Introduction
The C4.4A protein was initially found in a
metastatic rat pancreatic adenocarcinoma cell line (1,2). Rat
C4.4A cDNA was cloned and the glycosylphosphatidyl inositol
(GPI)-anchored membrane protein was found to have 30% homology to
the urokinase-type plasminogen activator receptor (3). The human homologue of rat C4.4A,
located on chromosome 19q13.1–q13.2, was subsequently cloned
(4). The C4.4A mRNA is present in
normal human placental tissue, skin, esophagus tissue and
leukocytes (4). Although the
physiological function of the C4.4A protein is largely unknown,
upregulation of C4.4A expression has been observed during the
wound-healing process of migrating keratinocytes and in the
urothelium (5,6).
We demonstrated that C4.4A expression at the
invasive front of colorectal cancer predicted disease recurrence
and the C4.4A expression was associated with tumor budding and EMT
change (7–9). Very recently we have shown that C4.4A
expression is associated with a poor prognosis of esophageal
squamous cell carcinoma (ESCC) (10). To investigate the mechanism of how
C4.4A influences the prognosis of ESCC, we performed PCR-array
loading in various extracellular matrix proteins and cell adhesion
molecules. The result indicated that C4.4A expression correlated
well with Tenascin-C (TNC) expression. TNC is an extracellular
matrix protein secreted from both tumor cells and myofibroblasts.
Strong expression of TNC occurs during development, starting at
gastrulation. In contrast, TNC is absent or greatly reduced in most
adult tissue, but it increased in some pathological conditions,
inclucing inflammation, wound healing and in a variety of
neoplasias. In many tumors, TNC expression correlated with
invasiveness and malignancy (11–19).
In this study, we attempted to examine the clinical relevance of
the TNC in relation to C4.4A expression in ESCC.
Materials and methods
Clinical tissue samples
Esophageal tissue samples (n=111) were collected
during surgery (1998–2007) at the Department of Surgery, Osaka
University (Osaka, Japan). Chemoradiotherapy for advanced
esophageal cancer is commonly used in Western countries (20,21)
and neoadjuvant chemo-therapy is a standard therapy before surgery
in stage II and III ESCC patients in Japan (22). Therefore, we collected samples from
previous ESCC patients (1998–2007) who did not receive preoperative
radiotherapy and/or chemotherapy, since our institute usually
employs preoperative chemotherapy using FAP (5-FU, adriamycin,
CDDP) or DCF (docetaxel, CDDP, 5-FU) in recent ESCC cases. Samples
were fixed in buffered formalin at 4°C overnight, processed through
graded ethanol solutions and embedded in paraffin. A piece of
tissue sample was frozen in liquid nitrogen and stored at −80°C
until protein extraction. The specimens were used appropriately and
under the approval of the ethics committee at the Graduate School
of Medicine, Osaka University.
Cell culture
The human colon cancer cell line HCT116 was obtained
from the American Type Culture Collection (Manassas, VA, USA). The
human esophageal cancer cell line TE8 was obtained from Tohoku
University (Miyagi, Japan). These cells were grown in DMEM, or
RPMI, respectively, supplemented with 10% fetal bovine serum (FBS),
100 U/ml penicillin and 100 μg/ml streptomycin, at 37°C in a
humidified incubator with 5% CO2 in the air.
Collagen gel culture
Collagen gel cultures were carried out using a
collagen gel kit (Nitta Gelatin, Osaka, Japan), according to the
manufacturer’s protocol. One milliliter of collagen solution,
containing 5×105 cells was overlaid on a pre-prepared
basal collagen layer in a 6-well dish and incubated at 37°C for 30
min. RPMI supplemented with 10% FBS was added after gelatinization.
The culture medium was changed every day.
siRNA for C4.4A
For small interfering RNA (siRNA) inhibition, the
following double-stranded RNA duplexes targeting human C4.4A were
used: 5′-GCUGUAACUCUGACCUCCG
CAACAA-3′/5′-UUGUUGCGGAGGUCAGAGUUACAGC-3′ (siRNA I, HSS120351,
Invitrogen) and 5′-CAACGUCACCU
UGACGGCAGCUAAU-3′/5′-AUUAGCUGCCGUCAAGGUG ACGUUG-3′ (siRNA II,
HSS178302, Invitrogen). Negative-control siRNAs were purchased in a
Stealth RNAi kit (Invitrogen). Cells were transfected with siRNA
using Lipofectamine™ RNAiMAX (Invitrogen) according to the
manufacturer’s protocols.
PCR-array
Total RNA was extracted using the RNAeasy mini kit
(Qiagen-Sample & Assay Technologies, Hilden, Germany). The ABI
PRISM 7500 Sequence Detector (Applied Biosystems, Foster City, CA,
USA) was used for PCR. RT2PCR Array loading
extracellular matrix proteins and cell adhesion molecules (code
#PAHS-013A-2, Table I) were
employed to analyze each sample (SABioscience, Frederick, MD, USA).
For each plate, results were normalized to the median value of a
set of housekeeping genes. A significant threshold of a 2-fold
change in gene expression corresponded to P<0.001. HCT116 cell
samples were prepared from the following 5 groups: i) parental
cells in 2D culture for 48 h, ii) parental cells in 3D type
I-collagen gel culture for 48 h, iii) 3D cultures for 48 h after
24-h treatment with C4.4A-siRNA I, iv) 3D cultures for 48 h after
24-h treatment with C4.4A-siRNA II and v) 3D cultures for 48 h
after 24-h treatment with negative control-siRNA.
 | Table I.The molecules mounted on PCR
array. |
Table I.
The molecules mounted on PCR
array.
| Cell adhesion
molecules |
| Transmembrane
molecules: | CD44, CDH1, HAS1,
ICAM1, ITGA1, ITGA2, ITGA3, ITGA4, ITGA5, ITGA6, ITGA7, |
| ITGA8, ITGAL, ITGAM,
ITGAV, ITGB1, ITGB2, ITGB3, ITGB4, ITGB5, MMP14, |
| MMP15, MMP16, NCAM1,
PECAM1, SELE, SELL, SELP, SGCE, SPG7, VCAM1 |
| Cell-cell
adhesion: | CD44, CDH1, COL11A1,
COL14A1, COL6A2, CTNND1, ICAM1, ITGA8, VCAM1 |
| Cell-matrix
adhesion: | ADAMTS13, CD44,
ITGA1, ITGA2, ITGA3, ITGA4, ITGA5, ITGA6, ITGA7, ITGA8, |
| ITGAL, ITGAM, ITGAV,
ITGB1, ITGB2, ITGB3, ITGB4, ITGB5, SGCE, SPP1, THBS3 |
| Other adhesion
molecules: | CNTN1, COL12A1,
COL15A1, COL16A1, COL5A1, COL6A1, COL7A1, COL8A1, |
| VCAN, CTGF, CTNNA1,
CTNNB1, CTNND2, FN1, KAL1, LAMA1, LAMA2, |
| LAMA3, LAMB1, LAMB3,
LAMC1, THBS1, THBS2, CLEC3B, TNC, VTN |
| Extracellular matrix
proteins |
| Basement membrane
constituents: | COL4A2, COL7A1,
LAMA1, LAMA2, LAMA3, LAMB1, LAMB3, LAMC1, SPARC |
| Collagens and ECM
structural: | COL11A1, COL12A1,
COL14A1, COL15A1, COL16A1, COL1A1, COL4A2, COL5A1, |
| constituents | COL6A1, COL6A2,
COL7A1, COL8A1, FN1, KAL1 |
| ECM
proteases: | ADAMTS1, ADAMTS13,
ADAMTS8, MMP1, MMP10, MMP11, MMP12, MMP13, |
| MMP14, MMP15,
MMP16, MMP2, MMP3, MMP7, MMP8, MMP9, SPG7, TIMP1 |
| ECM protease
inhibitors: | COL7A1, KAL1,
THBS1, TIMP1, TIMP2, TIMP3 |
| Other ECM
molecules: | VCAN, CTGF, ECM1,
HAS1, SPP1, TGFBI, THBS2, THBS3, CLEC3B, TNC, VTN |
Quantitative real-time PCR
cDNA was generated from 1 μg total RNA using
the high capacity RNA-to-cDNA kit (Applied Biosystems).
Quantitative real-time PCR was carried out using the LightCycler
(Idaho Technology, ID, USA) as described previously (23). Quantification data from each sample
were analyzed using the LightCycler analysis software.
The C4.4A primer sequences were 5′-TCACCTTGACG
GCAGCTA-3′, C4.4A sense and 5′-AGCCACTGAGCGTGA ACC-3′, C4.4A
antisense; and the probe was UPL No. 58 (Roche). The TNC primer
sequences were 5′-CTGAAGGTGG AGGGGTACAG-3′, TNC sense and
5′-AGAAGGATCTGCC ATTGTGG-3′, TNC antisense; and the probe was UPL
No. 56 (Roche). The amount of each transcript was normalized
against the expression of the housekeeping gene,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), from the same
sample using the primers 5′-AGCCACATCGCTCAGACAC-3′, sense and
5′-GCCCAATACGACCAAATCC-3′, antisense; and the probe UPL No. 60
(Roche).
Immunohistochemistry
A rabbit anti-human C4.4A GPI-related polyclonal
antibody (7–10) hereafter designated as the C4.4A-GPI
antibody, was used. The anti-human TNC mouse monoclonal antibody
(ab6393) was purchased from Abcam (Cambridge, UK). Tissue sections
(4 μm thick) were prepared from paraffin-embedded blocks.
After antigen retrieval treatment in 10 mM citrate buffer (pH 6.0)
at 95°C for 40 min, immunostaining was carried out using the
Vectastain ABC peroxidase kit (Vector Laboratories, Burlingame, CA,
USA), as we described previously (24,25).
The slides were incubated with appropriate antibodies overnight at
4°C at the following dilutions: C4.4A-GPI, 1:300 and TNC, 1:4,000.
Non-immunized rabbit IgG or mouse IgG (Vector Laboratories) was
used as a negative control and substituted for the primary antibody
to exclude possible false-positive responses from the secondary
antibody or from non-specific binding of IgG.
Transfection of plasmids
We stably transduced the C4.4A plasmid into HCT116
and TE8 cells with the lentivirus. Plasmids with the human C4.4A
NM_014400 (Origene Inc., Rockville, MD, USA) were transfected with
the pLenti6/ V5 Directional TOPO® Cloning kit
(Invitrogen). An empty pLenti6/V5 vector was used as a mock
control.
Statistical analysis
Statistical analysis was carried out using the JMP8
program (SAS Institute, Cary, NC, USA). The Kaplan-Meier method was
used to estimate tumor recurrence from CRC and the log-rank test
was used to determine the statistical significance. Associations
between discrete variables were assessed using the χ2
test. Mean values were compared using the Mann-Whitney U test.
P-values <0.05 were considered statistically significant.
Results
Molecular linkage of C4.4A to Tenascin-C
in 3D collagen cultures
To investigate a molecular linkage of C4.4A to
certain molecules, especially cell adhesion molecules and
extracellular matrix proteins, we used a custom PCR array analysis
(Table I). When C4.4A-positive
control HCT116 cells were cultured, C4.4A mRNA levels increased
2.6-fold at 48 h in 3D cultures when compared to 2D cultures (data
not shown). Thus, we searched the gene sets that were increased in
3D cultures and yet decreased when these cultures were treated with
C4.4A siRNA (siRNA I, siRNA II). We then focused on TNC, which
ranked at the top of the list (Table
II). Confirmation studies using qRT-PCR assays showed that
C4.4A siRNA treatments significantly reduced TNC mRNA in HCT116
cells in 3D cultures (Fig. 1A,
P<0.05). By contrast, lentivirus-mediated forced expression of
C4.4A in 3D cultures significantly enhanced TNC mRNA levels in
HCT116 (Fig. 1B, P<0.05) and
ESCC TE8 cells (Fig. 1C,
P<0.05).
 | Table II.Top 15 gene lists by analysis with
PCR array. |
Table II.
Top 15 gene lists by analysis with
PCR array.
| 3D:2D | siRNA I:neg
conta | siRNA II:neg
conta |
|---|
| TNC | 3.19 | 0.14 | 0.18 |
| HAS1 | 3.15 | 0.14 | 0.07 |
| MMP13 | 2.54 | 0.33 | 0.44 |
| ITGAM | 2.26 | 1.83 | 0.96 |
| ECM1 | 1.97 | 0.22 | 0.67 |
| MMP16 | 1.88 | 1.1 | 0.87 |
| ITGA4 | 1.87 | 0.63 | 0.8 |
| MMP10 | 1.83 | 0.15 | 0.90 |
| LAMA3 | 1.78 | 0.50 | 0.99 |
| MMP1 | 1.77 | 1.15 | 0.96 |
| COL4A2 | 1.73 | 1.87 | 0.42 |
| COL12A1 | 1.64 | 0.43 | 0.35 |
| NCAM1 | 1.52 | 0.76 | 0.96 |
| ADAMTS1 | 1.51 | 1.22 | 0.07 |
| ITGA8 | 1.50 | 0.69 | 0.54 |
Tenascin-C expression in ESCC tissue
samples
Based on the molecular linkage between C4.4A and TNC
in vitro, we examined the TNC protein expression in tumor
cells of ESCC by immunohistochemistry. We found 27 of 111 ESCCs
(24.3%) expressed TNC in the cytoplasm of the tumor cells (Fig. 2). When TNC-positive cases (n=27)
and TNC-negative cases (n=84) were compared for the various
clinical and pathological parameters including age, sex, histology,
T stage, lymphatic invasion, venous invasion, lymph node metastasis
and distant metastasis, TNC expression was associated with deeper
wall invasion (Table III,
P=0.028).
 | Table III.TNC expression and
clinicopathological parameters in esophagel SCC patients. |
Table III.
TNC expression and
clinicopathological parameters in esophagel SCC patients.
| TNC-positive
(n=27) | TNC negative
(n=84) | P-value |
|---|
| Age | 38–84 (median
64.7) | 48–80 (median
64.0) | NSa |
| Gender | | | |
| Male | 23 | 77 | NS |
| Female | 4 | 7 | |
| Histologyb | | | |
| Well, Mod | 24 | 60 | NS |
| Por | 3 | 24 | |
| T
classificationc | | | |
| T1, T2 | 10 | 52 | 0.028 |
| T3, T4 | 17 | 32 | |
| Lymphatic
invasion | | | |
| Positive | 23 | 63 | NS |
| Negative | 4 | 21 | |
| Venous
invasion | | | |
| Positive | 14 | 29 | NS |
| Negative | 13 | 55 | |
| Lymph node
metastasis | | | |
| Positive | 22 | 53 | NS |
| Negative | 5 | 31 | |
| Distant
metastasis | | | |
| Positive | 2 | 8 | NS |
| Negative | 25 | 76 | |
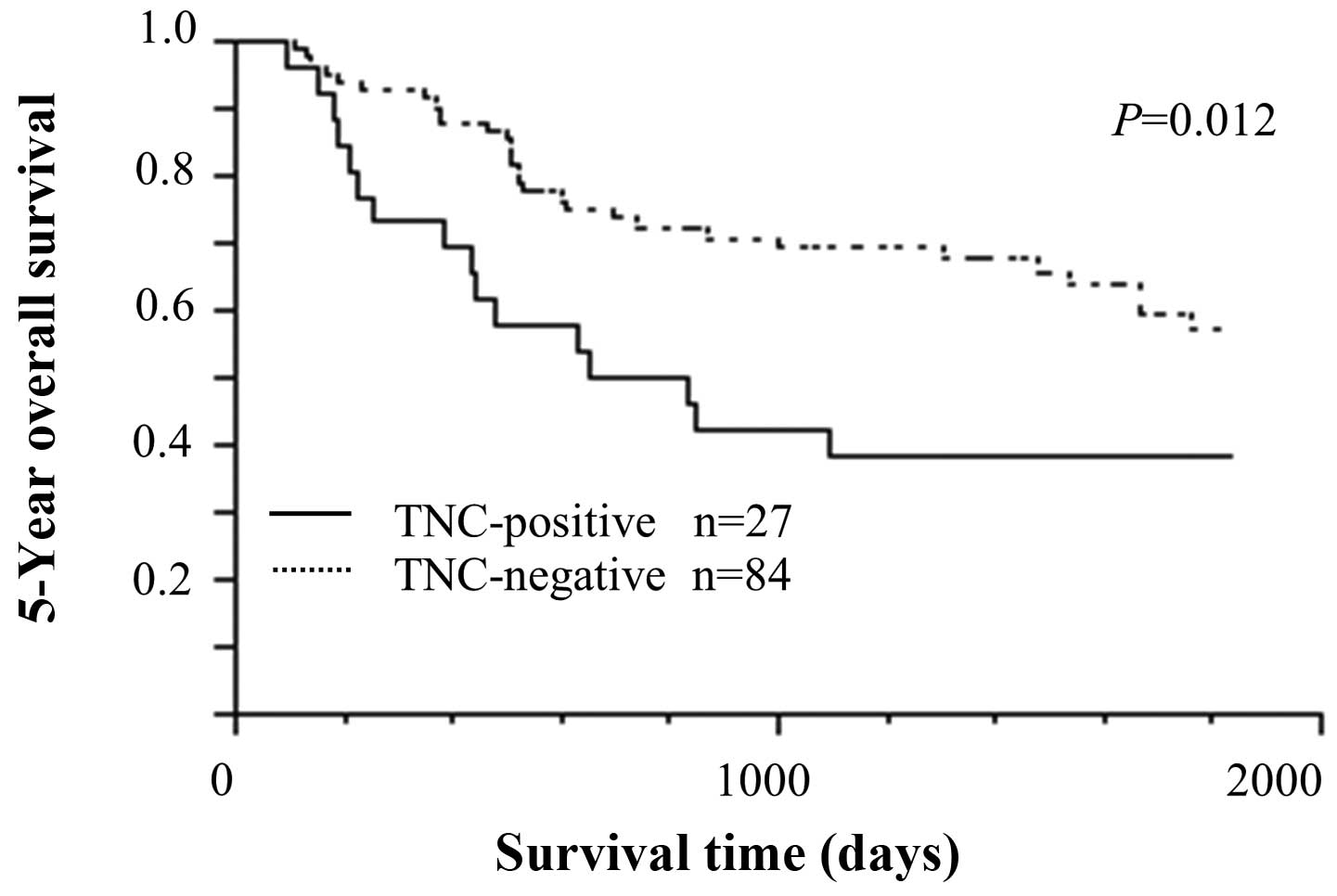
Survival analysis by TNC expression
Survival curves were drawn between TNC-positive and
TNC-negative ESCCs. The TNC-positive group had significantly poorer
prognosis than the TNC-negative group in 5-year OS (Fig. 3, P=0.012). The univariate analysis
indicated that TNC expression, tumor stage, lymph node metastasis
and venous invasion were significant predictors of a poor 5-year OS
(Table IV). We carried out a
multivariate analysis to further determine the most significant
prognostic factors. Lymph node metastasis was identified as an
independent prognostic factor (P=0.045). TNC expression was not an
independent prognostic factor (Table
IV).
 | Table IV.Five-year overall survival in ESCC
patients (n=111). |
Table IV.
Five-year overall survival in ESCC
patients (n=111).
| A, Univariate
analysis | Risk ratio | Confidence
interval | P-value |
|
| TNC (positive vs
negative) | 2.139 | 1.137–3.876 | 0.019 |
| Age (>65 vs
<65 years) | 1.285 | 0.717–2.300 | NS |
| Sex (male vs
female) | 1.690 | 0.616–6.977 | NS |
| T stage (T1/T2 vs
T3/T4) | 2.769 | 1.540–5.080 | <0.001 |
| Differentiation
(well/mod vs por) | 1.696 | 0.875–3.119 | NS |
| Lymph node
metastasis (positive vs negative) | 3.351 | 1.640–7.770 | <0.001 |
| Distant metastasis
(positive vs negative) | 1.857 | 0.759–3.906 | NS |
| Lymph invasion
(positive vs negative) | 2.060 | 0.980–5.043 | NS |
| Venous invasion
(positive vs negative) | 2.104 | 1.175–3.787 | 0.013 |
|
| B, Multivariate
analysis | Risk ratio | Confidence
interval | P-value |
|
| TNC (positive vs
negative) | 1.786 | 0.943–3.251 | NS |
| T stage (T1/T2 vs
T3/T4) | 1.846 | 0.988–3.545 | NS |
| Lymph node
metastasis (positive vs negative) | 2.172 | 1.015–5.323 | 0.045 |
| Venous invasion
(positive vs negative) | 1.294 | 0.807–2.691 | NS |
C4.4A expression in ESCC by the C4.4A-GPI
antibody
C4.4A protein expression was shown in the normal
epithelium of the esophagus by IHC using the C4.4A-GPI antibody.
Expression was on the plasma membrane, mainly at the parabasal
layer (Fig. 4A). In contrast,
tumor cells in the same tissue sample did not always express the
C4.4A protein (Fig. 4A). We
defined the esophageal carcinoma tissues as C4.4A-negative if
staining was not noted at all in the tumor cells on the tissue
sections. There were 45 C4.4A-negative esophageal tumors in the 111
cases tested (40.5%); while, 66 esophageal tumors (59.5%) provided
clear C4.4A staining on the plasma membrane (Fig. 4B) or cytoplasm (Fig. 4C). Sixty-six positive cases were
classified according to intracellular localization, i.e., 29
membrane staining alone, 11 cytoplasmic staining alone and 26 both
membrane and cytoplasmic staining. When expression of C4.4A and TNC
was compared, TNC expression was significantly associated with
C4.4A expression (Fig. 5,
P=0.007).
Furthermore, sub-group analysis revealed that only
C4.4A-positive/TNC-positive cases had a significantly worse
prognosis when compared to C4.4A-negative/TNC-negative cases
(Fig. 6, P=0.005).
Discussion
Esophageal cancer is the eighth most common cancer
worldwide and the sixth most common cause of death from cancer
(26). In Asian countries
esophageal squamous cell carcinoma (ESCC) is more prevalent than
adenocarcinoma and accounts for >90% of esophageal carcinomas.
ESCC is usually diagnosed at an advanced stage, which leads to a
5-year survival of only 10% (27).
To improve the unfavorable outcome of ESCC, it is essential to
explore the molecular basis of the underlying mechanism of this
disease.
HCT116 colon cancer cells represent a positive
control for the C4.4A protein (7,8). We
previously observed that the C4.4A protein was located in the
cytoplasm of HCT116 cells on collagen gels, but it was translocated
onto the plasma membrane when HCT116 cells were cultured in the
collagen matrix (7). We also found
that C4.4A mRNA levels increased 2.6-fold in 3D collagen cultures
(data not shown). Based on these findings, we hypothesized that
membranous C4.4A might exert certain roles when cells are grown in
the 3D collagen matrix. To examine this possibility, we performed a
PCR array analysis using HCT116 cell cultures grown for 48 h in
3D-collagen matrix after C4.4A-siRNA treatment. TNC was one of the
notable genes that increased in the 3D condition compared to the 2D
culture, but was reduced when C4.4A was knocked down in the 3D
condition.
TNC is an extracellular matrix protein composed of
six monomers linked at their N-termini with disulfide bounds to
form a 1080-1500-kDa hexamer and various solid tumors express high
level of TNC (12–19,28,29).
TNC is secreted from both tumor cells and myofibroblasts. The most
prominent effects of TNC are anti-adhesion and inhabitation of cell
attachment, both of which favor cancer cell motility and invasion
(30,31). Furthermore, TNC promotes malignant
transformation, uncontrolled proliferation, metastasis,
angiogenesis, drug resistance and escape from tumor
immunosurveillance (15,32,33).
Knockdown experiments for TNC in glioblastomas and breast cancer
cells caused inhibition of tumor growth, migration, invasion and
metastasis (12,18,34).
Recently, we have shown that C4.4A expression is
associated with a poor prognosis of ESCC (10), but the underlying mechanism of how
C4.4A influences the prognosis of ESCC remains unknown. Based on
the in vitro observation that TNC levels increased in
parallel with lentivirus-mediated introduction of the C4.4A gene in
TE8 esophageal cancer cells, we examined TNC and C4.4A expression
in 111 ESCCs. As results, we found that TNC expression was
significantly associated with C4.4A expression in clinical ESCC
samples (Fig. 5, P=0.007),
suggesting that there may be a functional role for the C4.4A to
induce TNC in vivo. Among the C4.4A positive ESCCs, only
double positive C4.4A and TNC group had poorer prognosis (Fig. 6).
In conclusion, we revealed for the first time that
the TNC-positive group (24.3%) had significantly poorer prognosis
than the TNC-negative group in 5-year OS. It is also postulated
that TNC may partly account for the C4.4A-related unfavorable
outcome in ESCC patients.
Acknowledgements
This study was supported by a Grant-in
Aid for Cancer Research from the Ministry of Education, Science,
Sports and Culture Technology, Japan, to H.Y. (grant no.
21390360).
References
|
1.
|
Matzku S, Wenzel A, Liu S and Zoller M:
Antigenic differences between metastatic and nonmetastatic BSp73
rat tumor variants characterized by monoclonal antibodies. Cancer
Res. 49:1294–1299. 1989.PubMed/NCBI
|
|
2.
|
Claas C, Herrmann K, Matzku S, Moller P
and Zoller M: Developmentally regulated expression of
metastasis-associated antigens in the rat. Cell Growth Differ.
7:663–678. 1996.PubMed/NCBI
|
|
3.
|
Rosel M, Claas C, Seiter S, Herlevsen M
and Zoller M: Cloning and functional characterization of a new
phosphatidyl-inositol anchored molecule of a metastasizing rat
pancreatic tumor. Oncogene. 17:1989–2002. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Wurfel J, Seiter S, Stassar M, et al:
Cloning of the human homologue of the metastasis-associated rat
C4.4A. Gene. 262:35–41. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Smith BA, Kennedy WJ, Harnden P, Selby PJ,
Trejdosiewicz LK and Southgate J: Identification of genes involved
in human urothelial cell-matrix interactions: implications for the
progression pathways of malignant urothelium. Cancer Res.
61:1678–1685. 2001.PubMed/NCBI
|
|
6.
|
Hansen LV, Gardsvoll H, Nielsen BS, et al:
Structural analysis and tissue localization of human C4.4A: a
protein homologue of the urokinase receptor. Biochem J.
380:845–857. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Konishi K, Yamamoto H, Mimori K, et al:
Expression of C4.4A at the invasive front is a novel prognostic
marker for disease recurrence of colorectal cancer. Cancer Sci.
101:2269–2277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Oshiro R, Yamamoto H, Takahashi H, et al:
C4.4A is associated with tumor budding and epithelial-mesenchymal
transition of colorectal cancer. Cancer Sci. 103:1155–1164. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Yamamoto H, Oshiro R, Ohtsuka M, et al:
Distinct expression of C4.4A in colorectal cancer detected by
different antibodies. Int J Oncol. 42:197–201. 2013.
|
|
10.
|
Ohtsuka M, Yamamoto H, Masuzawa T, et al:
C4.4A is associated with a poor prognosis of esophageal squamous
cell carcinoma. Ann Surg Oncol. Feb 24–2013.(Epub ahead of
print).
|
|
11.
|
Beiter K, Hiendlmeyer E, Brabletz T, et
al: beta-catenin regulates the expression of tenascin-C in human
colorectal tumors. Oncogene. 24:8200–8204. 2005.PubMed/NCBI
|
|
12.
|
Oskarsson T, Acharyya S, Zhang XH, et al:
Breast cancer cells produce tenascin C as a metastatic niche
component to colonize the lungs. Nat Med. 17:867–874. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Nagaharu K, Zhang X, Yoshida T, et al:
Tenascin C induces epithelial-mesenchymal transition-like change
accompanied by SRC activation and focal adhesion kinase
phosphorylation in human breast cancer cells. Am J Pathol.
178:754–763. 2011. View Article : Google Scholar
|
|
14.
|
O’Connell JT, Sugimoto H, Cooke VG, et al:
VEGF-A and Tenascin-C produced by S100A4+ stromal cells
are important for metastatic colonization. Proc Natl Acad Sci USA.
108:16002–16007. 2011.
|
|
15.
|
Orend G and Chiquet-Ehrismann R:
Tenascin-C induced signaling in cancer. Cancer Lett. 244:143–163.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
De Wever O, Nguyen QD, Van Hoorde L, et
al: Tenascin-C and SF/HGF produced by myofibroblasts in vitro
provide convergent pro-invasive signals to human colon cancer cells
through RhoA and Rac. FASEB J. 18:1016–1018. 2004.PubMed/NCBI
|
|
17.
|
Sarkar S, Nuttall RK, Liu S, Edwards DR
and Yong VW: Tenascin-C stimulates glioma cell invasion through
matrix metalloproteinase-12. Cancer Res. 66:11771–11780. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Hirata E, Arakawa Y, Shirahata M, et al:
Endogenous tenascin-C enhances glioblastoma invasion with reactive
change of surrounding brain tissue. Cancer Sci. 100:1451–1459.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Fukunaga-Kalabis M, Martinez G, Nguyen TK,
et al: Tenascin-C promotes melanoma progression by maintaining the
ABCB5-positive side population. Oncogene. 29:6115–6124. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Sjoquist KM, Burmeister BH, Smithers BM,
et al: Survival after neoadjuvant chemotherapy or chemoradiotherapy
for resectable oesophageal carcinoma: an updated meta-analysis.
Lancet Oncol. 12:681–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Stahl M, Walz MK, Stuschke M, et al: Phase
III comparison of preoperative chemotherapy compared with
chemoradiotherapy in patients with locally advanced adenocarcinoma
of the esophagogastric junction. J Clin Oncol. 27:851–856. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Ando N, Kato H, Igaki H, et al: A
randomized trial comparing postoperative adjuvant chemotherapy with
cisplatin and 5-fluorouracil versus preoperative chemotherapy for
localized advanced squamous cell carcinoma of the thoracic
esophagus (JCOG9907). Ann Surg Oncol. 19:68–74. 2012. View Article : Google Scholar
|
|
23.
|
Yamamoto H, Kondo M, Nakamori S, et al:
JTE-522, a cyclooxygenase-2 inhibitor, is an effective
chemopreventive agent against rat experimental liver fibrosis1.
Gastroenterology. 125:556–571. 2003.PubMed/NCBI
|
|
24.
|
Hayashi N, Yamamoto H, Hiraoka N, et al:
Differential expression of cyclooxygenase-2 (COX-2) in human bile
duct epithelial cells and bile duct neoplasm. Hepatology.
34:638–650. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Noura S, Yamamoto H, Ohnishi T, et al:
Comparative detection of lymph node micrometastases of stage II
colorectal cancer by reverse transcriptase polymerase chain
reaction and immunohistochemistry. J Clin Oncol. 20:4232–4241.
2002. View Article : Google Scholar
|
|
26.
|
Ferlay J, Shin H-R, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Holmes RS and Vaughan TL: Epidemiology and
pathogenesis of esophageal cancer. Semin Radiat Oncol. 17:2–9.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Pas J, Wyszko E, Rolle K, et al: Analysis
of structure and function of tenascin-C. Int J Biochem Cell Biol.
38:1594–1602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Jones FS and Jones PL: The tenascin family
of ECM glyco-proteins: structure, function and regulation during
embryonic development and tissue remodeling. Dev Dyn. 218:235–259.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Chiquet-Ehrismann R, Kalla P and Pearson
CA: Participation of tenascin and transforming growth factor-beta
in reciprocal epithelial-mesenchymal interactions of MCF7 cells and
fibroblasts. Cancer Res. 49:4322–4325. 1989.PubMed/NCBI
|
|
31.
|
Orend G and Chiquet-Ehrismann R: Adhesion
modulation by antiadhesive molecules of the extracellular matrix.
Exp Cell Res. 261:104–110. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Villuendas R, Steegmann JL, Pollan M, et
al: Identification of genes involved in imatinib resistance in CML:
a gene-expression profiling approach. Leukemia. 20:1047–1054. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Helleman J, Jansen MP, Ruigrok-Ritstier K,
et al: Association of an extracellular matrix gene cluster with
breast cancer prognosis and endocrine therapy response. Clin Cancer
Res. 14:5555–5564. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Calvo A, Catena R, Noble MS, et al:
Identification of VEGF-regulated genes associated with increased
lung metastatic potential: functional involvement of tenascin-C in
tumor growth and lung metastasis. Oncogene. 27:5373–5384. 2008.
View Article : Google Scholar : PubMed/NCBI
|