Introduction
Dicer, a highly conserved RNase III-type enzyme
found in the majority of eukaryotes (1), cleaves microRNA (miRNA) and small
interfering RNA (siRNA) precursors into approximately 22
nucleotides (2,3). It is the key enzyme for RNAi and
miRNA pathways. Contrary to other organisms, mammals have a single
species of Dicer, which is involved in processing precursor
double-stranded RNA (dsRNA) into miRNA and siRNA (4). Over the past several years, Dicer has
increasingly evoked interest in the field of cancer biology, based
on a number of studies demonstrating the aberrant expression of
DICER1 among different types of cancer (5–8).
Furthermore, the abnormal expression of DICER1 has been
associated with poor prognosis in cancer patients (9–12).
Despite growing evidence that Dicer plays a significant role in
tumorigenesis, the regulation of the expression of this gene
remains to be elucidated. As regards hematological malignancies,
the potential role of Dicer and the regulation of its expression
have not been widely investigated. Therefore, in this study, we
assessed DICER1 expression levels in acute myeloid leukemia
(AML) cells and investigated whether alterations in DICER1
expression play an important role in AML cell proliferation and
apoptosis. In addition, we investigated the mechanism behind the
dysregulation of DICER1 expression in AML cells.
Materials and methods
Patients, samples and reagents
A total of 42 untreated de novo AML patients
(non-M3) with ≥80% leukemia cells and 5 healthy volunteers were
included in this study. According to the French-American-British
(FAB) classification, the patient subtypes were as follows: 1
patient had M0, 2 had M1, 5 had M2, 9 had M4, 22 had M5 and 3 had
M6. At the time of recruitment, informed consent was obtained from
each subject, in accordance with the declaration of the hospital
authorities. This study was approved by the Research Ethics
Committee of China Medical University, Shenyang, China. Samples
were collected from patients at the Department of Hematology of
Shenyang General Military Hospital, Shenyang, China. Leukemia bone
marrow mononuclear cells (BMMCs) were enriched from the diagnostic
bone marrow samples of patients with de novo AML by
Ficoll-Hypaque (Sigma-Aldrich, St. Louis, MO, USA) density gradient
centrifugation. Purification of CD34+ hematopoietic
progenitor cells in normal BMMCs was performed by positive
selection using the MACS Direct CD34 Progenitor Cell Isolation kit
(Miltenyi Biotec, Bergish Gladbach, Germany) as the control.
Cell culture
The U937 and K562 human leukemia cell lines were
suspended in RPMI-1640 (Invitrogen, Carlsbad, CA, USA). Human
embryonic kidney 293 (HEK293) cells, were cultured in DMEM (Gibco,
Carlsbad, CA, USA). All media contained standard antibiotics and
10% fetal bovine serum (HyClone Laboratories, Logan, UT, USA). The
cells were incubated at 37°C in a humidified atmosphere of 5%
CO2.
Real-time quantitative PCR
Total RNA was isolated using TRIzol reagent
(Invitrogen). cDNA was prepared from 1–2 μg of total RNA
using SuperScript II Reverse Transcriptase (Invitrogen). cDNA (2
μl) was used for real-time PCR, which was performed to
detect DICER1 using SYBR-Green (Takara, Shiga, Japan),
according to the manufacturer’s instructions. Sequences of primers
were as follows: DICER1, 5′-GTACGA CTACCACAAGTACTTC-3′ and
5′-ATAGTACACCTGCCA GACTGT-3′; GATA1,
5′-CAGTAAACGGGCAGGTACTC-3′ and 5′-CATAAAGCCACCAGCTGGTC-3′; and
GAPDH, 5′-TGCACCACCAACTGCTTAG-3′ and 5′-GACGCAGGG
ATGATGTTC-3′. Normalization was performed with respect to
GAPDH levels. Real-time PCR consisted of an initial
denaturation at 95°C for 15 sec, followed by 40 cycles at 95°C for
5 sec and 60°C for 34 sec. Each sample was analyzed in triplicate,
using ABI 7500 System software. The 2−ΔΔCt method was
used to calculate relative changes in gene expression.
Western blot analysis
Cells were lysed in RIPA buffer and the protein
concentration was determined using the Bradford method with BSA
(Sigma-Aldrich) as the standard (Bio-Rad, Hercules, CA, USA). Equal
amounts of cell extract (50 μg) were subjected to
electrophoresis on a SDS-polyacrylamide gel and transferred onto a
PVDF membrane. The membrane was blocked and then incubated with
primary antibodies against DICER1 (Santa Cruz Biotechnology, Santa
Cruz, CA, USA), α-tubulin, GAPDH (BiYunTian Biotechnology Co.,
Ltd., Shanghai, China) and GATA1 (Abcam), followed by an
HRP-conjugated secondary antibody. The blots were developed using
chemiluminescence.
RNAi assays
The K562 and U937 human leukemia cells were
transfected with an shRNA expression vector. The DICER1-shRNA and
control-shRNA vectors were purchased from Shanghai GenePharma, Co.,
Ltd., Shanghai, China. The DICER1-shRNA sequence used was
5′-GCTCGAAATC TTACGCAAATATTCAAGAGATATTTGCGTAAGATTTC GAGCTT-3′. The
GATA1-shRNA sequence used was 5′-CTC
AATTCAGCAGCCTATTCTCGAGAATAGGCTGCTGAA TT GAG-3′. Transfection was
performed with Lipofectamine™ LTX and PLUS™ Reagents (Invitrogen)
according to the manufacturer’s instructions. The cells were
harvested 5 days after transfection and used for further
assays.
Cell growth assay
Cell proliferation was assessed using a standard
CCK-8 assay (Dojindo, Kunamoto, Japan). Absorbance was determined
at a test wavelength of 450 nm on a multi-well plate reader
(Microplate Reader; Bio-Rad, Hercules, CA, USA).
Apoptosis assay
Cell apoptosis was assayed by staining with Annexin
V-FITC (KeyGEN, Nanjing, China), according to the manufacturer’s
instructions and detected by flow cytometry on a FACSCalibur
(Becton-Dickinson, Franklin Lakes, NJ, USA).
Plasmid construction
The construction of a series of truncated constructs
containing the human DICER1 promoter was performed as
follows: A 1,062-bp fragment of the human DICER1 promoter
was generated through PCR, using as a template genomic DNA obtained
from human blood samples. The DICER1 promoter PCR primers
included a forward primer, starting at −975 bp (where +1 is the
transcription start site of DICER1), a reverse primer,
starting at +55 bp and several smaller forward primers: primer 975,
5′-TCCGGTACC TATCTCGATTCCGACTAGC-3′; primer 652, 5′-TCCGGT
ACCCAGCTTACAAACAGAGGGCCA-3′; primer 489,
5′-TCCGGTACCGAGGTGCTCAGAGGGAAGCTAA-3′ and primer 208,
5′-TCCGGTACCGATTAACCTTTCACCGCC AGG-3′. The sequence of the reverse
primer was 5′-GGCAAG CTTCGCAACCCTCATTAAAAGAGC-3′. The amplified
fragments were attached to the KpnI and HindIII
digestion sites of the pGL3-basic vector (Promega, Madison, WI,
USA). The human GATA1 expression vector (pcDNA3.1-GATA1,
pGATA1) was purchased from GenScript Corp. - Biology CRO,
Piscataway, NJ, USA.
Luciferase assay
Cells in 24-well plates were transfected in
triplicate using Lipofectamine LTX and PLUS Reagents (Invitrogen)
with 1 μg of specific plasmids. After 48 h of transfection,
cells were harvested in 100 μl of Passive Lysis buffer
(Promega) and the luciferase assay was performed using the Dual
Luciferase Assay system (Promega), according to the manufacturer’s
instructions. Relative luciferase activity was calculated as the
ratio of firefly luciferase activity to Renilla luciferase
activity.
Electrophoretic mobility shift assay
(EMSA)
Preparation of nuclear extracts from K562 and U937
cells was performed using an Active Motif Nuclear Extract kit
(Active Motif, Carlsbad, CA, USA) according to the manufacturer’s
instructions. The following double-stranded oligonucleotides were
used in the present study (wild-type and mutant binding sites are
underlined): Wild-type, 5′-tctcaTTATCTgggga-3′; and mutant,
5′-tctcatGCGTtgggga-3′. Oligonucleotide
labeling was performed using the Biotin 3′ End Labeling kit (Pierce
Biotechnology, Rockford, IL, USA). The LightShift Chemiluminescent
EMSA kit (Pierce Biotechnology) was used according to the
instructions provided. For competition assays, samples were
pre-incubated with a 100-fold excess of the unlabeled wild-type or
mutated oligonucleotide duplex competitors. For the supershift
reaction, 1 μg of anti-GATA1 antibody (Abcam) was
pre-incubated with the nuclear extracts for 30 min on ice.
Chromatin immunoprecipitation assay
(ChIP)
ChIP assays were performed as previously described
(13,14). Briefly, cells were cross-linked
with 1% formaldehyde for 10 min at 37°C. After washing with
ice-cold PBS, cells were lysed with lysis buffer containing
protease inhibitors and sonicated to obtain DNA fragments. A sample
of untreated DNA fragments was saved as the input DNA. The
remaining sample was diluted, treated with anti-GATA1 antibody
(Abcam) or rabbit IgG and immunoprecipitated by Protein A/G
Plus-Agarose (Santa Cruz Biotechnology). Each precipitated sample
was eluted and extracted with phenol-chloroform and subjected to
PCR. PCR was performed using DICER1 promoter-specific
primers amplifying the GATA1-binding regions (forward primer,
5′-CTGGGGATCCCGGTTGG-3′; reverse primer,
5′-GCATTGTTGCTCCTTCTG-3′).
Statistical analysis
Data were subjected to statistical analysis and are
presented as the means ± standard error of the mean (SEM).
Differences in mean values were analyzed using one-way analysis of
variance (ANOVA). For the comparison of DICER1 mRNA levels
between AML cases and normal donors, the Mann-Whitney U test was
used. All analyses were performed with SPSS Statistics version 18.0
software (SPSS Inc., Chicago, IL, USA). In the figures, P-values
indicating a statistically significant difference are marked with
asterisks (*P<0.05, **P<0.01).
Results
High expression of DICER1 in AML
patients
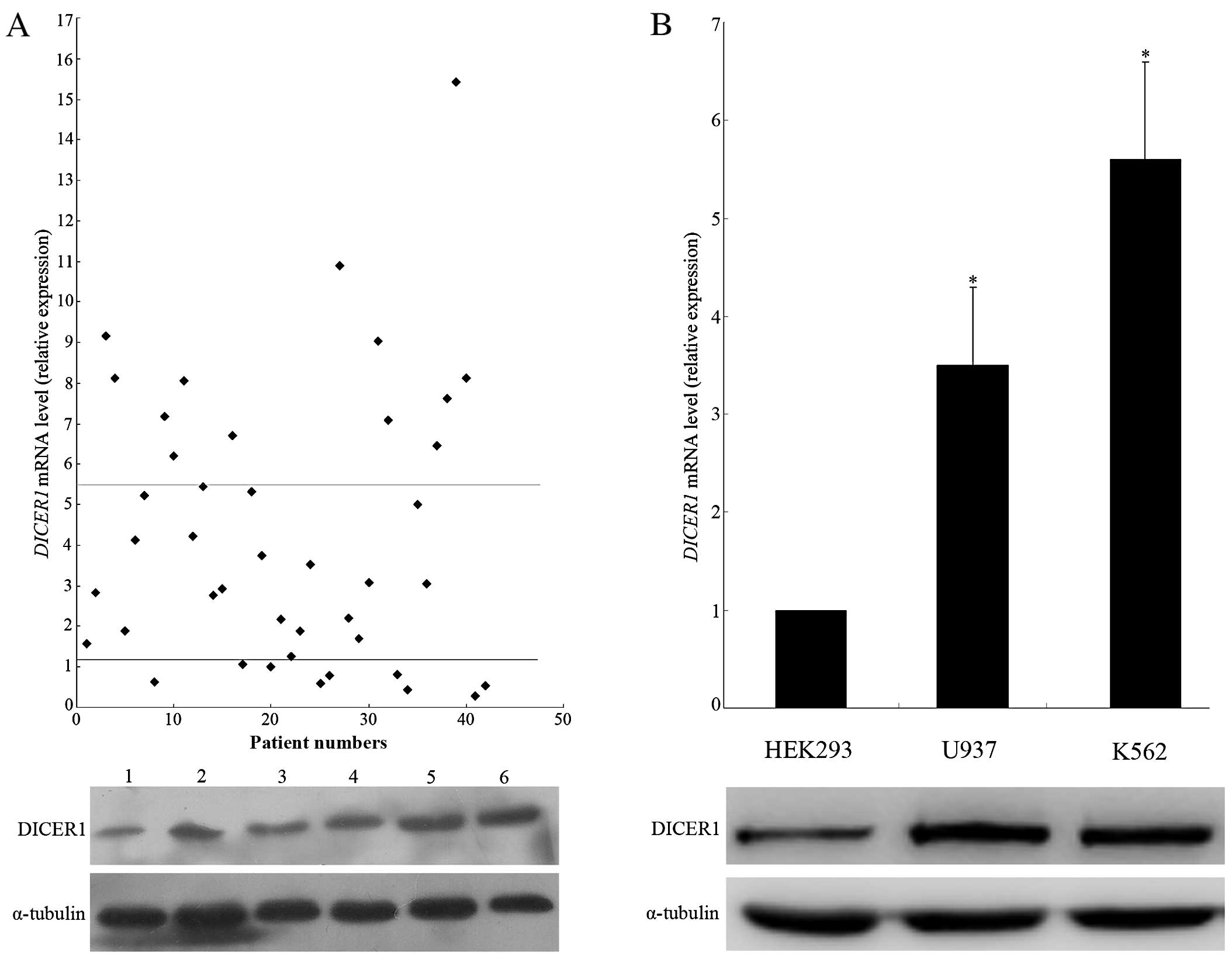
We examined bone marrow (BM) samples of 42 AML cases
and normal donors using real-time quantitative PCR for
DICER1 mRNA. Compared to the normal controls, we observed an
upregulation of DICER1 mRNA expression in the bone marrow of
AML patients with statistically significant differences (P<0.01,
Fig. 1A). The mean increased
DICER1 mRNA expression level in AML patients was 4.58-fold
higher compared to that in the normal controls and the increased
DICER1 mRNA level was defined as higher than the mean
expression level (1.2±0.81) of the normal controls. In the 19 cases
in which DICER1 mRNA expression was increased, the
DICER1 protein level was detected by western blot analysis.
The results showed that the expression of the DICER1 protein
was markedly elevated in a significant proportion of AML cases
(74%), compared to the normal controls. In addition, we assessed
DICER1 expression in the K562 and U937 human leukemia cell
lines, in which we observed a significantly increased expression of
DICER1 at the mRNA and protein levels, compared to HEK293
cells (Fig. 1B).
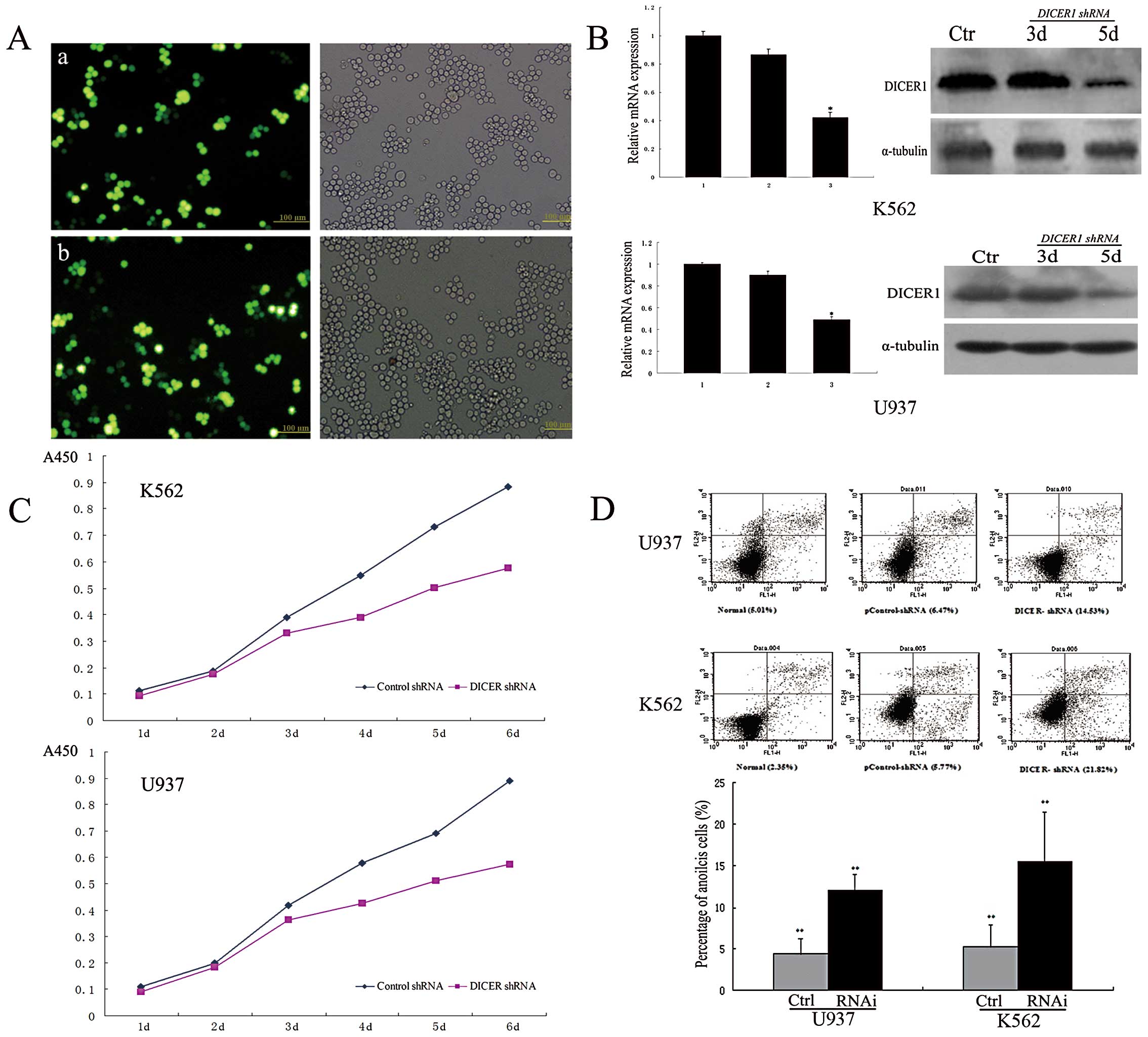
Silencing of DICER1 induces growth arrest
and promotes apoptosis in leukemia cell lines
To validate the functional role of DICER1 in
leukemia cells, we used DICER1-shRNA to knock down
DICER1 in 2 leukemia cell lines (K562 and U937). Real-time
quantitative PCR and western blot analysis confirmed DICER1
knockdown in both cell lines. As shown in Fig. 2C, cell proliferation experiments
revealed that DICER1 silencing inhibited the proliferation
of K562 and U937 cells. We further investigated the effects of
DICER1 silencing on cell apoptosis in both cell lines. To
that end, apoptotic cells were assessed by Annexin V-FITC staining,
followed by flow cytometry. Flow cytometry analysis demonstrated
that the silencing of DICER1 significantly induced apoptosis
compared to the controls (P<0.01, Fig. 2D). The percentages of early
apoptotic U937 cells in the blank, control-shRNA and DICER-shRNA
groups were 5.01±1.25, 6.47±2.06 and 14.53±1.81%, respectively;
similar results were observed in the K562 cells: 2.35±0.84,
5.77±1.62 and 21.82±2.36%, respectively.
Analysis of the regulation of DICER1
expression as well as that of its promoter
Previous studies have demonstrated that specific
types of tumors exhibit differential expression of
DICER1(5–8), suggesting the tissue-specific
regulation of its expression. In this study, we demonstrated that
DICER1 expression in AML patients was increased and that
DICER1 mRNA expression correlated with its protein
expression. Therefore, we hypothesized that in AML patients, the
upregulation of DICER1 may be associated with
hematopoietic-specific regulation. The regulation of DICER1
expression appeared to occur mainly at the transcriptional level.
We analyzed 1,300 bp (−1200 to +100) of the 5′-flanking region
sequence of DICER1 using TRANSFAC-transcription element
search software (TESS) and discovered 1 GATA1-binding element (−617
to −611) in this region (Fig. 3).
GATA1 is a zinc-finger transcription factor that plays an important
role in gene regulation during the development and differentiation
of hematopoietic cells. The tissue-specific expression of GATA1 is
mainly restricted to hematopoietic and Sertoli cells (15,16).
Previous studies have demonstrated that leukocytes from AML
patients express a significant amount of GATA1 (17–19)
and that the overexpression of GATA1 exerts a negative effect on
the event-free survival of AML patients (20,21).
These data led us to explore the possibility that GATA1 may
directly upregulate DICER1 in AML.
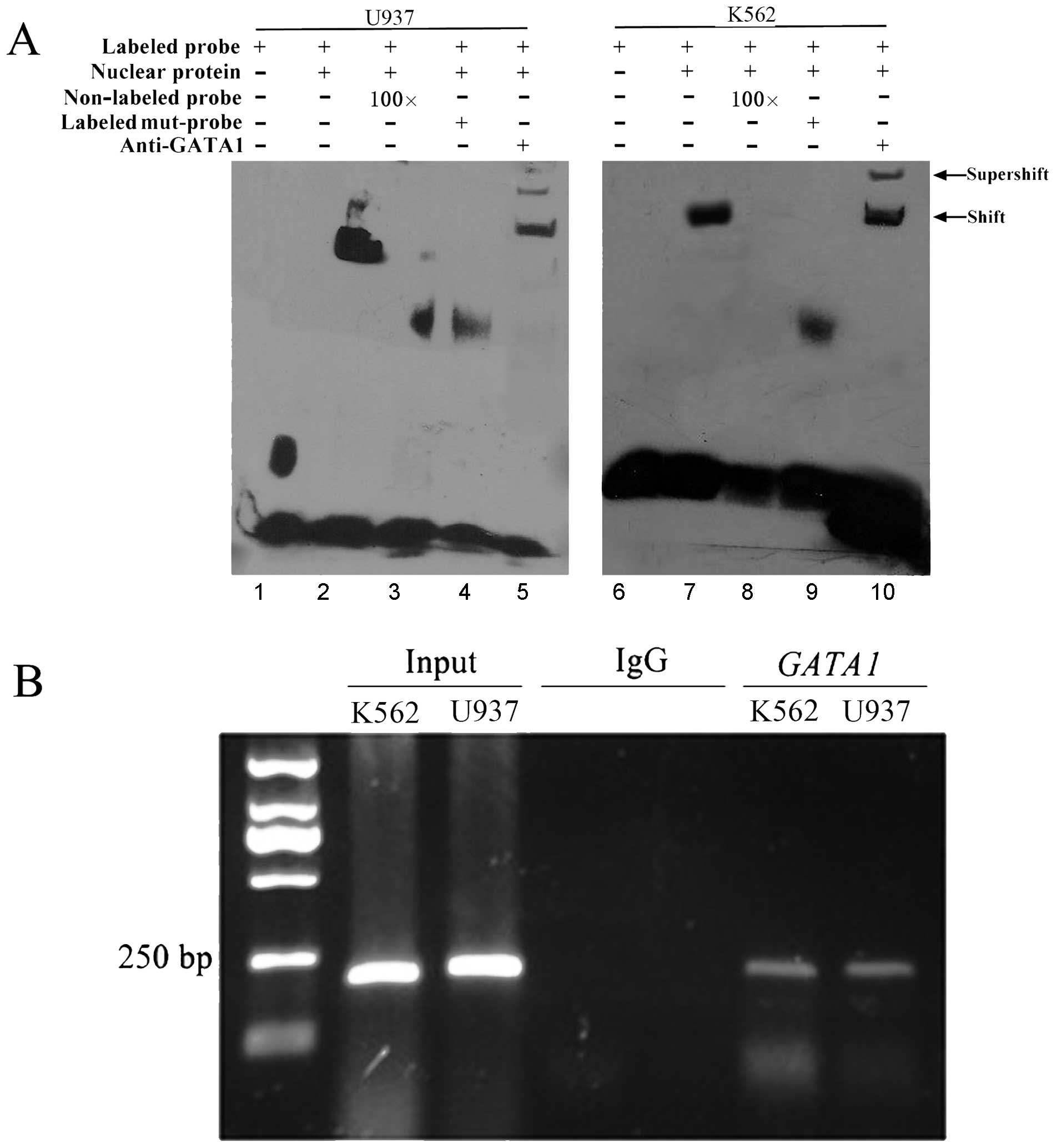
GATA1 binds to the DICER1 promoter
In order to determine whether GATA1 binds to the
DICER1 promoter, we performed EMSAs using nuclear protein
from K562 and U937 cells and the oligonucleotide probe containing
the GATA1 sites. As shown in Fig.
4A, nuclear protein from K562 and U937 cells bound to the
labeled wild-type oligonucleotide probe and binding was diminished
with the addition of 100X unlabeled wild-type and labeled mutant
oligonucleotide probes. Furthermore, the bands were supershifted by
specific GATA1 antibodies. The interaction between GATA1 and the
DICER1 promoter was further confirmed by ChIP assay. Primers
were used to amplify the GATA1-binding regions of the DICER1
promoter. We observed PCR products when using the GATA1
antibody-precipitated samples from the K562 and U937 cells, but not
in the negative control immunoprecipitation samples when using the
anti-rabbit IgG antibody (Fig.
4B).
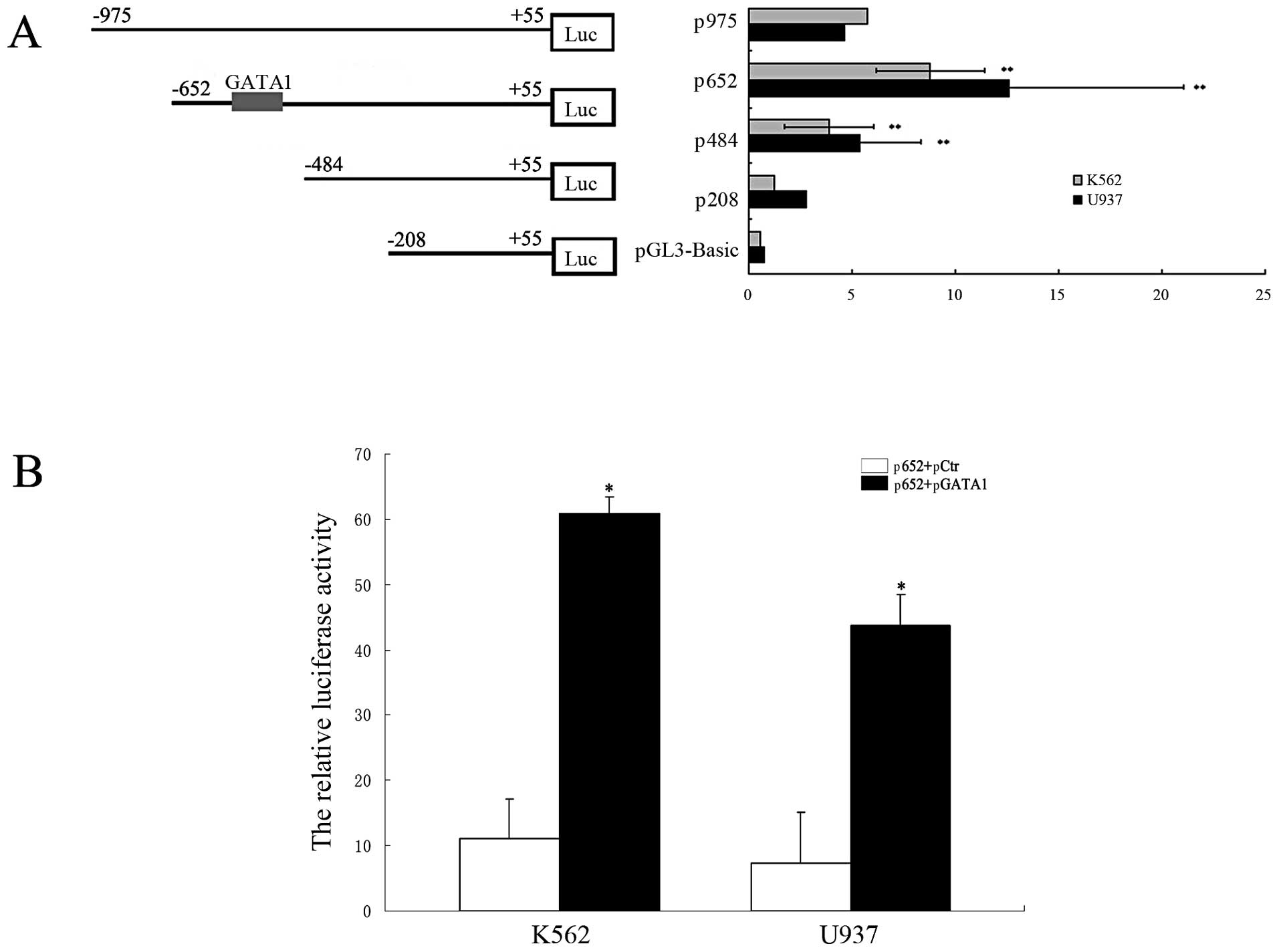
GATA1 transactivates the DICER1
promoter
EMSA and ChIP studies confirmed that GATA1 bound to
the DICER1 promoter; therefore, we sought to determine
whether GATA1 can transactivate the DICER1 promoter. We
cloned the human DICER1 promoter (1,062 bp) into a
luciferase reporter plasmid (pGL3-Basic) and generated a series of
truncated constructs (Fig. 5A,
left panel). The locations of the primers of these constructs are
shown in Fig. 3 (gray areas). The
generated fragments were transiently co-transfected into the K562
and U937 cells in conjunction with pRL-TK. Luciferase assay results
revealed an approximately 2-fold increase in the luciferase
activity of p652 compared to that of p484, indicating that the
region spanning between −652 and −484 bp contained one or more
positive regulatory elements. The GATA1-binding sites (−617 to
−611) are located in this region; therefore, to confirm that the
GATA1 sites contributed to DICER1 transcription, we
co-transfected a GATA1 expression vector (pcDNA3.1-GATA1,
pGATA1) and a p652 vector into the K562 and U937 cells. The
relative luciferase activity in the p652 + pGATA1 group was
significantly increased compared to that in the p652 + empty
pcDNA3.1 (pCtr) group on the 3rd day post-transfection (P<0.01,
Fig. 5B). These results indicate
that GATA1 positively regulates DICER1 promoter
expression.
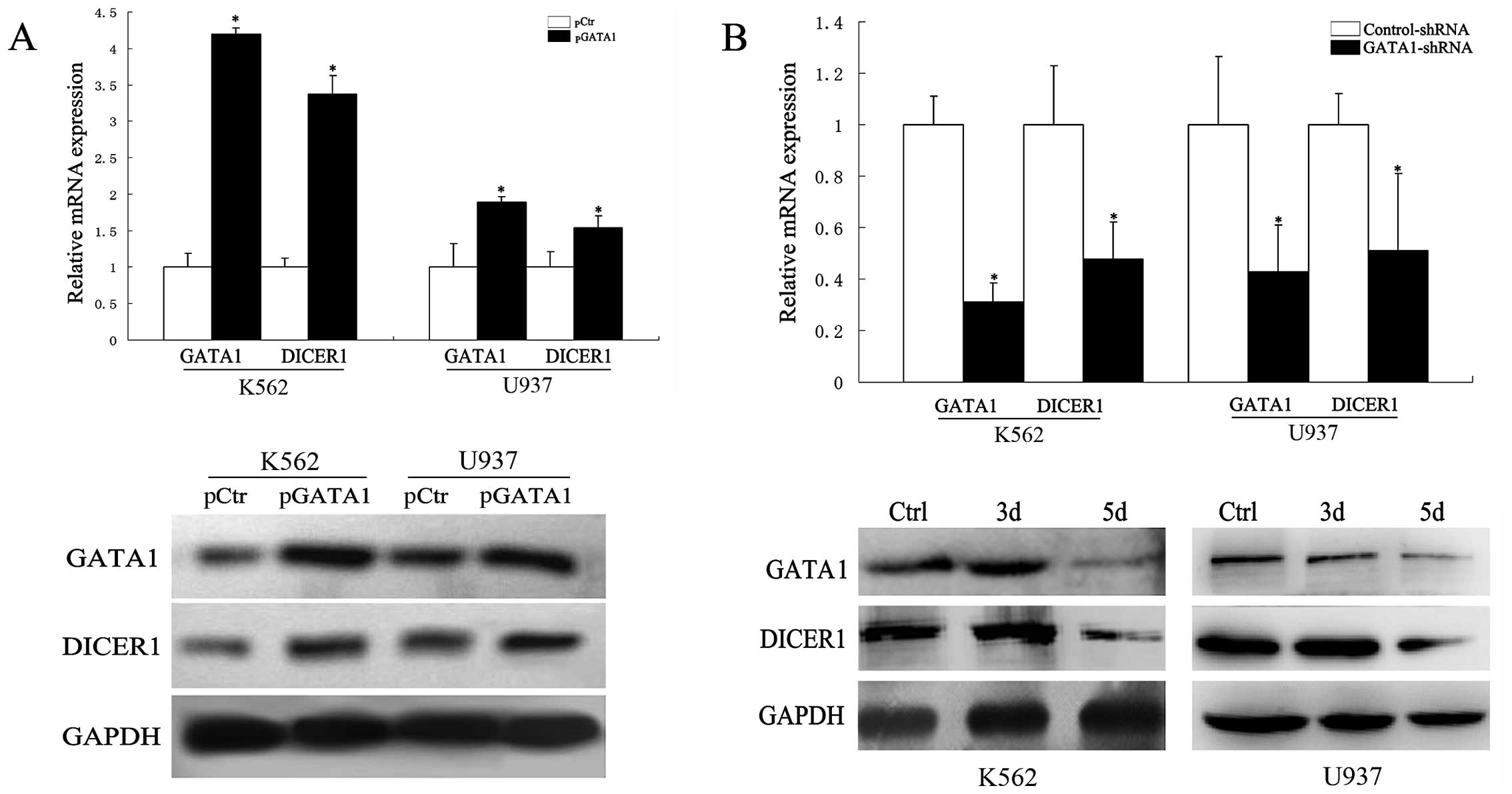
GATA1 upregulates DICER1 expression in
AML
Since our luciferase reporter, EMSA and ChIP assays
described above indicated that GATA1 upregulated DICER1
transcription, we attempted to verify that GATA1 regulated
DICER1 expression. To that end, K562 and U937 cells were
transfected with a GATA1 expression construct for 2 days. The
forced expression of GATA1 increased DICER1 expression at 2
days post-transfection (P<0.05, Fig. 6A). Furthermore, previous studies
have demonstrated GATA1 overexpression in AML (20,21).
Therefore, we transfected K562 and U937 cells with GATA1-shRNA and,
as shown in Fig. 6B, GATA1-shRNA
downregulated the mRNA and protein levels of DICER1, as
opposed to the control-shRNA. Taken together, these results lead to
the conclusion that the overexpression of DICER1 is induced
by GATA1 in AML.
Discussion
Alterations in DICER1 expression levels in
various types of cancer have been observed. In prostate cancer,
precursor lesions of lung adenocarcinomas and colorectal cancer,
DICER1 over-expression has been observed (5,6,22),
while in ovarian cancer, invasive lung adenocarcinoma and breast
cancer, decreased expression has been demonstrated (7–9). The
dysregulation of DICER1 expression appears to be related to
the aggressiveness and metastatic spread of cancer. For example, in
prostate and colorectal cancer, the overexpression of DICER1
has been shown to predict poor survival (5,11),
whereas the downregulation of its expression has been associated
with aggressiveness and the metastatic spread of breast and lung
cancer (8,9). These findings suggest that
DICER1 plays an important role in carcinogenesis. However,
the functional role of DICER1 in tumors is controversial.
Therefore, in this study we aimed to investigate the expression
levels of DICER1 in AML and the potential role of
DICER1 in leukemia cells. The results revealed that
DICER1 expression in AML patients was highly increased
compared to the normal donors. The functional assays demonstrated
that the silencing of DICER1 inhibited cell proliferation
and promoted apoptosis in leukemia cell lines. Our results are
similar to those reported by Zhou et al; their study
demonstrated that the silencing of DICER1 was associated
with significantly enhanced G0- to G1-phase accumulation and
significantly increased apoptosis in myeloma cell lines (23). Furthermore, studies on oral cancer
cells have demonstrated that the overexpression of DICER1
increases cell proliferation (24). These data indicate that high
DICER1 expression may induce leukemia cell proliferation and
inhibit apoptosis, mechanisms by which DICER1 may promote
leukemia progression. Additional studies are required in order to
further elucidate the function of DICER1 in tumor cells.
Although our study, as well as others have
demonstrated that specific types of tumors exhibit elevated levels
of DICER1, there is evidence that other types of cancer have
decreased DICER1 expression levels. These discrepancies may
be attributed to tissue-specific differences or progression of
disease. For example, in lung cancer, the upregulation of
DICER1 has been observed in precursor lesions and the
reduced expression at the more aggressive stages (8).
Unfortunately, knowledge of the regulatory
mechanisms underlying these alterations of DICER1 expression
levels in cancer is limited. A previous study demonstrated that the
regulation of Dicer expression was mainly post-transcriptional, as
cellular Dicer mRNA did not correlate well with protein expression
(25). However, in breast cancer
cell lines, a 72% concordance was observed between DICER1
mRNA and protein levels (9). In
this study, we detected DICER1 protein expression in 19 AML
cases, in which high DICER1 mRNA levels were observed. The
results demonstrated a 74% concordance between DICER1 mRNA
and protein levels, which suggested that DICER1 may be
transcriptionally upregulated in AML. Levy et al identified
a novel mechanism of DICER upregulation via its transcriptional
targeting by the melanocytic transcription factor,
microphthalmia-associated transcription factor (MITF), during
melanocyte differentiation (26).
In this study, we reported that DICER1 expression in AML was
regulated by the hematopoietic transcription factor, GATA1. GATA1
is a zinc-finger transcription factor that plays an important role
in gene regulation during development and differentiation of
hematopoietic cells. The tissue-specific expression of GATA1 is
mainly restricted to hematopoietic (erythroid cells,
megakaryocytes, eosinophils and mast cells) and Sertoli cells
(13,14). During normal hematopoiesis, GATA1
is not expressed in myeloid progenitor cells; however, previous
studies have shown that leukocytes from AML patients express a
significant amount of GATA1 (17–19).
In addition, the over-expression of GATA1 has been shown to exert a
negative effect on the event-free survival of AML patients
(20,21).
In the present study, we demonsrate that the
upegulation of GATA1 affects the transcription of DICER1 in
AML. This finding may provide a framework for understanding the
differential DICER1 expression among different types of
cancer. Elucidating the effects of DICER1 on leukemia cell
proliferation and apoptosis will aid in the understanding of its
role in the development of cancer.
Acknowledgements
We thank Professor Wei-Neng Fu of the
Department of Medical Genetics, China Medical University, for
providing technical support and helpful suggestions.
References
|
1
|
Cerutti H and Casas-Mollano JA: On the
origin and functions of RNA-mediated silencing: from protists to
man. Curr Genet. 50:81–99. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peng JJ, Yan F, Chen HR and Chen JP:
Progress of studies on Dicer structure and function. Yi Chuan.
30:1550–1556. 2008.(In Chinese).
|
|
3
|
Lee YS, Nakahara K, Pham JW, et al:
Distinct roles for Drosophila Dicer-1 and Dicer-2 in the
siRNA/miRNA silencing pathways. Cell. 117:69–81. 2004.
|
|
4
|
Filipowicz W, Jaskiewicz L, Kolb FA and
Pillai RS: Post-transcriptional gene silencing by siRNAs and
miRNAs. Curr Opin Struct Biol. 15:331–341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chiosea S, Jelezcova E, Chandran U, et al:
Up-regulation of dicer, a component of the MicroRNA machinery, in
prostate adenocarcinoma. Am J Pathol. 169:1812–1820. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chiosea S, Jelezcova E, Chandran U, et al:
Overexpression of Dicer in precursor lesions of lung
adenocarcinoma. Cancer Res. 67:2345–2350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Merritt WM, Lin YG, Han LY, et al: Dicer,
Drosha, and outcomes in patients with ovarian cancer. N Engl J Med.
359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karube Y, Tanaka H, Osada H, et al:
Reduced expression of Dicer associated with poor prognosis in lung
cancer patients. Cancer Sci. 96:111–115. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grelier G, Voirin N, Ay AS, et al:
Prognostic value of Dicer expression in human breast cancers and
association with the mesenchymal phenotype. Br J Cancer.
101:673–683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zighelboim I, Reinhart AJ, Gao F, et al:
DICER1 expression and outcomes in endometrioid endometrial
adenocarcinoma. Cancer. 117:1446–1453. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Faber C, Horst D, Hlubek F and Kirchner T:
Overexpression of Dicer predicts poor survival in colorectal
cancer. Eur J Cancer. 47:1414–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vaksman O, Hetland TE, Trope’ CG, et al:
Argonaute, Dicer, and Drosha are up-regulated along tumor
progression in serous ovarian carcinoma. Hum Pathol. 43:2062–2069.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ross J, Bottardi S, Bourgoin V,
Wollenschlaeger A, Drobetsky E, et al: Differential requirement of
a distal regulatory region for pre-initiation complex formation at
globin gene promoters. Nucleic Acids Res. 37:5295–5308. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Wang E, Dutta S, Lau JS, Jiang SW,
et al: Protein kinase C-mediated modulation of FIH-1 expression by
the home-odomain protein CDP/Cut/Cux. Mol Cell Biol. 27:7345–7353.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Harigae H: GATA transcription factors and
hematological diseases. Tohoku J Exp Med. 210:1–9. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shimizu R and Yamamoto M: Function and
gene expression regulation of GATA-1 and GATA-2 transcription.
Seikagaku. 79:941–952. 2007.(In Japanese).
|
|
17
|
Shimamoto T, Ohyashiki JH, Ohyashiki K, et
al: GATA-1, GATA-2, and stem cell leukemia gene expression in acute
myeloid leukemia. Leukemia. 8:1176–1180. 1994.PubMed/NCBI
|
|
18
|
Guerrasio A, Saglio G, Rosso C, et al:
Expression of GATA-1 mRNA in human myeloid leukemic cells.
Leukemia. 8:1034–1038. 1994.PubMed/NCBI
|
|
19
|
Shimamoto T, Ohyashiki K, Ohyashiki JH, et
al: The expression pattern of erythrocyte/megakaryocytes-related
transcription factors GATA-1 and stem cell leukemia gene correlates
with hematopoietic differentiation and is associated with outcome
of acute myeloid leukemia. Blood. 86:3173–3180. 1995.
|
|
20
|
Ayala RM, Martínez-López J, Albízua E, et
al: Clinical significance of Gata-1, Gata-2, EKLF, and c-MPL
expression in acute myeloid leukemia. Am J Hematol. 84:79–86. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ohyashiki K, Ohyashiki JH, Shimamoto T and
Toyama K: Pattern of expression and their clinical implications of
the GATA family, stem cell leukemia gene, and EVI1 in leukemia and
myelodysplastic syndromes. Leuk Lymphoma. 23:431–436. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Papachristou DJ, Korpetinou A,
Giannopoulou E, et al: Expression of the ribonucleases Drosha,
Dicer, and Ago2 in colorectal carcinomas. Virchows Arch.
459:431–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou Y, Chen L, Barlogie B, Stephens O, Wu
X, Williams DR, et al: High-risk myeloma is associated with global
elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl
Acad Sci USA. 107:7904–7909. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jakymiw A, Patel RS, Deming N, et al:
Overexpression of Dicer as a result of reduced let-7 microRNA
levels contributes to increased cell proliferation of oral cancer
cells. Genes Chromosomes Cancer. 49:549–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wiesen JL and Tomasi TB: Dicer is
regulated by cellular stresses and interferons. Mol Immunol.
46:1222–1228. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Levy C, Khaled M, Robinson KC, et al:
Lineage-specific transcriptional regulation of DICER by MITF in
melanocytes. Cell. 141:994–1005. 2010. View Article : Google Scholar : PubMed/NCBI
|