetuximab for squamous-cell carcinoma of the head
and neck. N Engl J Med. 354:567–578. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vermorken JB, Mesia R, Rivera F, et al:
Platinum-based chemotherapy plus cetuximab in head and neck cancer.
N Engl J Med. 359:1116–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|