Introduction
Glioma is the most common primary brain tumor, but
its prognosis has changed little over the past 30 years, despite
comprehensive and intensive treatment. The invasive growth pattern
of glioma cells is partially responsible for its poor response to
treatment. A major pathophysiological feature of gliomas is their
ability to invade diffusely into the surrounding brain tissues
(1,2), even across the brain lobes, producing
new growth foci and finally spreading throughout the central
nervous system. Limiting the invasive ability of gliomas thus
represents a potential approach for curing patients and prolonging
survival.
Growing evidence has implicated the secreted stromal
protein periostin (POSTN) in many tumors (3–6),
including breast cancer, prostate cancer and cholangiocarcinoma
(7). POSTN has been confirmed to
play roles in invasion/metastasis (8,9),
cell proliferation (10,11), migration (12) and angiogenesis (9,13),
but its function and mechanism in all grades of glioma is not
completely clear, although there are some relative reports in
glioblastoma (14–16). Here, we report the expression
pattern of POSTN using the expression values from microarrays of
220 frozen glioma tissues and investigated the expression of POSTN
in relation to glioma grade progression validated by 71 glioma
samples using immunohistochemistry. In addition, to underscore the
potential biological insights of POSTN 89 GBM samples were
subjected to whole genome gene profiling. Integrated analysis of
POSTN and whole genome gene expression patterns showed that
overexpression of POSTN was tightly correlated with the gene sets
related to cell invasion and proliferation. Functional assays
showed that POSTN might serve as a potential target for
anti-invasion and anti-proliferation therapies in glioma
patients.
Materials and methods
Tissue samples
Two hundred and twenty frozen glioma samples were
obtained from newly diagnosed patients treated by the CGGA Group.
Tumor histology of all patients was confirmed independently by two
neuropathologists based on the 2007 edition of WHO classification
of central nervous system tumors. All the patients in the study
received similar treatments. The inclusion criteria were as
follows: i) The patients were treated with conventional therapies
consisting of maximal surgical resection, followed by radiotherapy
and/or chemotherapy. ii) Patients who received radiotherapy or
chemotherapy before admission or died from non-glioma-related
diseases were excluded from this study. iii) All the patients had
follow-up and age >18 years. This study was approved by the
Research Ethics Committee of Beijing Tiantan Hospital. Written
informed consent was obtained from each patient.
RNA extraction and microarray
analysis
All the tissue samples were immediately snap-frozen
in liquid nitrogen after surgery. A frozen section from each tumor
was stained with hematoxylin and eosin for assessment of the
percentage of tumor cells before RNA extraction. Only samples with
>80% tumor cells were selected. Total RNA from frozen tumor
tissues was extracted from frozen tumor tissues by using a Total
RNA Isolation kit (Ambion, Austin, TX) according to the
manufacturer’s protocol. RNA concentrations were measured using a
NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Houston,
TX). The expression profiling of all the samples was tested with
the Agilent Whole Human Genome Oligo Microarray, which is in a
4x44k slide format: each block represents >41,000 probesets. All
the steps of sample-labeling, hybridization, washing and scanning
steps were conducted at the laboratory in the Cancer
Institute/Hospital of Peking Union Medical College, following the
manufacturer’s instructions. In brief, POSTN-labeled cRNA was
generated from 500 ng input total RNA by in vitro
transcription using Agilent’s Low RNA Input Linear Amplification
kit Plus and 1.65 μg cRNA from each labeling reaction was
hybridized to one block of the microarray. After hybridization,
slides were washed and then scanned with the Agilent G2565BA
Microarray Scanner System. Fluorescence intensities on scanned
images were extracted and preprocessed by Agilent Feature
Extraction Software (v9.1). Data normalization and filtering was
performed using the GeneSpring GX 11.0 (Agilent Technologies). Only
those genes with expression levels marked as present or marginal in
all of the chips (blocks) passed through the quality filtering.
Then, measurements were set to <0.01 and the chip data were
normalized to the 50th percentile of the intensity of each chip.
Significant analysis of microarrays (SAM) was implemented on
low-grade glioma (astrocytoma, oligodendroglioma and
oligoastrocytoma) and high-grade glioma (anaplastic gliomas and
glioblastomas). Genes that were significantly up- or downregulated
were identified using SAM (17).
SAM assigns a score to each gene on the basis of a change in gene
expression relative to the standard deviation of repeated
measurements. Analysis parameters (Delta) were set to result in
false discovery rate (FDR) <0.2.
Gene ontology analysis
To obtain more information on the biologic processes
related to POSTN expression in 89 glioblastomas, we performed
gene-expression profiling using Agilent Whole Human Genome Array
and Gene ontology analysis (GO) (david.abcc.ncifcrf.gov) (Table I).
 | Table IGene sets enriched in glioblastoma
samples with POSTN overexpression. |
Table I
Gene sets enriched in glioblastoma
samples with POSTN overexpression.
| Name | Count | Fold enrichment | p-value | FDR |
|---|
| GO:0006928-cell
motion | 94 | 2.00083707 | 6.19E-11 | 1.14E-07 |
|
GO:0051270-regulation of cell motion | 49 | 2.56694316 | 1.13E-09 | 2.10E-06 |
| GO:0016477-cell
migration | 60 | 3.444752445 | 8.02E-09 | 1.16E-05 |
|
GO:0042127-regulation of cell
proliferation | 128 | 1.64441988 | 1.11E-08 | 2.05E-05 |
|
GO:0043122-regulation of I-κB kinase/NF-κB
cascade | 31 | 2.929242977 | 8.69E-08 | 1.61E-04 |
|
GO:0030334-regulation of cell
migration | 41 | 2.452870574 | 1.18E-07 | 2.17E-04 |
| GO:0048870-cell
motility | 61 | 2.008949134 | 1.75E-07 | 3.24E-04 |
| GO:0008284-positive
regulation of cell proliferation | 74 | 1.807210993 | 6.04E-07 | 0.001116781 |
| GO:0007249-I-κB
kinase/NF-κB cascade | 21 | 3.370204285 | 1.45E-06 | 0.002671136 |
| GO:0008283-cell
proliferation | 69 | 1.600074053 | 9.30E-05 | 0.171734749 |
Validated samples and treatment
Surgical specimens of 71 glioma patients included 24
astrocytoma (WHO grade II), 9 anaplastic astrocytoma (AA, WHO grade
III) and 38 glioblastomas (WHO grade IV) were included into this
retrospective study at the Glioma Center of Beijing Tiantan
Hospital. The inclusion criteria were the same as for micro-array
samples. Progression-free survival (PFS) was defined as the time
from surgical resection to the first MRI-confirmed recurrence or
until death. Overall survival (OS) was defined as the period of
time from surgical resection to death. The glioma tissues and data
were provided by China Glioma Genome Atlas (CGGA, www.CGGA.org.cn). Standard treatment consisted of
surgical resection and postoperative radiotherapy, with adjuvant
chemotherapy. The extent of resection was assessed using
postoperative enhanced MRI within 72 h and graded as total or
subtotal resection. The patients received standard radiotherapy (60
Gy in 2-Gy fractions, with five fractions administered per week) of
the contrast-enhancing lesion (plus a 2-cm safety margin and the
area of preoperative edema). This was approved by the Ethics
Committee of Beijing Tiantan Hospital and was based on the criteria
of the Helsinki convention. Written informed consents was provided
by each participant.
Immunohistochemistry
Paraffin-embedded specimens were cut into
4-μm sections and baked at 65°C for 30 min. The sections
were deparaffinized with xylene and rehydrated. Sections were
submerged into EDTA (pH 8.0) and autoclaved for antigen retrieval,
then treated with 3% hydrogen peroxide, followed by incubation with
1% FBS. Anti-POSTN antibody (ab14041, rabbit polyclonal, USA, 1:200
dilutions) was added and incubated overnight at 4°C. For negative
controls, the primary antibody was replaced by normal mouse serum.
Horseradish peroxidase (HRP) labeled secondary antibody in the
MaxVision™ HRP-Polymer anti-rabbit IHC kit (KIT-5930 Maixin Biol,
Fu Zhou, China) was applied and incubated for 30 min at room
temperature, followed by 5-min incubation at room temperature with
DAB provided in the kit for color development. The sections were
finally counterstained with haematoxylin and mounted with Permount
(BIOS, Beijing, China). Results were visualized and photographed
under a light microscope (Olympus BX-51; Olympus Optical). The
degree of immunostaining of sections was viewed and scored
separately by two independent investigators. The scores were
determined by combining the proportion of positively stained tumor
cells and the intensity of staining. Scores from the two
investigators were averaged for further comparative evaluation of
the POSTN expression. The intensity of staining was recorded on a
scale of 0 (no staining), 1 (weak staining, light yellow), 2
(moderate staining, yellowish brown) and 3 (strong staining,
brown).
Cell lines and cell culture
Human glioblastoma cell lines U87 and LN229 were
obtained from the Institute of Biochemistry and Cell Biology,
Chinese Academy of Science, Shanghai, China. The cells were
maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA)
supplemented with 10% fetal bovine serum and incubated at 37°C in
5% CO2. Upon 80% confluency, cells were starved in DMEM
with 1% FBS for 24 h and maintained in this low serum condition for
the course of treatment.
POSTN gene knockdown by siRNA
Specific oligos (21-bp) targeting the POSTN gene
were selected (Shanghai GenePharma Co., Ltd.). SiRNA2, the most
efficiency was screened to knock down POSTN from three siRNAs
(Table II). Logarithmically
growing cells were seeded at a density of 105 cells per
6-cm dish and transfected with 5 μmol POSTN siRNA using
Lipofectamine 2000 (Invitrogen) according to the manufacturer’s
instructions. Forty-eight hours later, cells were used for in
vitro functional assay as described below.
 | Table IISiRNA and negative control
strand. |
Table II
SiRNA and negative control
strand.
| Sense strand | Antisense
strand |
|---|
| siRNA1 |
GCCAUCACAUCGGACAUAUTT |
AUAUGUCCGAUGUGAUGGCTT |
| siRNA2 |
GGUCCUAAUUCCUGAUUCUTT |
AGAAUCAGGAAUUAGGACCTT |
| siRNA3 |
GCCCUGGUUAUAUGAGAAUTT |
AUUCUCAUAUAACCAGGGCTT |
| Negative
control |
UUCUCCGAACGUGUCACGUTT |
ACGUGACACGUUCGGAGAATT |
Transwell invasion assay
The transwell invasion assay was done in 24-well
cell culture chambers using transwell inserts (Corning Life
Sciences, NY, USA) with 8-μm pore membrane precoated with
Matrigel (BD Bioscience, San Jose, CA, USA). U87 and LN229 cells
were plated at the density of 1×104 per upper well in
200 μl culture medium (DMEM, no FBS), control group, siRNA
control group and siRNA group especially. The lower chamber was
filled with 500 μl medium (DMEM, 12% FBS). The cells were
allowed to invade for 24 h, after which, the non-invading cells
with Matrigel matrix were removed from the upper surface of the
membrane by scrubbing with a cotton-tipped swab. Cells on the lower
surface of the filter were fixed for 30 min in methanol and glacial
acetic acid mixture (3:1), air-dried briefly and stained with
crystal violet. The mean number of invaded cells was counted from
five preselected microscopic fields at magnification ×200, all
experiments were performed in triplicate.
MTT assays
LN229 and U87 cells were plated in 96-well plates in
medium containing 10% FBS at ∼3,000 cells per well 24 h after
transfected siRNA. For quantitation of cell viability, cultures
were stained after 4 days in MTT assays. In brief, 20 μl of
5 mg/ml MTT (thiazolyl blue) solution was added to each well and
incubated for 4 h at 37°C. The medium was removed from each well
and the resulting MTT formazan was solubilized in 150 μl of
DMSO. Each solution was measured spectrophotometrically at 490
nm.
Colony formation assay
LN229 and U87 cells were transfected with siRNA or a
negative control for 48 h and then plated into six-orifice plate
(1,000 per orifice) and transfected with siRNA one more time on day
6. Until day 12, plates were washed with PBS and stained with
crystal violet. The number of colonies with >30 cells was
counted. The colonies were manually counted using a microscope.
Western blot analysis
After cell treatment, cell lysates were prepared via
lysis buffer, electrophoresed onto SDS-polyacrylamide gels and
transferred to polyvinylidene difluoride membranes. Membranes were
probed with rabbit antibodies against POSTN and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (A-3; Santa Cruz
Biotechnology) at dilutions of 1:1,000. Blots were detected with
horseradish peroxidase-labeled anti-rabbit antibodies (1:5,000
dilution), developed using enhanced chemiluminescence (ECL)
reagents (Amersham Pharmacia, UK). The primary antibody against
MMP-9 (Oncogene, Boston, MA), was used to examine protein
expression.
Statistical analysis
Quantitative results represent mean ± standard
deviation. For the microarray data, if multiple probes were used
for a single candidate gene, then the reference sequence of each
probe was searched in GenBank and probes detecting exons were
chosen for further analysis. Overall survival (OS) time was
calculated from the date of diagnosis until death or the last
follow-up contact. OS of the low- and high-gene expression groups
were compared using the Kaplan-Meier plotting method and log-rank
tests. For POSTN expression analyses using immunohistochemistry
analysis in the CGGA cohort, the difference between high and low
POSTN-expressing tissues was assessed using χ2 test.
Gene ontology (GO) analysis was performed using DAVID (18). All analyses were two-tailed.
p-values of <0.05 were considered statistically significant.
Analyses were performed using Matlab 2009b, GraphPad Prism
(GraphPad Software, La Jolla, CA) and SPSS version 13.0 (SPSS Inc.,
Chicago, IL).
Differences of tumor cell invasion and expression of
POSTN and MMP-9 between treated and control groups were analyzed by
t-test. Cox regression was used to correlate POSTN expression with
PFS and OS of high-grade glioma patients. Factors with
corresponding p-values of <0.1 in the univariate analysis was
applied for further multivariate Cox regression model.
Results
POSTN is relative to glioma grade
progression and confers a poor prognosis of high-grade glioma
patients based on large cohorts
The expression of POSTN was measured in a series of
220 glioma samples (97 low-grade gliomas, 34 anaplastic gliomas and
89 GBMs) via microarrays. POSTN was significantly upregulated as
identified using significance analysis of microarrays (SAM). As
shown in Fig. 1A, high-grade
gliomas demonstrated a significant increase in POSTN transcript
levels compared to the mean expression levels observed in low-grade
gliomas (p<0.0001). The correlation between POSTN expression and
overall survival was measured through Kaplan-Meier survival curve
analysis with a log-rank comparison. POSTN expression was inversely
correlated with overall survival in the high-grade glioma samples
(p=0.0157) (Fig. 1B).
POSTN is tightly associated with the
biological processes of cell invasion and proliferation
A total of 3,677 probes significantly correlated
(R>0.4 or <−0.4, p<0.05) with POSTN expression levels
(Fig. 2; 2,249 probes with
positive correlation and 1,428 with negative correlation). Arrows
indicate genes that are discussed in the text include MMP-11,
MMP-2, MMP-25, MMP-14, MMP-9, MMP-7, MMP-1, MMP-19 and NFκB1. Gene
ontology analysis (GO) showed that the POSTN positive correlative
gene sets related to cell invasion and proliferation were
significantly enriched in the cases with POSTN overexpression
(Table I). Using a p<0.001, 38
biologic processes mainly related to cell migration, cell
proliferation, cell motility and extracellular matrix organization
were significantly enriched in the high-grade glioma samples with
POSTN overexpression.
Validated patient demographics
Seventy-one mainland Han Chinese glioma patients who
were receiving standard treatment were included in this study. The
patient population consisted of 40 males and 31 females with a
median age of 45 years (range 18–71). There were 24 patients with
astrocytoma (A, WHO grade II), 9 with anaplastic astrocytoma (AA,
WHO grade III) and 38 with glioblastoma multiforme (GBM; WHO grade
IV). Of the 71 cases, 52 underwent total tumor resection and 19
underwent subtotal tumor resection. Preoperative KPS scores ranged
from 50–100 (median 80). Fifty-six patients died after a median
follow-up of 32.4 months (range 16–71 months). The median
progression-free survival (PFS) was 406 days [95% confidence
interval (CI) (347–465 days] and the median overall survival (OS)
was 501 days (95% CI, 487–623 days).
Examination of POSTN expression level and
its association with survival
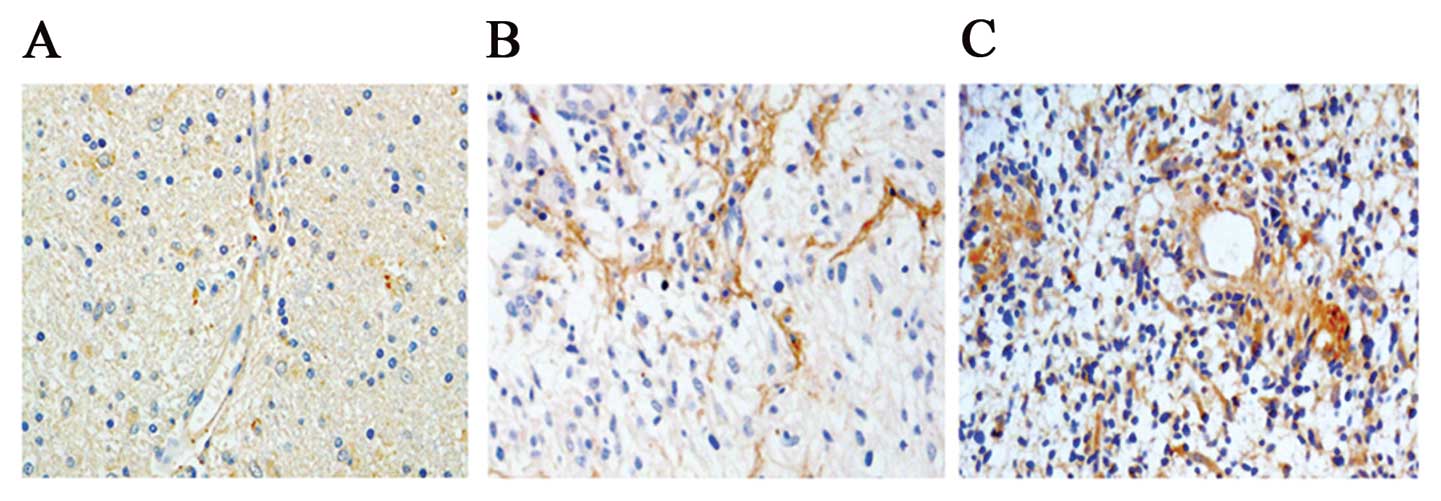
Immunohistochemical staining showed POSTN expression
ranged from low to high in WHO grades II, III and IV gliomas and
was intensely stained (score >2) in 69.01% (49/71) of cases.
POSTN was expressed in both tumor cells and stroma, but expression
was higher in the stroma than in the cells (Fig. 3). We analyzed the correlation
between POSTN protein expression and histological staging of
gliomas. As summarized in Table
III, POSTN expression was significantly higher in high-grade
gliomas compared to low-grade gliomas (p<0.01), suggesting that
POSTN expression level was associated with glioma progression.
Survival analysis showed that patients with high POSTN expression
had significant shorter overall survival (OS, p=0.032) and
progression-free survival (PFS, p=0.009) than those with low
expression respectively (Fig.
4).
 | Table IIIPOSTN expression is correlated with
glioma progression. |
Table III
POSTN expression is correlated with
glioma progression.
| Degree of POSTN
immunoreactivity (%)
|
|---|
|
Characteristics | Low | High |
|---|
| Low-gradea | 13/24 (54.2) | 11/24 (45.8) |
| High-gradea | 9/47 (19.1) | 38/47 (80.9) |
| IIIb | 3/9 (33.3) | 6/9 (66.7) |
| IVb | 6/38 (15.8) | 32/38 (84.2) |
High POSTN expression predicted poor prognosis in
high-grade gliomas. In 47 high-grade glioma patients,
immunohistochemical detection revealed high POSTN expression in
tissue samples from 38 patients (80.9%), while samples from nine
patients (19.1%) displayed low expression (Table III). POSTN expression was
correlated with PFS and OS by univariate analysis (p=0.009 and
0.032, respectively) (Tables IV
and V). Patients with high POSTN
expression had median PFS and OS times of 382 and 460 days,
respectively, compared to 683 and 1,099 days, respectively, in
patients with low POSTN expression. Preoperative KPS score was also
correlated with PFS (p=0.005) and OS (p=0.002). There were no
significant associations between age, gender or extent of resection
and PFS or OS (Tables IV and
V). The multivariate Cox
proportional hazards model, after adjusting for age, KPS score and
extent of resection, identified high POSTN expression as an
independent unfavorable prognostic factor for PFS [hazard ratio
(HR), 3.762; p=0.009] and OS (HR, 2.816; p=0.037).
 | Table IVVariables related to progression-free
survival (PFS) in 47 high-grade glioma with combined treatment:
univariate and multivariate analysis. |
Table IV
Variables related to progression-free
survival (PFS) in 47 high-grade glioma with combined treatment:
univariate and multivariate analysis.
| | Univariate analysis
| Multivariate
analysis
|
|---|
| Variable | No. of
patients | Median PFS
(days) | 95% CI (days) | p-value | Relative risk | 95% CI | p-value |
|---|
| Gender | | | | | | | |
| Male | 28 | 387 | 293–481 | 0.426 | | | |
| Female | 19 | 396 | 205–587 | | | | |
| Age (years) | | | | | | | |
| ≤50 | 25 | 388 | 321–455 | | | | |
| >50 | 22 | 511 | 300–722 | 0.780 | | | |
| KPS | | | | | | | |
| <80 | 15 | 277 | 320–334 | | | | |
| ≥80 | 32 | 511 | 339–683 | 0.005 | 0.262 | 0.105–0.656 | 0.004 |
| Extent of
resection | | | | | | | |
| Subtotal | 19 | 361 | 188–534 | | | | |
| Total | 28 | 511 | 278–744 | 0.202 | | | |
|
POSTN-expression | | | | | | | |
| Low | 9 | 683 | 550–816 | | | | |
| High | 38 | 382 | 326–438 | 0.009 | 3.762 | 1.390–10.186 | 0.009 |
 | Table VVariables related to OS in 47
high-grade glioma with combined treatment: univariate and
multivariate analysis. |
Table V
Variables related to OS in 47
high-grade glioma with combined treatment: univariate and
multivariate analysis.
| | Univariate analysis
| Multivariate
analysis
|
|---|
| Variable | No. of
patients | Median OS
(days) | 95% CI (days) | p-value | Relative risk | 95% CI | p-value |
|---|
| Gender | | | | | | | |
| Male | 28 | 387 | 324–449 | | | | |
| Female | 19 | 525 | 399–651 | 0.386 | | | |
| Age (years) | | | | | | | |
| ≤50 | 25 | 501 | 367–635 | | | | |
| >50 | 22 | 396 | 290–502 | 0.434 | | | |
| KPS | | | | | | | |
| <80 | 15 | 348 | 244–452 | | | | |
| ≥80 | 32 | 525 | 405–645 | 0.002 | 0.312 | 0.142–0.682 | 0.004 |
| Extent of
resection | | | | | | | |
| Subtotal | 19 | 412 | 206–618 | | | | |
| Total | 28 | 460 | 272–648 | 0.214 | | | |
|
POSTN-expression | | | | | | | |
| Low | 9 | 1099 | NR | | | | |
| High | 38 | 396 | 339–453 | 0.032 | 2.816 | 1.062–7.463 | 0.037 |
POSTN expression correlates with
expression of matrix metalloproteinase-9 (MMP-9)
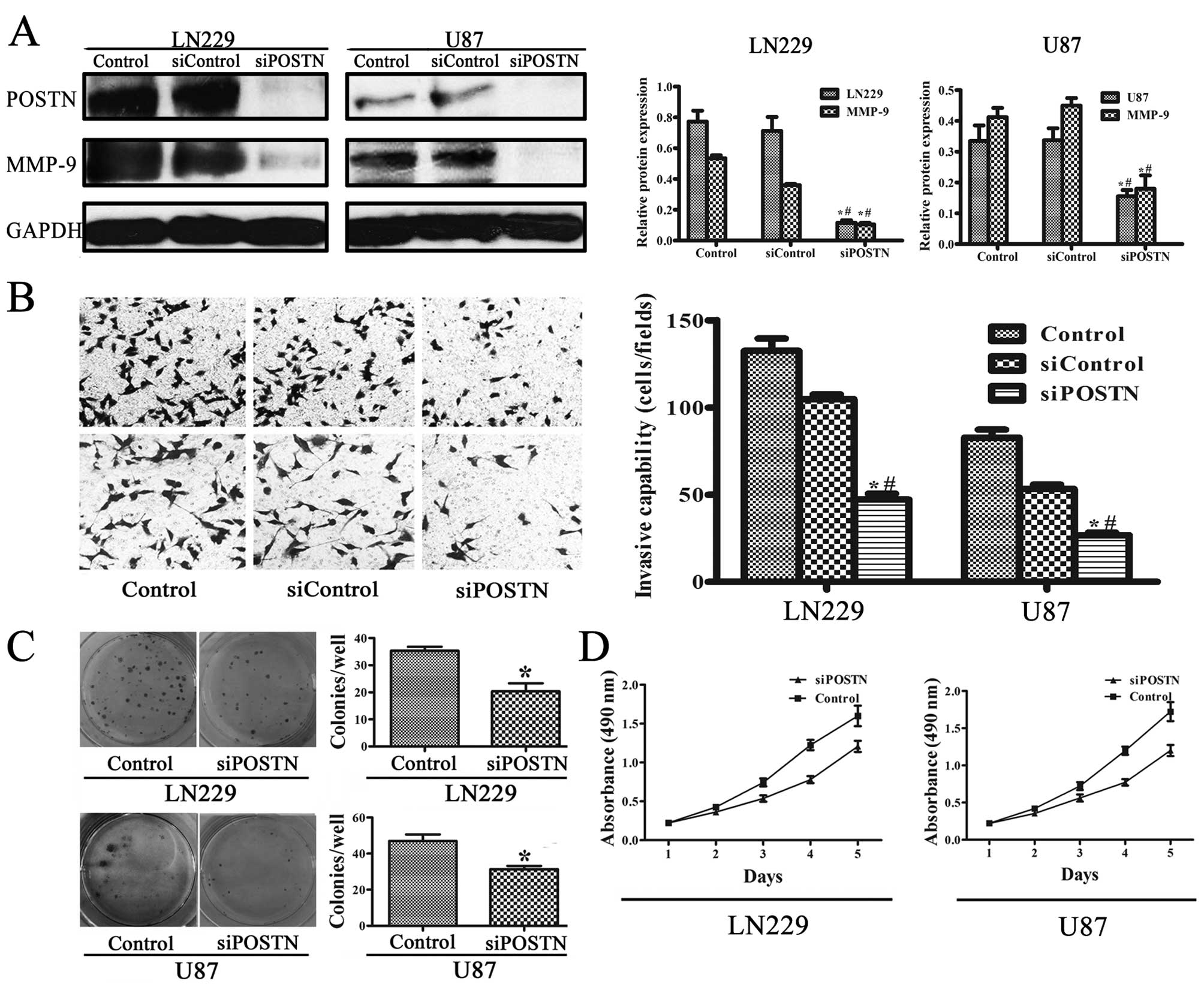
Western blot analysis identified POSTN expression in
LN229 and U87 human glioblastoma cell lines (Fig. 5A). A previous study showed that
siRNA could effectively inhibit POSTN (19). The expression of POSTN in the
control group was ∼6-fold that in the siPOSTN (transfected siRNA)
group in LN229 cells (p<0.001) and ∼2-fold that in U87 cells
(p<0.001). These results indicate that POSTN siRNA inhibited
constitutively activated POSTN. To confirm the effect of POSTN on
the regulation of intracellular molecules involved in aggressive
cell behavior, we examined its influence on MMP-9 expression.
High-POSTN-expressing cells expressed higher levels of MMP-9
compared to siPOSTN cells (Fig.
5A). MMP-9 expression in the control group was ∼5-fold that in
the siPOSTN group in LN229 cells (p<0.001) and ∼2-fold that in
U87 cells (p<0.001).
POSTN promotes invasion and proliferation
of LN229 and U87 cell lines
To explore whether the cell invasion was dependent
on POSTN activity, we measured cell invasion after POSTN knockdown
in LN229 and U87 cell lines. In Fig.
5B, transwell invasion shows that the siRNA significantly
attenuated the effect of POSTN on cell invasion (p<0.05). Colony
formation assays showed that knockdown POSTN in LN229 and U87 cells
led to an decrease in focus numbers (p<0.05, Fig. 5C). Consistent with the colony
formation assay results, MTT assay showed a significant decrease in
proliferation was observed in siPOSTN LN229 and U87 cell lines
compared to cells transfected with control (Fig. 5D), suggesting that POSTN enhances
proliferation of LN229 and U87 cell lines.
Discussion
In the present study, we investigated the expression
level of POSTN in two independent cohorts of almost 300 glioma
patients in total. We further compared the expression levels of
POSTN in low-grade (astrocytoma, oligodendrocytoma and
oligoastrocytoma) and high-grade (anaplastic gliomas and
glioblastomas) human gliomas and demonstrated a significant
increase in POSTN expression from low- to high-grade tumors. POSTN
was an independent prognostic factor predicting PFS and OS in
high-grade gliomas, indicating a significant correlation between
POSTN expression and clinical outcome in glioma patients validated
by 71 independent samples. POSTN functional analyses were performed
in LN229 and U87 cell lines. MMP-9 was an effector of POSTN
signaling in glioma cells. As far as we know, this is the first
report on the expression difference, function and mechanism of
POSTN in all grades of glioma, while previous reports only focus on
glioblastoma (14–16).
Glioma is the most common and diffusely infiltrating
brain tumor, with characteristic invasive patterns including
preferential invasion along white-matter tracts, butterfly lesions
after crossing the corpus callosum, perineuronal satellitosis,
perivascular growth and subpial spread (20). The invasive characteristics of the
most malignant glioma cells probably result from the activation of
a genetic program controlling cell proliferation, remodeling of the
extracellular matrix, cell invasion and migration and the formation
of new blood vessels (21),
allowing these cells to spread throughout the brain. This
invasiveness of glioma cells is a major cause of therapeutic
failure, especially in GBM (2,22,23).
Although most recurrences occur close to the tumor resection site,
distant recurrences >2 cm are also common (24,25),
indicating that the invasive phenotype is acquired early in
tumorigenesis. Gliomas grow invasively and proliferate quickly, so
clarification of the molecular mechanisms, especially those
associated with cellular migration and invasion, is thus crucial to
allow better predictions of glioma patient prognosis and response
to therapies. POSTN has been found to increase aggressive cell
behavior and exogenous expression of POSTN promoted cells to
undergo epithelial-mesenchymal transition (19). Preventing glioma cells from
invading adjacent brain parenchyma and more distant sites is thus a
vital step in improving treatment.
Although POSTN expression has been detected in some
cancers, it is not a general characteristic of tumors. For example,
no POSTN expression was detected in cholangiocarcinoma cell lines,
while there was high expression in fibroblasts (26). In contrast, POSTN was detected in
cancer cells from head and neck, colon and ovary cancers and has
been proposed to induce tumorigenic activity of cancer cells via an
autocrine mechanism (8,12). Another report showed that both
stroma and tumor cells produced POSTN in melanoma, but POSTN was
not expressed in blood cancers (27). Contie et al(4) confirmed that in addition to
secretion, breast cancer cells also captured POSTN from the
surrounding stroma. We detected POSTN expression in tumor cells and
the stroma in gliomas (Fig. 3). It
has been confirmed that fibroblasts can secrete POSTN (3,26),
but there are no fibroblasts in the brain and it is possible that
glioma cells secrete POSTN via an autocrine mechanism, as confirmed
by the results of western blotting in U87 and LN 229 glioma cell
lines (Fig. 5A). POSTN expression
in most parts of gliomas may account for their invasive growth
pattern. In non-small cell lung cancer, POSTN was highly expressed
only at the periphery of the tumor, with no expression within the
tumor tissue (28). In contrast,
POSTN is expressed at both the periphery and within gliomas and has
been detected in both glioma cells and mesenchyme, as verified by
glioma tissue immunohistochemical staining (Fig. 3). These results suggest that POSTN
promotes the invasion of glioma cells and that these changes exist
throughout the glioma tissue, in contrast to tumors with
non-invasive growth patterns.
In the present study, two hundred and twenty samples
with large variability in POSTN transcript levels were subjected to
whole genome gene profiling. We observed a positive correlation of
POSTN expression (Fig. 2) with a
series of tumor genes on invasion and proliferation, including MMP
superfamily and NFκB1 (29,30).
Gene ontology analysis (GO) showed also that the POSTN positive
correlative gene sets related to cell migration and proliferation
were significantly enriched in the cases with POSTN overexpression
(Table I). MMP-9, a member of MMP
superfamily, elevated expression and activity allow tumor cells to
degrade and penetrate extracellular matrix (31,32).
MMP-9 is known to be an important factor in tumor invasion
(33,34) and in addition to the direct effects
of POSTN on cell behavior, knockdown of POSTN was associated with a
subsequent decrease in MMP-9 expression. These results suggest that
POSTN expression may reflect the microenvironment within glioma
tissues and we therefore conclude that POSTN promotes the invasive
ability of glioma cells at least partly via MMP-9, highlighting
MMP-9 as an effector of POSTN signaling in glioma cells (Fig. 5A).
In conclusion, our study demonstrated that POSTN
expression correlated with glioma patient survival, as well as to
glioma grade and is an independent prognostic factor in high-grade
glioma patients. The results of this study demonstrate the
important functional and molecular mechanisms of POSTN in tumor
invasion and proliferation. POSTN promoted glioma cell invasiveness
in vitro, accompanied by MMP-9 expression. The correlation
between POSTN expression and increased glioma invasion and
proliferation suggest that it could be used as a prognostic
biomarker for poor outcome and it may also represent a future
therapeutic target.
Acknowledgements
We would like to thank Dr Susan
Furness for her critical reading and Professor Chen (Beijing Sanbo
Brain Hospital) for IHC technical support. This study was supported
by grants National Key Project of Science and Technology Supporting
Programs of China (no. 2007BAI05B08) and National High Technology
Research and Development Program (2012AA02A508). Additional support
was provided through grant International Science and Technology
Cooperation Program (2012DFA30470).
References
|
1.
|
Zhang J, Sarkar S and Yong VW: The
chemokine stromal cell derived factor-1 (CXCL12) promotes glioma
invasiveness through MT2-matrix metalloproteinase. Carcinogenesis.
26:2069–2077. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Wang Y, Chen L, Bao Z, et al: Inhibition
of STAT3 reverses alkylator resistance through modulation of the
AKT and beta-catenin signaling pathways. Oncol Rep. 26:1173–1180.
2011.PubMed/NCBI
|
|
3.
|
Malanchi I, Santamaria-Martinez A, Susanto
E, et al: Interactions between cancer stem cells and their niche
govern metastatic colonization. Nature. 481:85–89. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Contie S, Voorzanger-Rousselot N, Litvin
J, Clezardin P and Garnero P: Increased expression and serum levels
of the stromal cell-secreted protein periostin in breast cancer
bone metastases. Int J Cancer. 128:352–360. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Sasaki H, Yu CY, Dai M, et al: Elevated
serum periostin levels in patients with bone metastases from breast
but not lung cancer. Breast Cancer Res Treat. 77:245–252. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Ben QW, Jin XL, Liu J, Cai X, Yuan F and
Yuan YZ: Periostin, a matrix specific protein, is associated with
proliferation and invasion of pancreatic cancer. Oncol Rep.
25:709–716. 2011.PubMed/NCBI
|
|
7.
|
Ruan K, Bao S and Ouyang G: The
multifaceted role of periostin in tumorigenesis. Cell Mol Life Sci.
66:2219–2230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Kudo Y, Ogawa I, Kitajima S, et al:
Periostin promotes invasion and anchorage-independent growth in the
metastatic process of head and neck cancer. Cancer Res.
66:6928–6935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Siriwardena BS, Kudo Y, Ogawa I, et al:
Periostin is frequently overexpressed and enhances invasion and
angiogenesis in oral cancer. Br J Cancer. 95:1396–1403. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Erkan M, Kleeff J, Gorbachevski A, et al:
Periostin creates a tumor-supportive microenvironment in the
pancreas by sustaining fibrogenic stellate cell activity.
Gastroenterology. 132:1447–1464. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Tai IT, Dai M and Chen LB: Periostin
induction in tumor cell line explants and inhibition of in vitro
cell growth by anti-periostin antibodies. Carcinogenesis.
26:908–915. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Gillan L, Matei D, Fishman DA, Gerbin CS,
Karlan BY and Chang DD: Periostin secreted by epithelial ovarian
carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5)
integrins and promotes cell motility. Cancer Res. 62:5358–5364.
2002.PubMed/NCBI
|
|
13.
|
Shao R, Bao S, Bai X, et al: Acquired
expression of periostin by human breast cancers promotes tumor
angiogenesis through up-regulation of vascular endothelial growth
factor receptor 2 expression. Mol Cell Biol. 24:3992–4003. 2004.
View Article : Google Scholar
|
|
14.
|
Zinn PO, Mahajan B, Sathyan P, et al:
Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in
glioblastoma multiforme. PLoS One. 6:e254512011. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Gottlieb A, Varshavsky R, Linial M and
Horn D: UFFizi: a generic platform for ranking informative
features. BMC Bioinformatics. 11:3002010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Mikheeva SA, Mikheev AM, Petit A, et al:
TWIST1 promotes invasion through mesenchymal change in human
glioblastoma. Mol Cancer. 9:1942010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
19.
|
Yan W and Shao R: Transduction of a
mesenchyme-specific gene periostin into 293T cells induces cell
invasive activity through epithelial-mesenchymal transformation. J
Biol Chem. 281:19700–19708. 2006. View Article : Google Scholar
|
|
20.
|
Claes A, Idema AJ and Wesseling P: Diffuse
glioma growth: a guerilla war. Acta Neuropathol. 114:443–458. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Dauer DJ, Ferraro B, Song L, et al: Stat3
regulates genes common to both wound healing and cancer. Oncogene.
24:3397–3408. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Senft C, Priester M, Polacin M, et al:
Inhibition of the JAK-2/STAT3 signaling pathway impedes the
migratory and invasive potential of human glioblastoma cells. J
Neurooncol. 101:393–403. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Stupp R, Mason WP, van den Bent MJ, et al:
Radiotherapy plus concomitant and adjuvant temozolomide for
glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Giese A, Bjerkvig R, Berens ME and
Westphal M: Cost of migration: invasion of malignant gliomas and
implications for treatment. J Clin Oncol. 21:1624–1636. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Wick W, Stupp R, Beule AC, et al: A novel
tool to analyze MRI recurrence patterns in glioblastoma.
Neurooncology. 10:1019–1024. 2008.PubMed/NCBI
|
|
26.
|
Utispan K, Thuwajit P, Abiko Y, et al:
Gene expression profiling of cholangiocarcinoma-derived fibroblast
reveals alterations related to tumor progression and indicates
periostin as a poor prognostic marker. Mol Cancer. 9:132010.
View Article : Google Scholar
|
|
27.
|
Tilman G, Mattiussi M, Brasseur F, van
Baren N and Decottignies A: Human periostin gene expression in
normal tissues, tumors and melanoma: evidences for periostin
production by both stromal and melanoma cells. Mol Cancer.
6:802007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Sasaki H, Dai M, Auclair D, et al: Serum
level of the periostin, a homologue of an insect cell adhesion
molecule, as a prognostic marker in nonsmall cell lung carcinomas.
Cancer. 92:843–848. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Park MH and Min do S: Quercetin-induced
downregulation of phospholipase D1 inhibits proliferation and
invasion in U87 glioma cells. Biochem Biophys Res Commun.
412:710–715. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Mut M, Amos S and Hussaini IM: PKC alpha
phosphorylates cytosolic NF-kappaB/p65 and PKC delta delays nuclear
translocation of NF-kappaB/p65 in U1242 glioblastoma cells. Turk
Neurosurg. 20:277–285. 2010.PubMed/NCBI
|
|
31.
|
Lakka SS, Gondi CS, Yanamandra N, et al:
Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma
cell line via RNA interference reduces tumor cell invasion, tumor
growth and angiogenesis. Oncogene. 23:4681–4689. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Kupferman ME, Fini ME, Muller WJ, Weber R,
Cheng Y and Muschel RJ: Matrix metalloproteinase 9 promoter
activity is induced coincident with invasion during tumor
progression. Am J Pathol. 157:1777–1783. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Kang CS, Pu PY, Li YH, et al: An in vitro
study on the suppressive effect of glioma cell growth induced by
plasmid-based small interference RNA (siRNA) targeting human
epidermal growth factor receptor. J Neurooncol. 74:267–273. 2005.
View Article : Google Scholar
|
|
34.
|
Friedberg MH, Glantz MJ, Klempner MS, Cole
BF and Perides G: Specific matrix metalloproteinase profiles in the
cerebrospinal fluid correlated with the presence of malignant
astrocytomas, brain metastases and carcinomatous meningitis.
Cancer. 82:923–930. 1998. View Article : Google Scholar
|