Introduction
Liver cancer in men is the fifth most frequently
diagnosed cancer worldwide but the second most frequent cause of
cancer death. An estimated 748,300 new liver cancer cases and
695,900 cancer deaths occurred worldwide in 2008 (1). Half of these cases and deaths were
estimated to occur in China (2).
Among primary liver cancers, hepatocellular carcinoma (HCC)
represents the major histological subtype, accounting for 70–85% of
the total liver cancer burden worldwide (3).
TF-1 apoptosis-related gene 19 (TFAR19) was first
cloned as an upregulated gene from TF-1 cells undergoing apoptosis
following serum withdrawn by Liu et al(4) in the Peking University Center for
Human Disease Genomics in 1999, then designated as programmed cell
death 5 (PDCD5) by International Human Gene Nomination Committee
(GenBank accession no. AF014955). PDCD5 gene is located on
chromosome 19q12-q13.1 (5) and the
integrated PDCD5 protein contains 125 amino acid (aa) residues
(6). Decreased expression of PDCD5
has been characterized for human tumors including breast cancer
(7), gastric cancer (8), astrocytic glioma (9) and chronic myelogenous leukemia
(10). The scientists in the
Peking University Center for Human Disease Genomics established an
ELISA method for detecting soluble PDCD5 protein (11). Wang et al(12) illustrated a significant elevation
of PDCD5 concentrations in both plasma and synovial fluid of knee
rheumatoid arthritis (RA) patients compared to osteoarthritis (OA)
patients and the control plasma concentrations. The serum PDCD5
levels in breast cancer, gastrointestinal tract cancer and lung
cancer patients were separately compared with healthy persons by
Wang et al(13). However,
the significance of serum PDCD5 and its changes in various cancers
await further investigations.
Xu et al(14) demonstrated that PDCD5 interacts
with Tip60, enhances the histone acetylation and p53 K120
acetylation and promotes the expression of Bax, consequently
accelerating apoptosis. In their recent study, novel evidence
demonstrated that PDCD5 is a p53 regulator during gene expression
and cell cycle (15). Li et
al(16) found that PDCD5
promotes cisplatin-induced apoptosis of glioma cells via activating
mitochondrial apoptotic pathway. Han et al(17) confirmed that PDCD5 shows a greater
inhibitory effect on the Ras/Raf/MEK/ERK signaling pathway in the
human osteosarcoma cell line MG-63. However, the molecular
mechanisms underlying PDCD5 functions during cell growth,
proliferation and apoptosis remain largely unclear.
In this study, we first demonstrate that PDCD5
expression is correlated with clinicopathological features and
patient survival and it may be a useful predictor of prognosis in
patients with HCC after surgical resection. Second, we compared the
PDCD5 levels in peripheral blood serum from cancer patients with
matched normal volunteers. The susceptibility of HCC cells to human
PDCD5 was also investigated in vitro.
Materials and methods
Cell culture and tumor specimens
Human liver cancer cell lines, HepG2 and Hep3B, were
cultured in DMEM (Hyclone, Logan, UT, USA) containing 10% fetal
bovine serum (Invitrogen Gibco, Carlsbad, CA, USA) and incubated in
a 5% CO2 incubator at 37°C. Liver cancer tissue
specimens and their adjacent normal liver tissues were derived from
56 patients undergoing surgical resection of primary hepatocellular
carcinoma without prior chemotherapeutic treatment or radiotherapy.
The adjacent non-malignant tissue was ≥1-cm distance from the tumor
margin. Resections were performed at Department of General Surgery,
First Affiliated Hospital of China Medical University from January
2008 to December 2012. All patients approved the use of tumor
tissues for clinical research and China Medical University Ethics
Committee approved the research protocols. Basic patient
information is summarized in Table
I.
 | Table IPDCD5 expression associated with
demographic and biological parameters in 56 hepatocellular
carcinoma samples. |
Table I
PDCD5 expression associated with
demographic and biological parameters in 56 hepatocellular
carcinoma samples.
| | PDCD5
expression |
|---|
| |
|
|---|
| Clinicopathological
features | n | − | + | ++ | +++ | PR (%) | χ2 | P-value |
|---|
| Gender | | | | | | | 2.57 | 0.462 |
| Female | 20 | 12 | 4 | 1 | 3 | 40.0 | | |
| Male | 36 | 28 | 4 | 2 | 2 | 22.2 | | |
| Age (years) | | | | | | | 4.87 | 0.181 |
| <55 | 26 | 15 | 5 | 2 | 4 | 42.3 | | |
| ≥55 | 30 | 25 | 3 | 1 | 1 | 16.7 | | |
| Tumor number | | | | | | | 8.89 | 0.030 |
| Multiple | 38 | 27 | 6 | 0 | 5 | 28.9 | | |
| Solitary | 18 | 13 | 2 | 3 | 0 | 27.8 | | |
|
Differentiation | | | | | | | 2.06 | 0.559 |
|
Differentiated | 27 | 20 | 4 | 2 | 1 | 25.9 | | |
|
Undifferentiated | 29 | 20 | 4 | 1 | 4 | 31.0 | | |
| Portal
invasion | | | | | | | 3.14 | 0.370 |
| − | 17 | 14 | 1 | 0 | 2 | 17.6 | | |
| + | 39 | 26 | 7 | 3 | 3 | 33.3 | | |
| Lymph node
metastasis | | | | | | | 11.07 | 0.011 |
| − | 22 | 11 | 7 | 2 | 2 | 50.0 | | |
| + | 34 | 29 | 1 | 1 | 3 | 14.7 | | |
| Tumor size
(cm) | | | | | | | 5.62 | 0.131 |
| <5 | 25 | 15 | 5 | 3 | 2 | 40.0 | | |
| ≥5 | 31 | 25 | 3 | 0 | 3 | 19.4 | | |
| HBV infection | | | | | | | 8.77 | 0.030 |
| − | 18 | 10 | 4 | 0 | 4 | 44.4 | | |
| + | 38 | 30 | 4 | 3 | 1 | 21.1 | | |
Quantitative real-time PCR
Total RNA was isolated using an RNeasy Mini kit
(Biomed, Beijing, China). First strand cDNA was reverse transcribed
with 1 μg of total RNA, using Takara Reverse Transcription kit
(Takara, Dalian, China) and oligo(dT) 15 primers (Takara). The
resultant cDNA was then used for quantitative PCR reactions. The
PDCD5 primers were: sense: 5′-CTGAGGAGACAGAGGCTGGC-3′ and
antisense: 5′-TTTCTGCTTCCCTGTGCTTTG-3′. The housekeeping genes,
GAPDH and β-actin, were used as internal controls for
normalization of the results. The GAPDH primers were: sense:
5′-AGAAGGCTGGGGCTCATTTG-3′ and antisense:
5′-AGGGGCCATCCACAGTCTTC-3′. The β-actin primers were: sense:
5′-CTCCCTGGAGAAGAGCTACGA-3′ and antisense:
5′-GTGGGACTTCCAGAACTGCA-3′. Amplification of PDCD5,
GADPH and β-actin was performed with 1 cycle at 95°C
for 10 min and 40 cycles of 95°C for 15 sec and 60°C for 60 sec.
Calculation of the relative expression of each transcript was
performed using the 2−ΔΔCt method (18).
RNA isolation and reverse
transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was isolated from HepG2 and Hep3B cells
using the RNeasy Mini kit (Biomed, Beijing, China). First strand
cDNA was reverse transcribed with 1 μg total RNA, using the Takara
Reverse Transcription kit (Takara) and oligo(dT)-15 primers
(Takara) according to the manufacturer′s instructions. The primers
of PDCD5 and GAPDH were used as described above. PCR
amplification of cDNA was performed in 20 μl mixtures. Finally,
products were resolved by 1.5% agarose gel electrophoresis and
visualized by ethidium bromide staining and a UV imaging system
(UVP, LLC, Upland, CA, USA).
Immunohistochemical staining (IHC)
IHC of 4-μm sections of paraffin-embedded specimens
was performed using the anti-PDCD5 polyclonal antibody (Beijing
Biosea Biotechnology, Beijing, China). Briefly, after
deparaffinization and hydration, the endogenous peroxidase activity
was quenched by a 30-min incubation in a mixture of 0.3% hydrogen
peroxide solution in 100% methanol. The sections were blocked for 2
h at room temperature with 1.5% blocking serum in PBS and incubated
with anti-PDCD5 antibody (1:200 dilution) at 4°C in a moist chamber
overnight, followed by incubation with Envision reagent (Dako,
Carpinteria, CA, USA) and color development in
3,3′-diaminobenzidine tetrahydrochloride (DAB, Sigma-Aldrich,
Carlsbad, CA, USA). The slides were then lightly counterstained
with hematoxylin, dehydrated with ethanol, cleaned with xylene and
mounted. Adjacent non-cancer tissues were used as controls.
Sections treated without primary antibodies were used as negative
controls. The positive percentage of counted cells was graded
semi-quantitatively according to a four-tier scoring system:
negative (−), 0–5%; weakly positive (+), 6–25%; moderately positive
(++), 26–50%; and strongly positive (+++), 51–100%.
Preparation of blood samples and
measurement of PDCD5 and AFP in serum
Peripheral blood was obtained from 58 patients with
HCC and 58 healthy volunteers. Samples were clotted for 30 min and
then centrifuged for 10 min at 1000 × g. All samples were then
frozen and stored at −80°C until required for assay. Concentration
of PDCD5 in serum was assayed using enzyme-linked immunosorbent
assay (ELISA) kit for PDCD5 (USCN Life Science Inc., Houston, TX,
USA). Concentration of AFP in serum was measured using an ELISA kit
from Diagnostic Automation/Cortez Diagnostics Inc (Calabasas, CA,
USA).
Plasmid and transfection
The plasmid, pcDNA3.1-PDCD5, was kindly provided by
Mr. Xiao-Rui Han (China Medical University, Shenyang). Transfection
of the plasmid into HepG2 cells or Hep3B was performed using
Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s
instructions. Cell lines after transfection were named HepG2-T and
Hep3B-T. Cell lines transfected with pcDNA3.1 was used as a mock
and named HepG2-M and Hep3B-M.
Immunofluorescence
Transfected cells were washed with PBS, fixed in 4%
paraformaldehyde, permeabilized in 1% Triton X-100 for 5 min and
blocked with 5% bovine serum albumin in PBS containing 0.5% Triton
X-100 for 1 h. PDCD5 expression was detected using anti-PDCD5
(Beijing Biosea Biotechnology) antibody for 1 h at room
temperature. Cells were washed with PBS and incubated with
appropriate secondary fluorophore-conjugated antibody for 1 h at
room temperature, washed with PBS and mounted using
SlowFade® Gold Antifade reagent (Invitrogen). Secondary
antibody used for detection of PDCD5 was Alexa Fluor®
594 Donkey Anti-Goat IgG (H+L) (Invitrogen).
Evaluation of cell viability by MTT
assay
Cell viability was assayed using
3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazoliumbromide (MTT)
assays (Sigma). Briefly, cells were plated in 96-well plates (1,500
cells per well). After 24 h, 0.5 mg/ml MTT was added to each well.
Four hours later, cells were lysed with dimethyl sulfoxide (DMSO)
and absorbance rates were measured at 550–560 nm using a microplate
reader (Bio-Rad, Hercules, CA, USA). Proteasome inhibitor, MG132,
purchased from Calbiochem (La Jolla, CA, USA), was used as a
control in the following experiments.
TUNEL assay
For apoptosis detection, HepG2 cells, HepG2-M cells,
HepG2-T cells, HepG2 treated with MG132, Hep3B cells, Hep3B-M
cells, Hep3B-T cells and Hep3B cells treated with MG132 were washed
in PBS, fixed, permeabilized and subjected to TUNEL labeling using
an In Situ Cell Death Detection kit (Keygen, Nanjing, China)
according to the manufacturer’s protocol. After counterstaining
with DAPI (1 μg/ml), photographic images were taken using an
Olympus CX71 fluorescence microscope (Olympus, Tokyo, Japan).
TUNEL-positive nuclei were stained green and all other nuclei were
stained blue (19).
Cell apoptosis assay
Cells (5×105) were collected without EDTA
and washed with PBS. A 500-μl binding buffer, 5 μl Annexin V-FITC
and 5 μl propidium iodide (PI) were added into the suspension in
this order and mixed at room temperature in dark for 10 min. The
examination was performed by flow cytometry within 1 h.
Cell cycle assay
Cells seeded on 6-well plates were treated and then
collected. After being washed with PBS three times, the cell
suspension was fixed with 70% ethanol, incubated with RNAse A at
37°C. After staining with 400 μl PI, the suspension was evaluated
by flow cytometry.
In vitro wound healing assay
Cells were grown in a 6-well dish. A confluent
monolayer of cells was scratched with a 200-μl pipette tip to
create a wound. Cells were washed twice with PBS and then
supplemented with medium and incubated for 4 h at 37°C. Cell
migration into the wounded area was monitored microscopically.
Images were captured at the interface of the unwounded and wounded
areas.
26S proteasome activity assay
26S proteasome function was assayed as described
previously (20). The assay is
based on detection of the fluorophore 7-amino-4-methylcoumarin
(AMC) after cleavage from the labeled substrate Suc-LLVY-AMC
(Boston Biochem, Boston, MA, USA). These fluorogenic proteasome
substrates were added to the cell lysate at a final concentration
of 80 μM in 1% DMSO. ATP-dependent cleavage activity was monitored
continuously by detection of free 7-amido-4-methylcoumarin using a
microplate reader (Bio-Rad) at 380/460 nm, at 37°C.
Western blot analysis
The total protein was extracted from human liver
cancer cell lines and liver tissues using RIPA buffer (Sigma). The
protein concentration was determined with BCA Protein Assay kit
(Pierce, Rockford, IL, USA). For performing nitrocellulose membrane
blotting, 60 μg cell lysates were separated on a 10% SDS-PAGE. The
blotted membranes were blocked with 5% BSA in TBST for 2 h at room
temperature and incubated with each primary antibody overnight. The
primary antibodies were PDCD5 (Abcam, Cambridge, UK) and β-actin
(Santa Cruz Biotechnology, Santa Cruz, CA, USA). The reaction was
followed by probing with peroxidase-coupled secondary antibodies at
dilutions ranging from 1:1,000 to 1:2,000 (Amersham Biosciences,
Needham, MA, USA) and binding results were visualized by enhanced
chemiluminescence (Amersham Pharmacia, Piscataway, NJ, USA).
Statistical analysis
Statistical analyses were performed using SPSS 15.0
software (SPSS, Chicago, IL, USA). Comparisons were made using
χ2 tests, the Wilcoxon signed-rank test and the t-test.
Overall survival was analyzed using the Kaplan-Meier method and the
significance of differences in survival rates was estimated using
the log-rank test. Values of P<0.05 were considered
significant.
Results
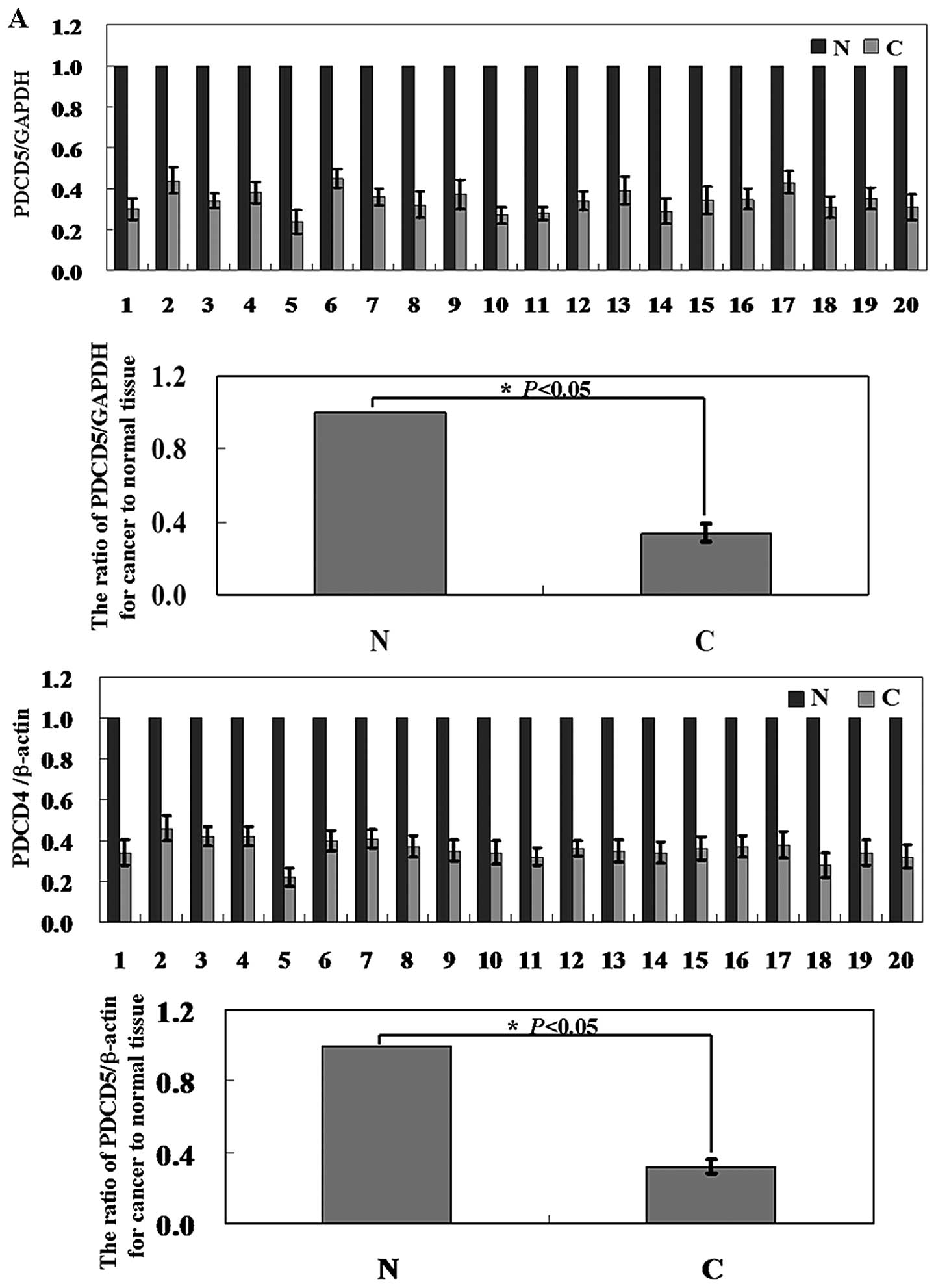
PDCD5 is downregulated in HCC tissues
compared with normal tissues
To initially confirm the roles of PDCD5 expression
in human liver cancer development and progression, we assayed the
expression of PDCD5 in 56 pairs of human hepatocellular carcinomas
and corresponding adjacent normal tissues. Levels of PDCD5 mRNA and
protein in the hepatoma tissue were significantly lower than those
in corresponding adjacent non-cancer tissue (Fig. 1A and B; P<0.05). In Fig. 1C, the results of
immunohistochemical staining showed that positive staining was seen
in the cytoplasm of the normal cells, in contrast, almost no
positive cells were seen in cancer tissue. Moreover, the PDCD5
expression was correlated statistically with HBV infection
(P=0.030), tumor number (P=0.030) and lymph node metastasis
(P=0.011). There was no significant correlation between PDCD5
expression and age, sex, tumor number, tumor size, differentiation
and portal invasion (Table I,
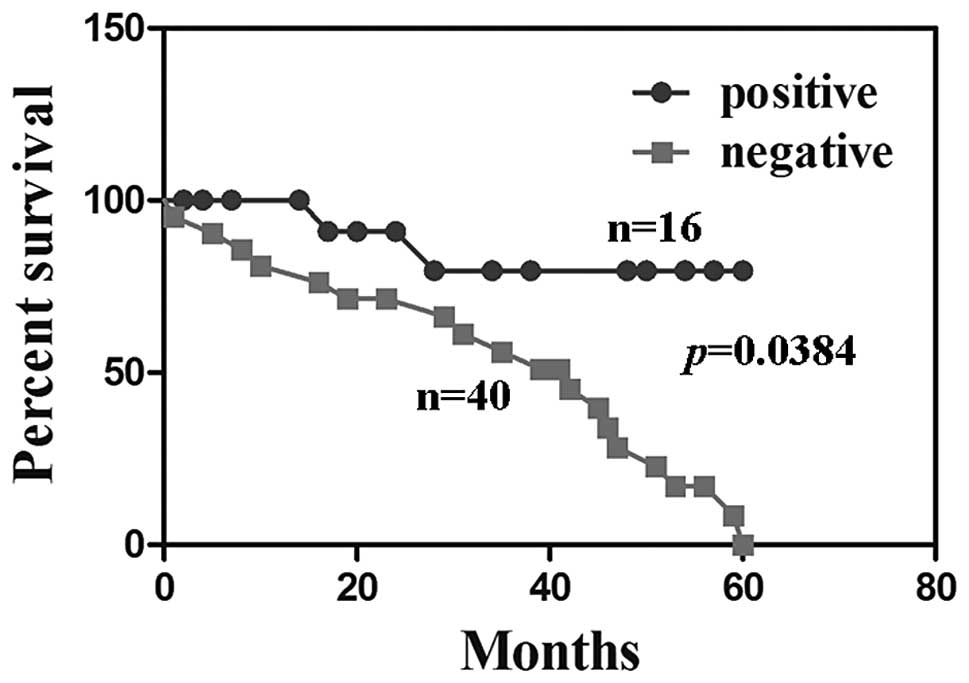
P>0.05). HCC patients with PDCD5 expression were associated with
a significantly better survival rate than those without PDCD5
expression (Fig. 2, P=0.038).
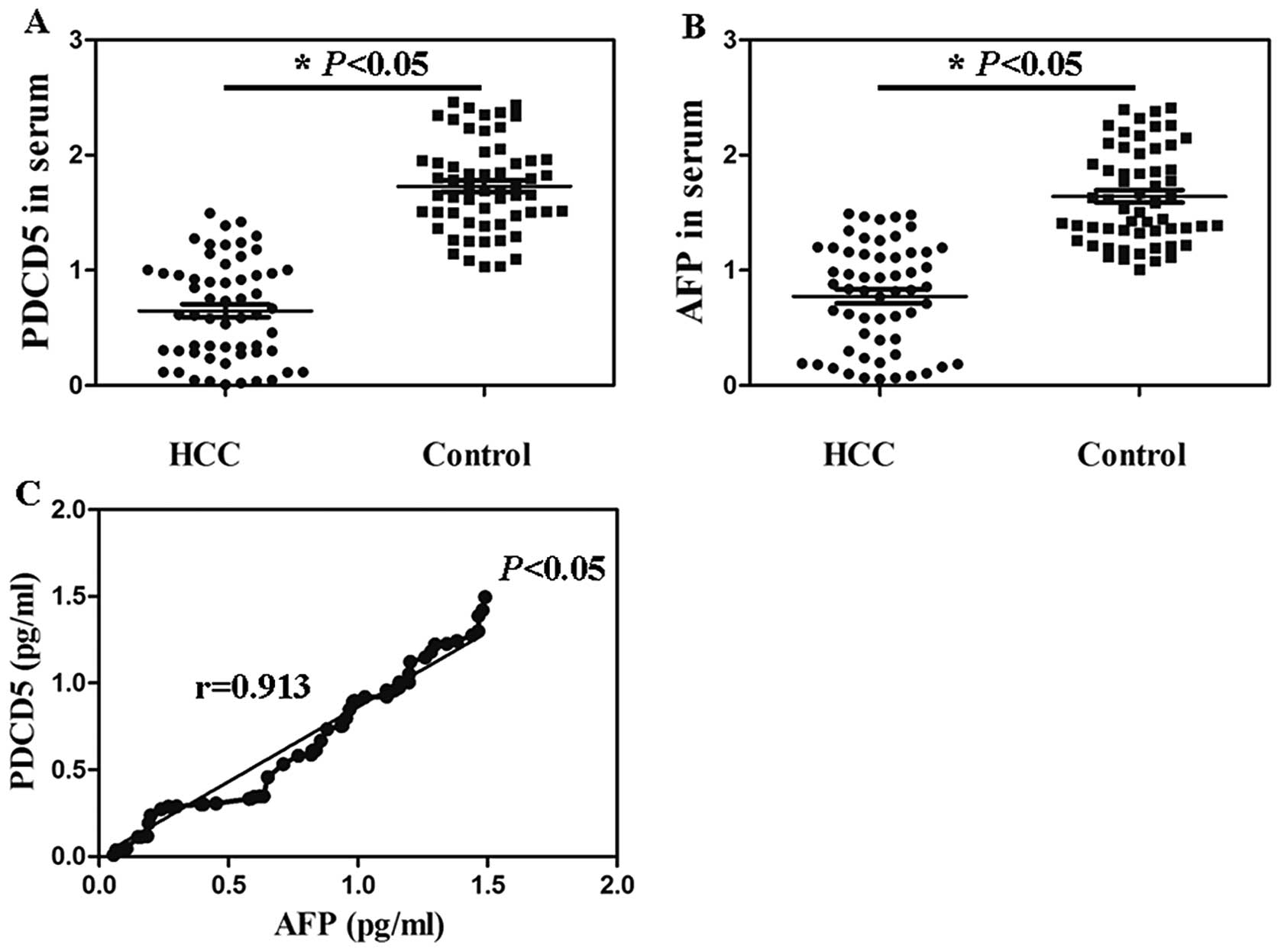
The serum levels of PDCD5 and AFP
detection in patients with HCC
The serum PDCD5 contents in 58 HCC patients ranged
from 4.41 pg/ml to 1.72 ng/ml with a median of 421.83±12.52 pg/ml.
There was statistical significance compared with the serum PDCD5
level in healthy persons (Fig. 3A,
P<0.05). Serum AFP levels in the HCC patients (6.24 pg/ml to
1.53 ng/ml) were significantly higher than that in the control
group (32.67 pg/ml to 2.46 ng/ml) (Fig. 3B, P<0.05). The serum levels of
PDCD5 and AFP in 58 HCC patients were significantly positively
correlated (r=0.913) (Fig. 3C,
P<0.05).
Establishment of PDCD5-overexpressing
HepG2 and Hep3B cells
To investigate further the phenomenon and mechanism
of the PDCD5-mediated regulation of liver cancer cell growth and
metastasis, cell lines, HepG2 and Hep3B, were selected for PDCD5
gene enhancement. After pcDNA3.1-PDCD5 was transfected into the two
hepatoma cell lines, PDCD5 mRNA and protein levels were
significantly increased compared to untransfected cells by using
RT-PCR and western blotting, respectively (Fig. 4A and B). Immunofluorescence
analysis showed PDCD5 was localized in the cytoplasm of transfected
cells (Fig. 4C). These results
collectively suggested that the transfection was successful.
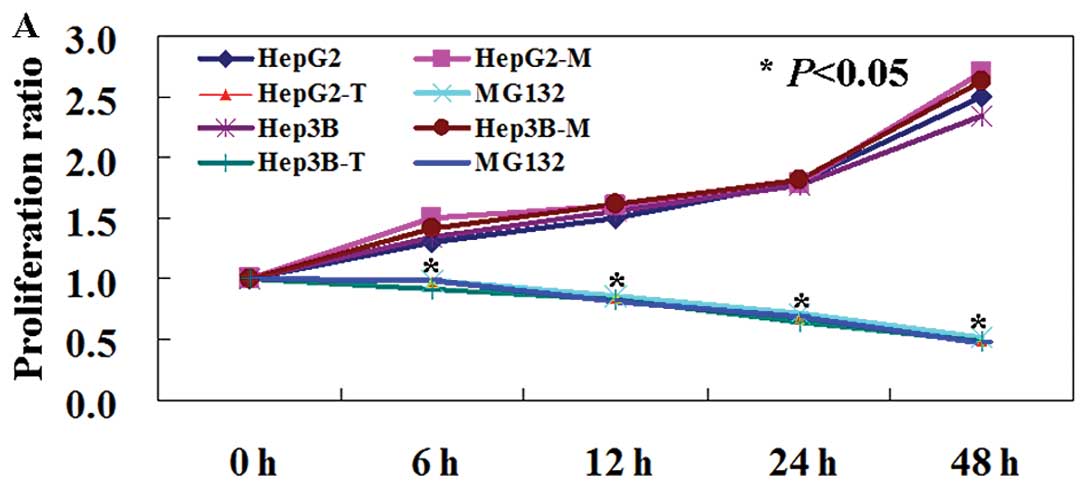
PDCD5 displays antitumor activity in
liver cancer cells
Since PDCD5 mRNA and protein levels are decreased in
liver cancer tissues and cells, we determined whether its exogenous
expression would reduce tumorigenicity of liver cancer cells. The
MTT assay showed that the proliferation rate of PDCD5-expressing
cells was decreased compared to untransfected and mock-transfected
cells (Fig. 5A, P<0.05). TUNEL
assay confirmed that the number of apoptotic cells in the
transfected group was significantly more than untransfected group
and mock-transfected group (Fig.
5B). To quantify apoptotic cells, Annexin V-FITC and PI double
staining was performed. In cells expressing PDCD5, the apoptotic
ratio was 3–6 times higher than that of untransfected and
mock-transfected cells. We determined that the percentage of
apoptosis in PDCD5-expressing HepG2 cells was 3.86%, significantly
higher compared to untransfected (0.73%) and mock-transfected
(1.12%) HepG2 cells (Fig. 5C,
P<0.05). The apoptotic percentage of PDCD5-expressing Hep3B
cells was 3.75% compared to untransfected (1.23%) and
mock-transfected (1.34%) Hep3B cells (Fig. 5C, P<0.05). PI staining of cells
revealed that PDCD5-expressing cells were arrested in G1
phase (Fig. 5D, P<0.05). We
determined the mobility change in PDCD5-expressing cells using the
wound healing assay. The percentage of wound closure of
PDCD5-positive cells was decreased when compared to untransfected
and mock-transfected cells (Fig.
5E, P<0.05).
PDCD5 inhibits proteasome activity in
liver cancer cells
In order to determine whether PDCD5 could impact on
the proteasome, we compared the response of MG132 treated liver
cancer cells with PDCD5 treated cells. The proteasome activity of
both PDCD5-expressing HepG2 cells and PDCD5-expressing Hep3B cells
was lower than that of untransfected and mock-transfected cells
(Fig. 5F, P<0.05). HepG2 cells
and Hep3B cells after MG132 treatment also showed increased
apoptosis, G1 arrest and decreased mobility. Thus, the
results showed that MG132 has the same effect as PDCD5 in liver
cancer cells (Fig. 5, P<0.05).
These data collectively indicate that PDCD5 suppressed HCC cells
via inhibition of proteasome activity.
Discussion
Several pieces of evidence have suggested that the
expression of PDCD5 protein is downregulated in some human tumors,
such as breast cancer (7), gastric
cancer (8) and astrocytic glioma
(9). In the study of Xu et
al(21), gene expression
profiles of HCC and non-HCC, based on cDNA arrays or RT-PCR,
reflected a number of genes with relatively high expression in
non-HCC, including PDCD5. However, the clinical significance
of PDCD5 expression in HCC has not been well characterized. In this
study, we confirmed both PDCD5 mRNA and protein were significantly
higher in normal matched tissues than that in HCC tissues. Low
expression of PDCD5 in chondrosarcoma samples was significantly
correlated with anatomical location and histological grade, but not
with age and gender (22). The
expression of PDCD5 in gliomas was correlated significantly with
the pathological grade. However, we found that the PDCD5 expression
was correlated statistically with HBV infection, tumor number and
lymph node metastasis. The discrepancy might be attributable to
clinicopathological characteristics of the subjects and grouping
methods. Previous studies have documented that PDCD5 expression can
be predictive for the prognosis of patients with several tumors
(8,9,22).
We performed Kaplan-Meier analysis and confirmed PDCD5 protein was
associated with the overall survival of patients with HCC.
Therefore, it is implied that PDCD5 may be a favorable biomarker
for the prognosis of patients with HCC. Since the sample size in
this study may be considered small, further investigation of a
larger patient population is necessary to confirm its clinical
value and prognostic evaluation in HCC. α-fetoprotein (AFP), is
still the only serological marker presently available for routine
screening in most parts of the world. High levels of AFP in fully
developed hepatocarcinoma or in serum of the host are associated
with more aggressive behavior and increased anaplasia (23,24).
In this study, the concentrations of PDCD5 and AFP in serum were
lower in HCC patients than that in healthy controls and confirmed
that the levels of PDCD5 and AFP were significantly positively
correlated. A strong correlation between PDCD5 and IL-17 levels in
serum of RA patients has been determined in a previous study
(25). Wang et al(13) found decreased expression of PDCD5
in peripheral blood of breast cancer, lung cancer and
gastrointestinal cancer, however, the PDCD5 levels in cancer
patients are not statistically different from that of normal
persons. The level of PDCD5 in serum may be used as a novel marker
for HCC.
Previous studies have demonstrated that PDCD5 is an
apoptosis-promoting molecule that is upregulated in cells
undergoing apoptosis (4,14,26,27).
Likewise, blocking the activity of PDCD5 by introducing an
anti-PDCD5 antibody or siRNA against PDCD5 could suppress etoposide
or Bax- induced apoptosis of tumor cells (28,29).
In MTT assay, the proliferation ratio of transfected cells was
reduced compared to untransfected ones in our study.
Correspondingly, transfected cells exhibited a higher apoptotic
ratio than untransfected cells by TUNEL assay and Annexin V/PI
double staining. Collectively, these results suggest that PDCD5 may
have a suppressive role in HCC cells.
The mechanisms involved in PDCD5 expression are
still being investigated. PDCD5 positively regulates Tip60, a
transcriptional coregulator, which in turn, promotes p53
acetylation, leading to enhanced p53-dependent apoptosis (14). Another study showed that apoptotic
potential of PDCD5 is linked with CK2 phosphorylation (30). It has also been shown that PDCD5
can enhance TAJ/TROY-induced paraptosis-like cell death (26). Han et al(17) found that PDCD5 mediates antitumor
effects in the osteosarcoma cell line by suppressing the
Ras/Raf/MEK/ERK signaling pathway. An et al(31) found that myocardial high PDCD5
overexpression results in dilated cardiomyopathy and heart failure
accompanied by dramatically enhanced autophagy. Interesting, the
major finding in the present study is that PDCD5 overexpression is
involved in proteasome inhibition. This is supported by the fact
that, PDCD5 showed the same antitumor activity as the proteosome
inhibitor, MG132. The proteasome activity of transfected cells was
lower than untransfected ones. Proteasome inhibition has gained
interest as anticancer therapy (32). Proteasome inhibition disturbs the
critical intracellular balance between proapoptotic and
antiapoptotic signals shifting it towards tumor growth inhibition,
apoptosis and decreased metastasis (33).
In conclusion, we have demonstrated that a decreased
expression of PDCD5 correlated significantly with HBV infection,
tumor number and lymph node metastasis. Although previous studies
have provided insightful mechanisms of PDCD5 involved in intrinsic
or extrinsic apoptotic pathways, our results showed evidence that
PDCD5 is involved with proteasome inhibition. PDCD5 may serve as a
novel therapeutic target for the treatment of hepatocellular
cancer.
Acknowledgements
We thank Dr Miao Yu for technical assistance.
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 6:69–90. 2011. View Article : Google Scholar
|
|
2
|
Bosch FX, Ribes J, Diaz M, et al: Primary
liver cancer: worldwide incidence and trends. Gastroenterology.
127:S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Perz JF, Armstrong GL, Farrington LA, et
al: The contributions of hepatitis B virus and hepatitis C virus
infections to cirrhosis and primary liver cancer worldwide. J
Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu H, Wang Y, Zhang Y, et al: TFAR19, a
novel apoptosis related gene cloned from human leukemia cell line
TF21, could enhance apoptosis of some tumor cells induced by growth
factor withdrawal. Biochem Biophys Res Commun. 254:203–210. 1999.
View Article : Google Scholar
|
|
5
|
Spinola M, Meyer P, Kammerer S, et al:
Association of the PDCD5 locus with lung cancer risk and prognosis
in smokers. J Clin Oncol. 24:1672–1678. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu D, Feng Y, Cheng Y, et al: Human
programmed cell death 5 protein has a helical-core and two
dissociated structural regions. Biochem Biophys Res Commun.
318:391–396. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hedenfalk I, Duggan D, Chen Y, et al:
Gene-expression profiles in hereditary breast cancer. N Engl J Med.
344:539–548. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Y, Zhao M, Li W, et al: Expression of
programmed cell death 5 gene involves in regulation of apoptosis in
gastric tumor cells. Apoptosis. 11:993–1001. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li H, Wang Q, Gao F, et al: Reduced
expression of PDCD5 is associated with high-grade astrocytic
gliomas. Oncol Rep. 20:573–579. 2008.PubMed/NCBI
|
|
10
|
Ruan G, Qin Y, Chen S, et al: Abnormal
expression of the programmed cell death 5 gene in acute and chronic
myeloid leukemia. Leuk Res. 30:1159–1165. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng YM, Zhang YM and Jing GZ: Soluble
expression in Escherichia coli, purification and characterization
of a human TF-1 cell apoptosis-related protein TFAR19. Protein Expr
Purif. 25:323–329. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Guan Z and Ge Z: Plasma and
synovial fluid programmed cell death 5 (PDCD5) levels are inversely
associated with TNF-α and disease activity in patients with
rheumatoid arthritis. Biomarkers. 18:155–159. 2013.PubMed/NCBI
|
|
13
|
Wang Y, Wang GH and Zhang QY:
Determination of PDCD5 in peripheral blood serum of cancer
patients. Chin J Cancer Res. 23:224–228. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu L, Chen Y, Song Q, et al: PDCD5
interacts with Tip60 and functions as a cooperator in
acetyltransferase activity and DNA damage-induced apoptosis.
Neoplasia. 11:345–354. 2009.PubMed/NCBI
|
|
15
|
Xu L, Hu J, Zhao Y, et al: PDCD5 interacts
with p53 and functions as a positive regulator in the p53 pathway.
Apoptosis. 17:1235–1245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H, Zhang X, Song X, et al: PDCD5
promotes cisplatin-induced apoptosis of glioma cells via activating
mitochondrial apoptotic pathway. Cancer Biol Ther. 13:822–830.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han XR, Sun Y and Bai XZ: The antitumor
role and mechanism of integrated and truncated PDCD5 proteins in
osteosarcoma cells. Cell Signal. 24:1713–1721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berman DM, Karhadkar SS, Maitra A, et al:
Widespread requirement for Hedgehog ligand stimulation in growth of
digestive tract tumours. Nature. 425:846–851. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Markaryan A, Nelson EG, Tretiakova M, et
al: Technical report: immunofluorescence and TUNEL staining of
celloidin embedded human temporal bone tissues. Hear Res. 241:1–6.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fekete MR, McBride WH and Pajonk F:
Anthracyclines, proteasome activity and multi-drug-resistance. BMC
Cancer. 5:1142005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu XR, Huang J, Xu ZG, et al: Insight into
hepatocellular carcinogenesis at transcriptome level by comparing
gene expression profiles of hepatocellular carcinoma with those of
corresponding non-cancer liver. Proc Natl Acad Sci USA.
98:15089–15094. 2001. View Article : Google Scholar
|
|
22
|
Chen C, Zhou H, Xu L, et al: Prognostic
significance of downregulated expression of programmed cell death 5
in chondrosarcoma. J Surg Oncol. 102:838–843. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dudich E, Semenkova L, Gorbatova E, et al:
Growth - regulative activity of human alpha-fetoprotein for
different types of tumor and normal cells. Tumour Biol. 19:30–40.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang XW and Xu B: Stimulation of tumor -
cell growth by alpha-fetoprotein. Int J Cancer. 75:596–599. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang JF, Guan ZP, Zhang SL, et al:
Programmed cell death 5 correlates with disease activity and
interleukin-17 in serum and synovial fluid of rheumatoid arthritis
patients. Chin Med J (Engl). 126:296–299. 2013.PubMed/NCBI
|
|
26
|
Wang Y, Li X, Wang L, et al: An
alternative form of paraptosis like cell death, triggered by
TAJ/TROYand enhanced by PDCD5 overexpression. J Cell Sci.
117:1525–1532. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Sun R, Han W, et al: Nuclear
translocation of PDCD5 (TFAR19): an early signal for apoptosis?
FEBS Lett. 509:191–196. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rui M, Chen YY, Zhang YM, et al: Transfer
of anti-TFAR19 monoclonal antibody into HeLa cells by in situ
electroporation can inhibit the apoptosis. Life Sci. 71:1771–1778.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen LN, Wang Y, Ma DL, et al: Short
interfering RNA against the PDCD5 attenuates cell apoptosis and
caspase-3 activity induced by Bax overexpression. Apoptosis.
11:101–111. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Salvi M, Xu D, Chen Y, et al: Programmed
cell death protein 5 (PDCD5) is phosphorylated by CK2 in vitro and
in 293T cells. Biochem Biophys Res Commun. 387:606–610. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
An L, Zhao X, Wu J, et al: Involvement of
autophagy in cardiac remodeling in transgenic mice with cardiac
specific over-expression of human programmed cell death 5. PLoS
One. 7:e300972012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Adams J: The development of proteasome
inhibitors as anticancer drugs. Cancer Cell. 5:417–421. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu WK, Cho CH, Lee CW, et al: Proteasome
inhibition: a new therapeutic strategy to cancer treatment. Cancer
Lett. 293:15–22. 2010. View Article : Google Scholar : PubMed/NCBI
|