Introduction
Lung cancer is the most common cancer worldwide and
the leading cause of cancer-related mortality (1). Non-small cell lung cancer (NSCLC)
accounts for ~80% of all lung cancer cases and has a 5-year overall
survival rate of <15% (2,3).
Approximately 40% of patients diagnosed with NSCLC have
unresectable stage III disease or inoperable disease (4). Therefore, additional efforts are
needed to understand and identify molecular targets for gene
therapy (5,6).
The cluster of differentiation 59 (CD59), also
called protectin, is a type of complement regulated proteins
(CRPs). CD59 inhibits the complement cytolytic activity by binding
to C8 and C9, which blocks the assembly of the membrane attack
complex (MAC) (7). CD59 is
overexpressed in most solid malignancies and presents at low levels
in normal tissues. CD59 overexpression may assist malignant cells
to escape immunologic surveillance and complement-mediated
cytolysis, limiting the effect of complement-fixing monoclonal
antibodies (7–10). Immune escape of tumor cells is a
primary cause of failed immunotherapy. Blocking of CD59 function on
the surface of tumor cells might allow effective
complement-mediated clearance of tumor cells that improve the
effect of complement-activating antitumor antibodies (11,12).
Therefore, CD59 is a promising therapeutic target for antitumor
gene therapy.
RNA interference (RNAi) is an economical, fast and
highly efficient technique for silencing gene expression (13–15).
Recently, siRNA-encoding plasmids delivered by virus has been
rapidly developed and widely applied in mammalian cells (16,17).
In this study, we examined the expression of CD59 in NSCLC and
constructed a CD59 small interfering RNA (siRNA) lentiviral vector.
We assessed its effect on the proliferation and apoptosis of lung
cancer cells and further characterized the functional role of CD59
during lung cancer tumorigenesis.
Materials and methods
Expression of CD59 in tissue specimens by
immunohistochemistry (IHC)
Twenty primary NSCLC specimen and corresponding
surgical margin specimens were obtained from the People’s Hospital
of Guangxi Zhuang Autonomous Region. A mouse polyclonal antibody
for CD59 was obtained from Santa Cruz Biotechnology (SC-133171) to
assess the expression of the CD59 protein. Briefly,
paraffin-embedded tissue sections (5-μm thick) were deparaffinized
with xylene and then dehydrated in sequential diluted ethanol
before rinsing in PBS. Sections were heated for 10 min in 0.01 M
citrate buffer (pH 6.0) twice to unmask the antigens. Endogenous
peroxidase activity was then blocked with 3% hydrogen peroxide for
20 min at room temperature. Before being incubated with CD59
antibody (dilution, 1/100 in 0.01 M PBS) at 4°C overnight, sections
were incubated with 5% normal goat serum in 1% BSA in PBS for 30
min to block non-specific IgG binding. A biotinylated goat
antimouse IgG was used for further incubation and a
strepavidin-biotin complex system (SABC) with diaminobenzidene as
chromogen was used for color development. The sections were weakly
counterstained with hematoxylin before mounting and then examined
under a light microscope. PBS (0.01 M) was used to replace primary
antibody to serve as negative staining controls.
Immunohistochemical staining was evaluated by two independent
pathologists.
Construction of small interfering RNA
targeted CD59 expression vector, production of lentivirus and
transfection into NCI-H 157 cells
The pSUPER vector was digested by BglII and
HindIII restriction enzyme and annealed oligos, siCD59:
5′-GATCCCCGCGTGTCTCATTACCAAAGttcaagagaCTT TGGTAATGAGACACGTTTTA-3.
siCD59-C: 5′-AGCTTA AAAAGCGTGTCTCATTACCAAagtctcttgAACTTTGGTA
ATGAGACACGCGGG-3′ were ligated with this vector. The recombinants
were identified by PCR, restriction endonuclease analyses and DNA
sequencing, respectively.
A packaging cell line Phoenix A and human non-small
lung cancer cell line NCI-H157 cells (18) were cultured in DMEM and RPMI-1640
supplemented with 10% FBS (Gibco, Carlsbad, CA, USA) and
penicillin/streptomycin, incubating in a humidified incubator
(37°C, 5% CO2). For retroviral production, Phoenix A
cells were transfected with this recombinant using Lipofectamine
2000 (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions. Culture supernatants were collected
after 48 h. NCI-H157 cells were infected with siCD59 and siCD59-C
in the presence of 8 g/ml polybrene, follow by clonal selection
with G418 (400 mg/l) to generate stable clones.
Detection of the expression of CD59 mRNA
by real-time quantitative PCR (RT-qPCR) test
Total RNA from different groups of NCI-H157 cells
was isolated using TRIzol reagent (Invitrogen). Two microgram of
total RNA was reverse-transcribed in a 20-μl reaction using
reverse-transcription system (Promega, Madison, WI, USA). Primers
were designed based on sequences of human CD59 and β-actin. The
forward primer of CD59 was 3′-ACACCATTGCTGGGGACCTC-5′ and the
reverse primer was 3′-GCTGAATCTTAAAGTCAGGCAA AGG-5′. The forward
primer of β-actin was 3′-CACACCCGC CACCAGTTCGC-5′ and the reverse
primer was 3′-AGCACAG GGTGCTCCTCAGGG-5′. The amplified sequence was
356 and 332 bp, respectively. Thermo cycling was carried out as
follows: CD59; 94°C for 5 min, then 40 cycles of 94°C for 45 sec,
52°C for 45 sec and 72°C for 45 sec, followed by 72°C for 7 min or
β-actin 94°C for 5 min, then 40 cycles of 94°C for 40 sec, 58°C for
35 sec and 72°C for 45 sec, followed by 72°C for 7 min. PCR
products were quantified by using Quantity One 6.4.0 (Bio-Rad,
Hercules, CA, USA) Software. CD59 levels were normalized to
β-actin.
Determination of CD59 protein expression
by western blotting
Different groups of NCI-H157 cells were harvested at
the indicated time-points, washed twice with cold
phosphate-buffered saline (PBS), lysed in fresh cell lysis buffer
for 2 h on ice and centrifuged at 12,000 g for 15 min at 4°C to
remove insoluble materials. Protein concentrations were determined
by BCA assay. Protein (20 μg) was separated by using 8 and 5%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred to PVDF membranes. The membrane was incubated with
anti-CD59 (1:500) and anti-β-actin (1:200) antibodies (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA), respectively, followed
by incubation with horseradish peroxidase-conjugated goat
anti-mouse secondary antibody. Western blots were developed by
chemiluminescence and quantified using Quantity One 6.4.0 (Bio-Rad)
software. CD59 levels were normalized to β-actin.
Determination of the NSCLC cell
proliferation by MTT assay
Different groups of NCI-H157 cells were cultured for
3 days in 96-well plates and then incubated in 5% MTT at 37°C for 4
h. DMSO (100 μl/well) was added and the light absorption value at
490 nm was measured.
Determination of the NSCLC cell apoptosis
by flow cytometry
Different groups of NCI-H157 cells were seeded in
6-well culture plates. Each group contained five culture flasks.
After 24 h, the cells were harvested and washed in cold PBS.
Annexin V and PI staining were carried out using the Annexin V-FITC
Apoptosis Detection kit (BD Biosciences, USA), according to the
manufacturer’s protocol. After a 20-min incubation in the dark at
room temperature, the cells were immediately analyzed by
FACSCalibur Flow Cytometry (BD Biosciences).
Determination of the NSCLC cell
resistance to complement cracking ability by LDH release test
Different groups of NCI-H157 cells were treated with
complement inactivated 8% fresh normal human serum (NHS) for 60 min
at 37°C. Triton X-100 (0.1%) in RPMI-1640 was used as the 100%
lysis control and RPMI-1640 alone was used for the 0% lysis
control. Following incubation, 40 μl of sample supernatant was
taken for LDH assay. To each well 100 μl solution C was added. Just
before analysis, 10 μl solution B was added. The absorbance at 440
nm was calculated, reflecting the activity of LDH present and the
following equation applied: LDH leakage rate.
Detection of the expression of caspase-3
in cells by immunohistochemistry (IHC)
Different groups of NCI-H157 cells (2×104
ml) were seeded onto glass coverslips for 48 h. After incubation
with fresh NHS for 6 h, the cover slips were washed thrice with
PBS, 2% PBS-paraformaldehyde solution was added for 15 min at room
temperature and 0.4% Triton X-100 was added at room temperature for
15 min. After washing thrice with PBS, the cells were treated with
30% H2O2. The caspase-3 protein expression
levels were measured by IHC staining. In brief, cells were
incubated in blocking buffer at room temperature for 20 min,
followed by the rabbit antihuman caspase-3 antibody (1:500 Abcam,
Cambridge, MA, USA) at 4°C overnight followed by an additional
washing step (3X) with PBS. Secondary antibody labeled with HRP
(mouse anti-rabbit) was added and incubated at 37°C for 1 h
followed by washing (3X) with PBS. SABC was added at 37°C for 20
min and DAB at room temperature for 5–30 min and the results were
observed under a light microscope. Positive cells were stained
brownish yellow. Caspase-3 protien levels were determined by
positive index = positive percentage (%) × mean optical density ×
100.
Determination of Bcl-2 and Fas protein by
western blotting
Total protein extraction of different groups of
NCI-H157 was performed, as described above. Twenty microgram (20
μg) was separated using 8 and 5% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and transferred to PVDF
membranes. The membrane was incubated with anti-bcl-2 (1:500),
anti-Fas (1:10,000) and anti-β-actin (1:200) antibodies (Santa
Cruz), followed by incubation with horseradish
peroxidase-conjugated goat anti-mouse secondary antibody. Western
blots were developed by chemiluminescence and bands were quantified
by using Quantity One 6.4.0 (Bio-Rad) software. CD59 levels were
normalized to β-actin.
Xenograft studies
Athymic male nude mice (5- to 6-week-old) were
purchased from Laboratory Animal Centre of Guangxi Medical
University and housed under pathogen-free conditions. All animal
experiments were reviewed and approved by the Institutional Animal
Care and Use Committee of the Guangxi Medical University. Thirty
mice were randomly divided into three groups of 10 each: control
group, siCD59 groups and siCD59-ctr groups. Each mouse of every
group was subcutaneously respectively injected with
2×105 luciferase-expressing Luc-NCI-H157 cell, siCD59
and siCD59-ctr cells (Invitrogen) into the right flank. Tumor
growth/regression was monitored every 5 day by in vivo
imaging after intraperitoneal injection of firefly luciferin (150
mg/kg) to the mice using an non-invasive imaging system (Roper,
USA). Each mouse cohort was also monitored for 60 days to determine
the tumor volume and the survival rate.
Determination of CD59 mRNA and CD59,
Bcl-2 and Fas protein expression in vivo
Mice were sacrificed at the end of the experiments
by CO2 asphyxiation. Tumors were excised and immediately
snap-frozen in liquid nitrogen. Total RNA and protein extraction of
tumor tissue were frozen in liquid nitrogen. CD59 mRNA expression
was determined by RT-PCR and CD59, Bcl-2, Fas protein was
determined by western blotting.
Statistical analysis
Data were analyzed using GraphPad Prism 5.0
statistical software. Quantitative data were expressed as the means
± standard deviation. Significant difference was determined by
P-values <0.05.
Results
Expression of CD59 in lung cancer
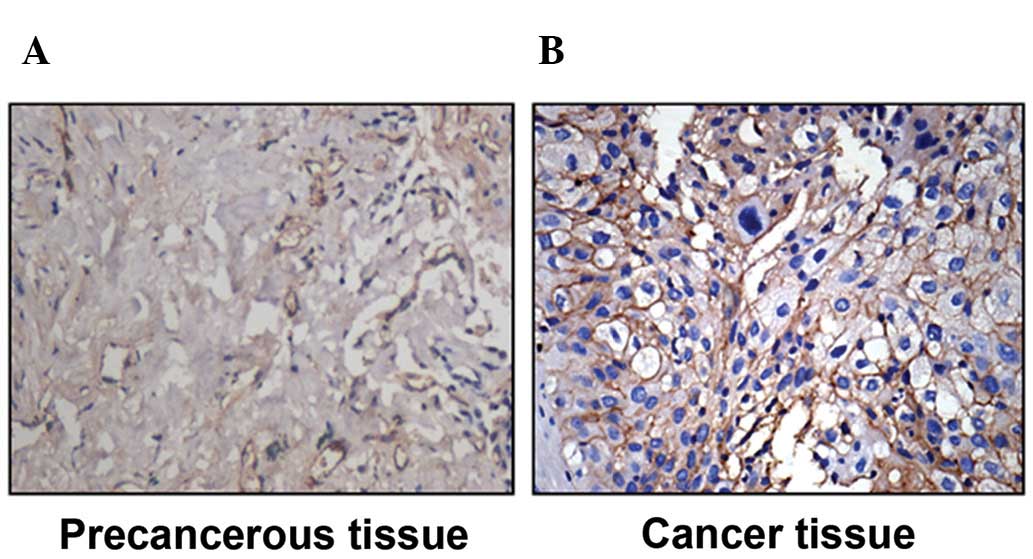
CD59 expression was significantly higher in tissues
from non-small cell lung carcinoma (NSCLC) than in preneoplastic
tissue (67.6 vs. 4.3%, P<0.05) (Fig. 1).
CD59-siRNA efficiently suppressed CD59
expression
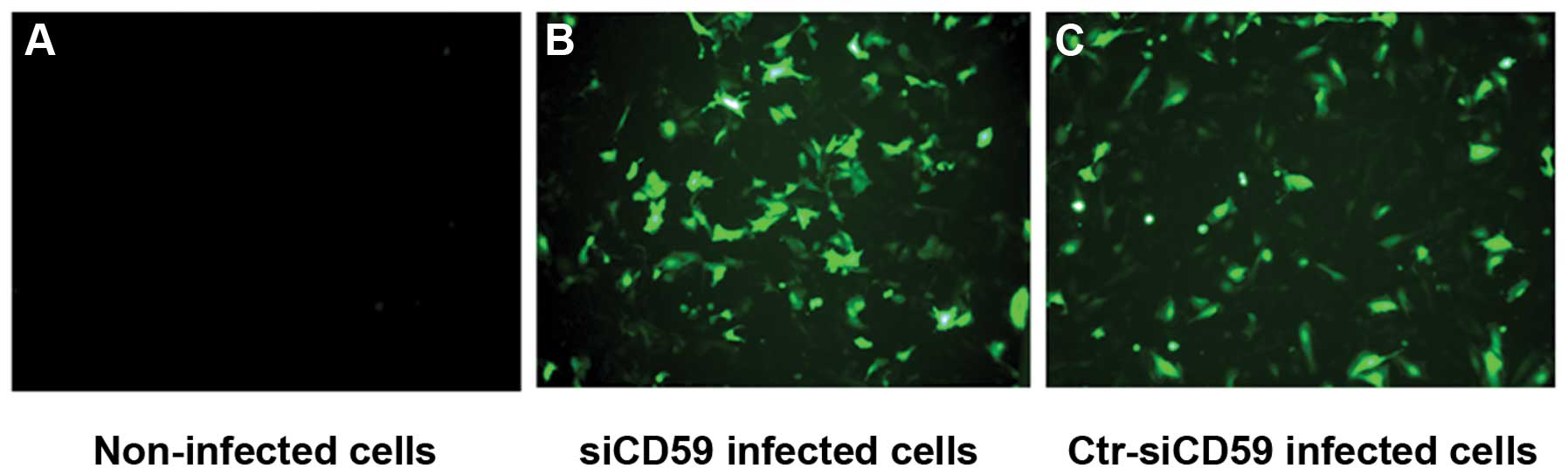
Recombinant retroviruses siCD59 and siCD59-control
(siCD59-C) both contain a green fluorescent protein (GFP) reporter
gene, which allowed for measuring infection efficiency in NCI-H157
cells. Forty-eight hours post-infection, the infection efficiency
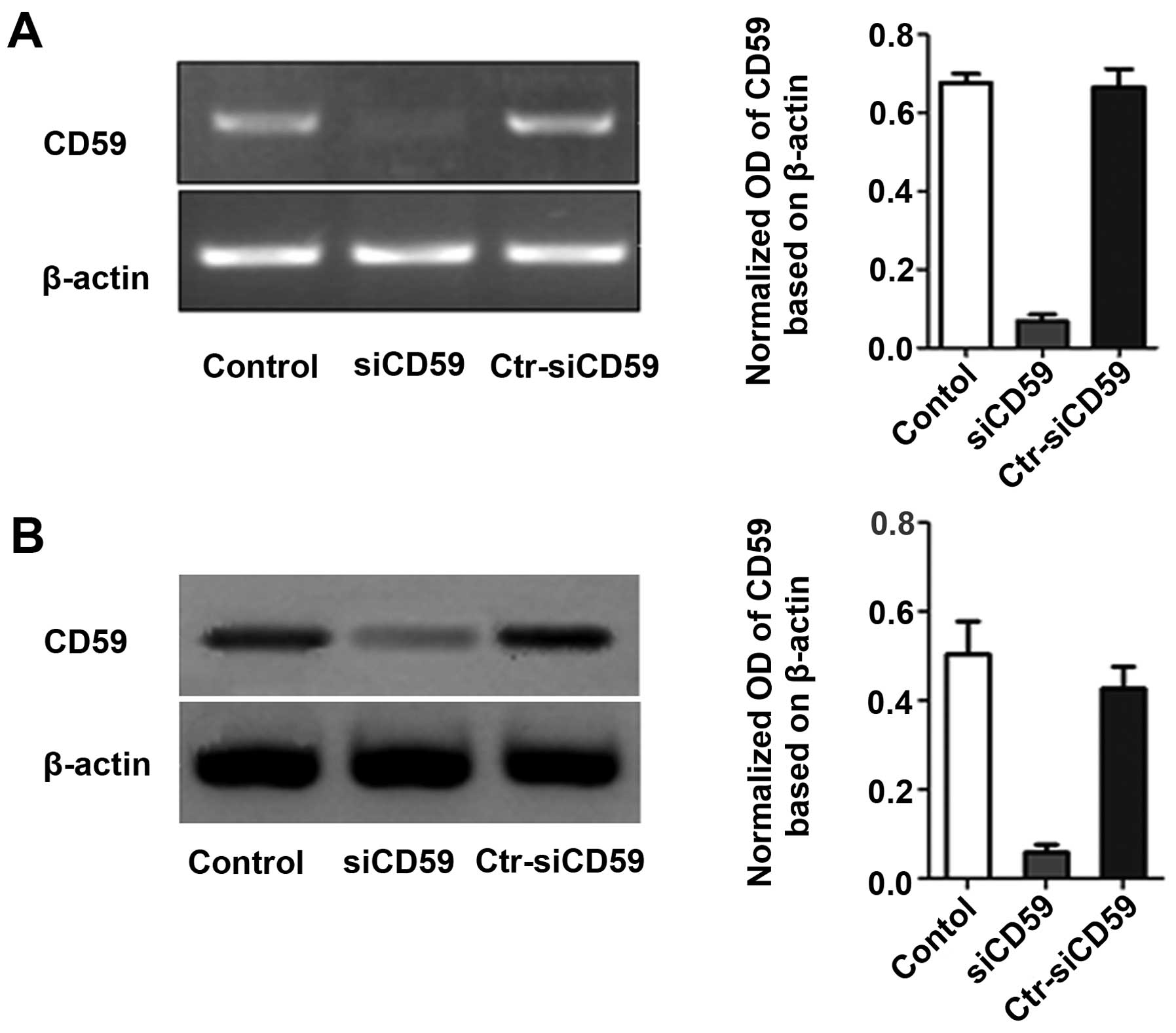
was ~70% in both siCD59 and siCD59-C infected cells (Fig. 2). CD59 mRNA and protein levels were
decreased significantly in siCD59 infected cells compared to
siCD59-C infected cells (Fig.
3).
Effect of CD59-siRNA on lung cancer cell
proliferation and apoptosis
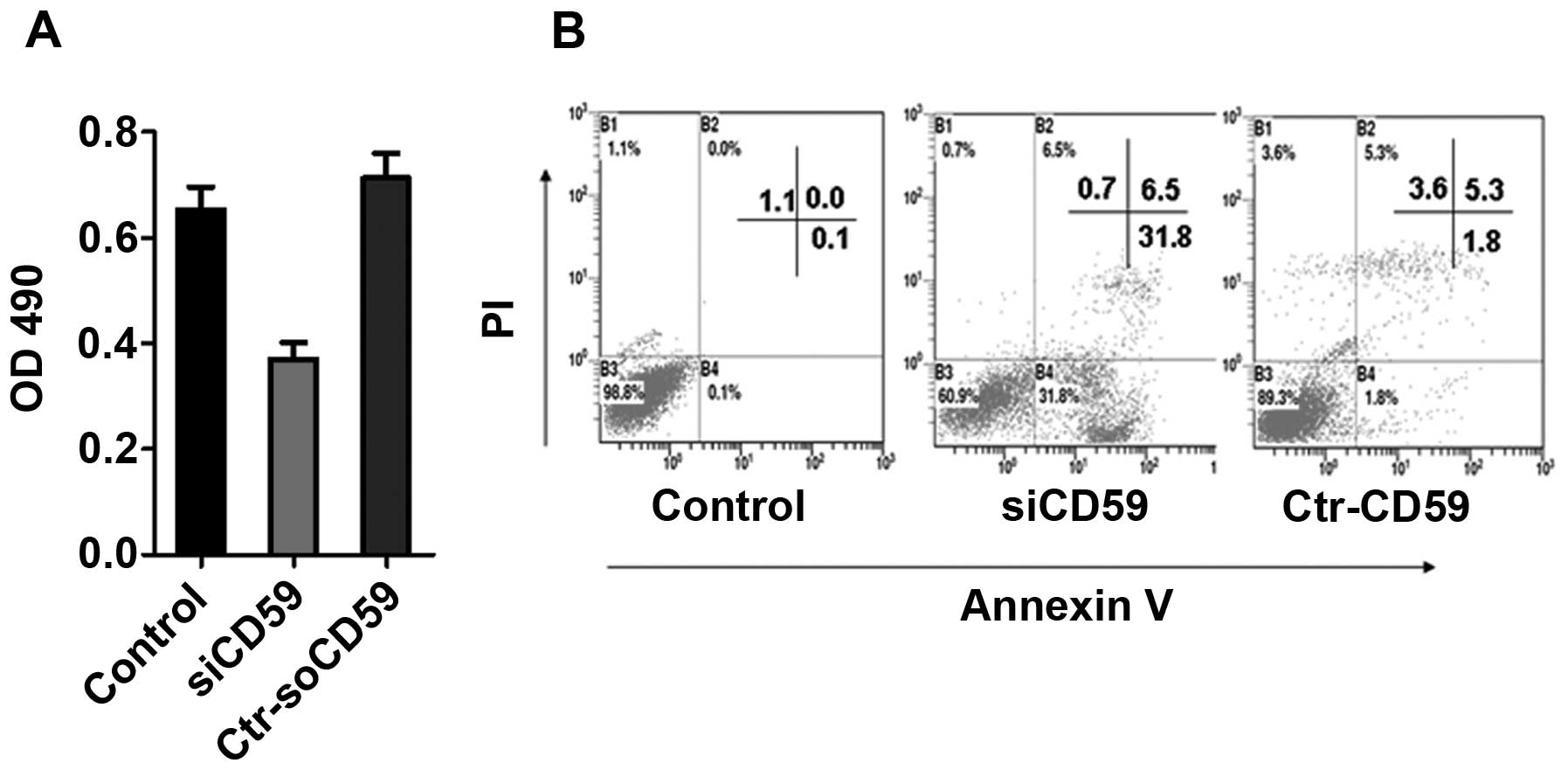
To determine the effect of CD59 knockdown on the
growth of NCI-H157 cells, an MTT assay was performed on NCI-H157
cells, siCD59 and siCD59-C transfected NCI-H157 cells.
SiCD59-transfected NCI-H157 cells displayed a significant decrease
in growth rate compared to siCD59-C-transfected cells and NCI-H157
cells (Fig. 4A). To determine the
effects of CD59 knockdown on cell apoptosis, flow cytometry were
used. Knockdown of CD59 increased apoptosis of NCI-H157 cells
compared to NCI-H157 cells and siCD59-ctr cells (31.8% P>0.1%,
P>0.05; 31.8% P>0.18%, P>0.05) (Fig. 4B).
CD59-siRNA reduced cell viability and
increased cell damage when treated with complement
The viability and cellular DNA damage of
siCD59-NCI-H157 transfected cells was also reduced and increased,
respectively, compared to the siCD59-C cells and NCI-H157 cells
(Table I).
 | Table ILDH test. |
Table I
LDH test.
| Groups | LDH release
rate |
|---|
| Cells only | 9.6±1.3 |
| Cells + normal
serum | 17.4±3.7 |
| siCD59 + normal
serum | 63.5±5.3 |
| Ctr-siCD59 + normal
serum | 16.3±4.1 |
CD59-siRNA affects expression of
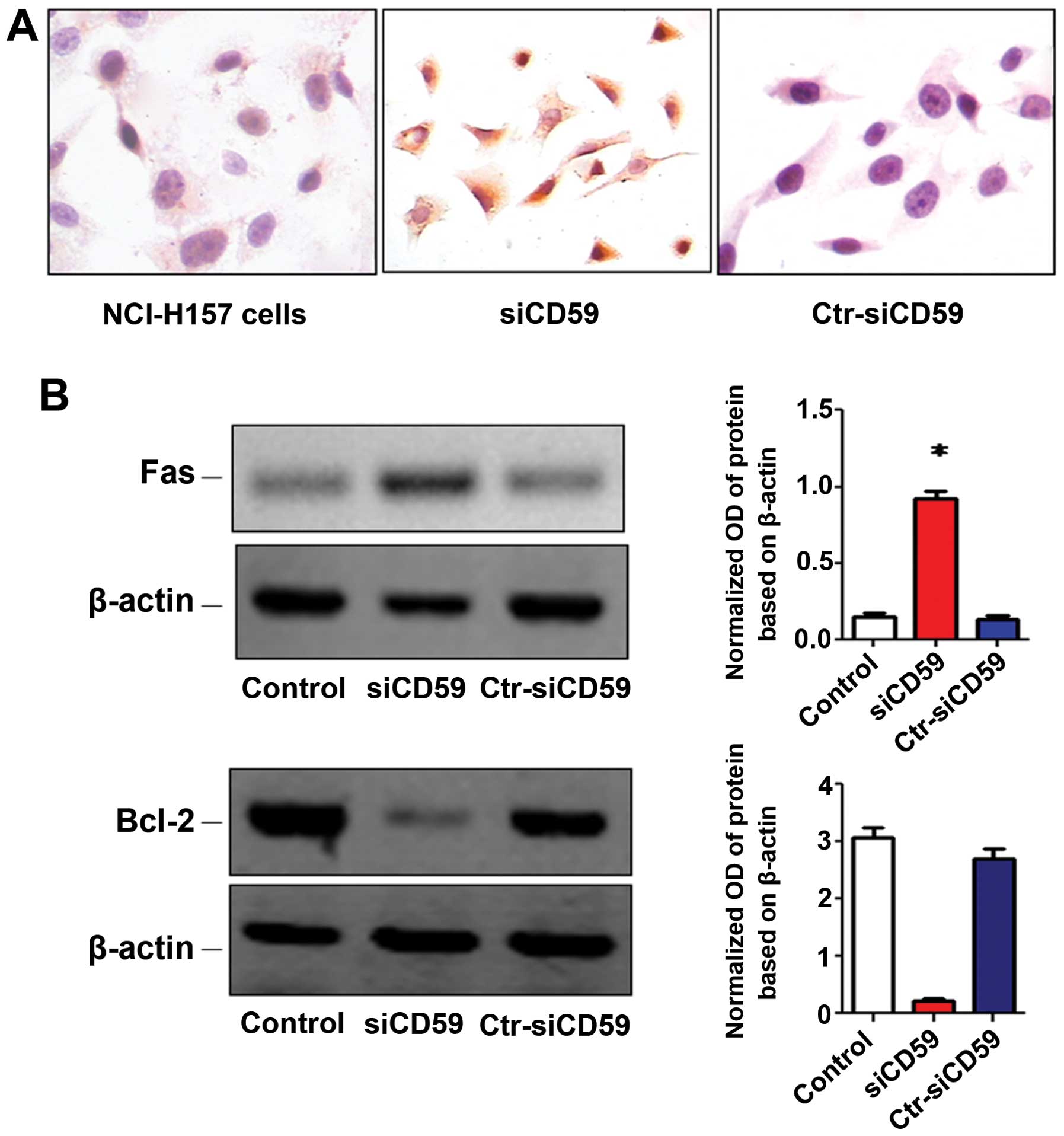
caspase-3, Fas and Bcl-2
Shi et al(6)
demonstrated that apoptosis could modulate the expression of CD59.
IHC and western blot analysis showed increased caspase-3 (Fig. 5A) and Fas (Fig. 5B) levels while Bcl-2 (Fig. 5B) levels were decreased in
siCD59-transfected NCI-H157 cells and NCI-H157 cells. These results
indicate that CD59 regulates apoptosis in non-small cell lung
cancer cells.
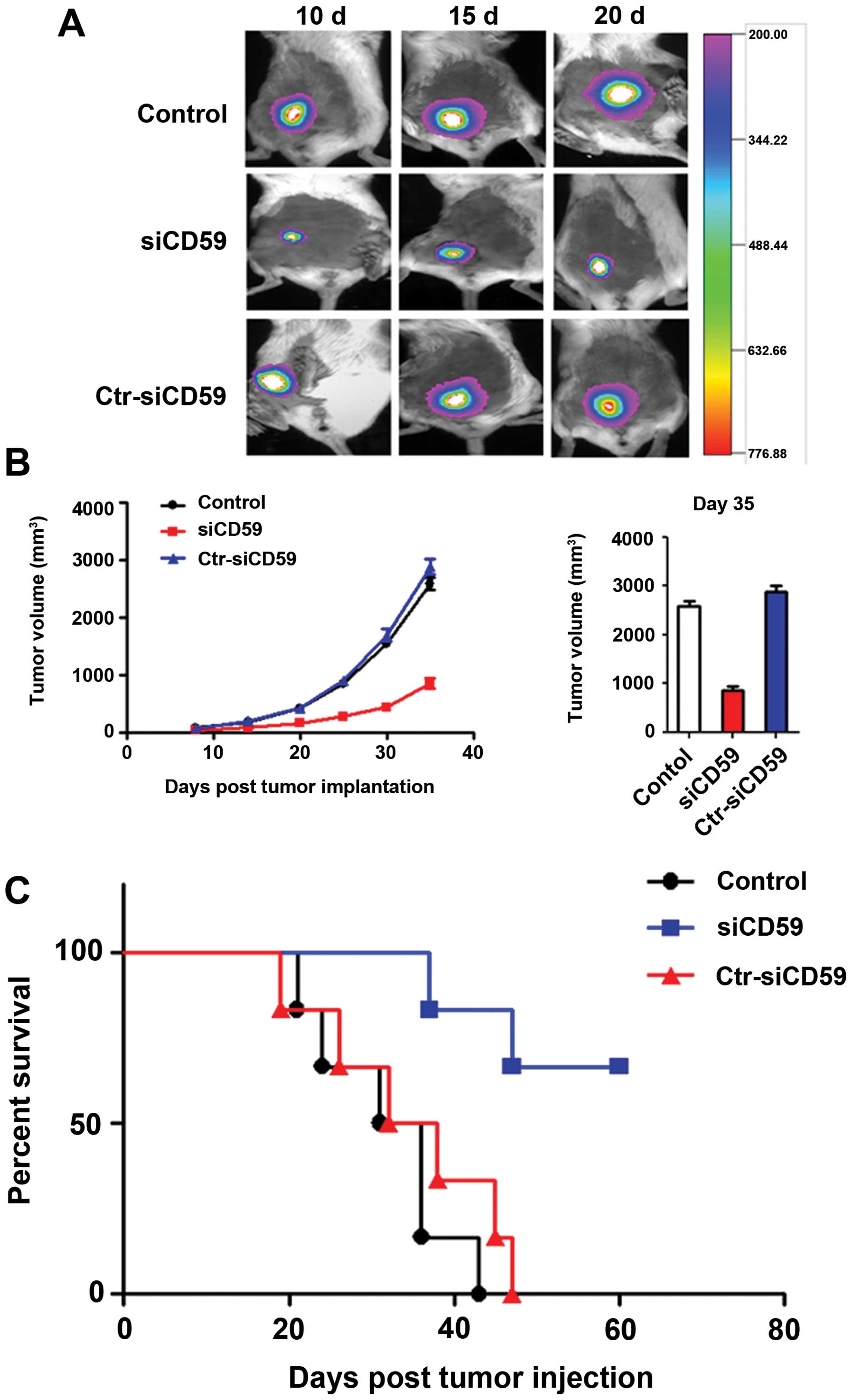
Knockdown of CD59 suppresses lung cancer
cell growth in vivo
To test whether loss of CD59 can suppress lung
cancer progression, we performed a xenograft study whereby
different groups of NCI-H157 cells were subcutaneously injected
into mice. Tumors were allowed to reach a size such that
~108 photons/sec/cm2 were emitted following
luciferin processing. Subsequently, luciferase signals in tumors of
mice were detected in 10, 15 and 20 days after post-injection.
Compared to s-CD59-C tumors and NCI-H157 cells, significant
decrease in tumor burden was observed upon CD59 knockdown (Fig. 6A and B). The survival rate of
tumor-bearing mice at 60 days was 70, 0 and 0% in
siCD59-transfected tumors group, siCD59-C tumors group and NCI-H157
tumors group (Fig. 6C).
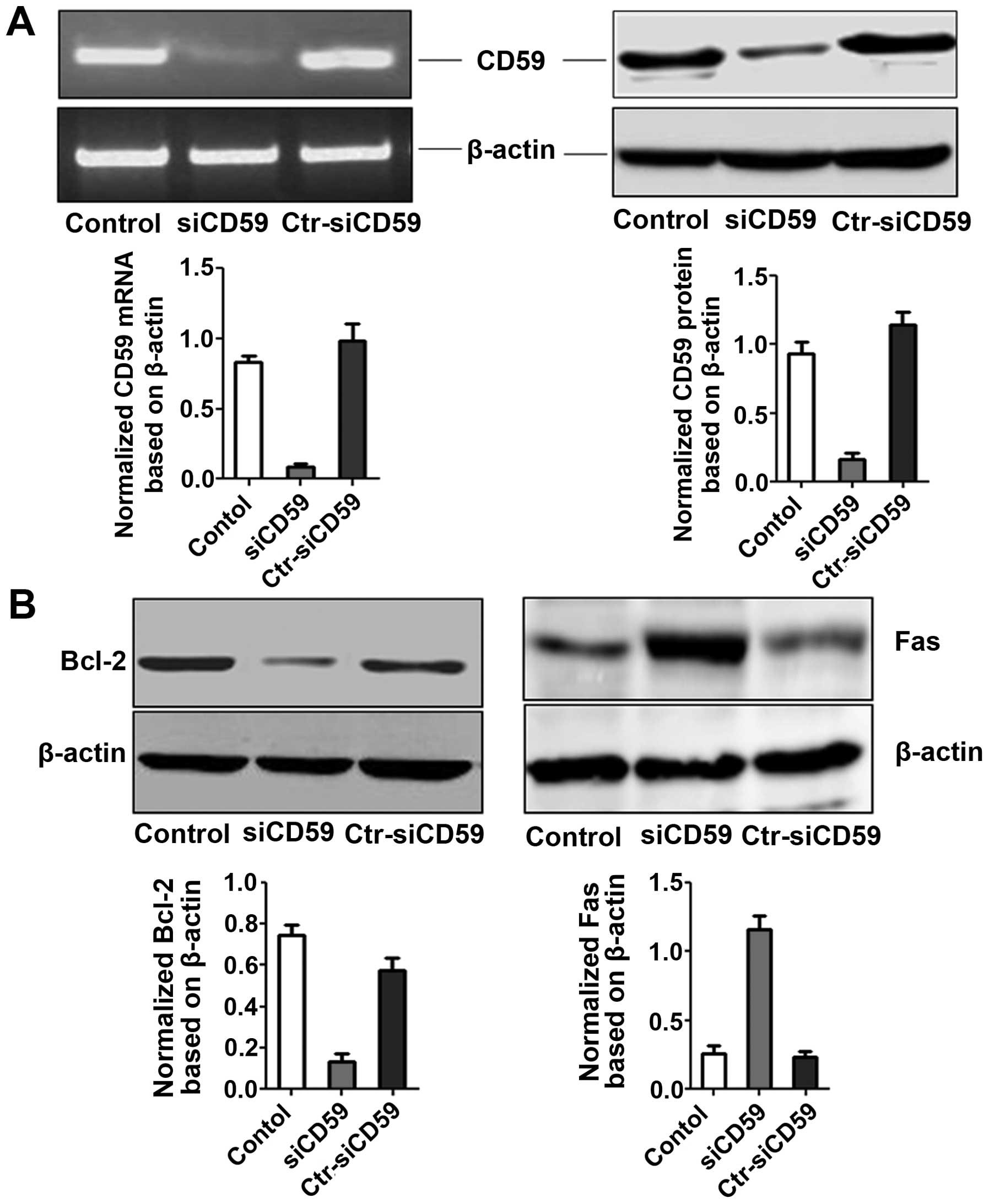
In vivo expression of CD59 mRNA and CD59,
Bcl-2 and Fas protein
Total RNA and protein were extracted from tumor
tissues to determine CD59 levels. Robust decrease in CD59 levels at
both the RNA and protein levels were observed in siCD59 cells
compared to siCD59-C cells and NCI-H157 cells (Fig. 7A). Furthermore, Fas and Bcl-2
expression was significantly increased and decreased, respectively,
in siCD59 cells compared to siCD59-C and NCI-H157 cells (Fig. 7B).
Discussion
Human CD59 is an 18–20 kDa protein anchored through
glycanphosphatidylinositol (GPI) to the cell membrane (19). CD59 belongs to the members of Ly6
superfamily (20). The functions
of CD59 protein are mainly involved in the following three aspects.
First, CD59 functions as an inhibitor of the C5b-9 membrane attack
complex (MAC) of human complement (21). Second, CD59 acts as the second
signal stimulant, inducing the activation of T lymphocytes and
taking part in the regulatory course of immunoreactions (22). Third, CD59 is the ligand of CD2
that can conglutinate with CD59. CD59-CD2 complex activates T cells
and then guides adhesion of T and T cells or T and other tissue
cells and further regulates the growth of tissue cells (23).
Previous studies have shown that CD59 is highly
expressed in many types of tumors, including breast cancer
(24–27), colorectal cancer (26,28,29),
ovarian cancer (30), malignant
gliomas (31), malignant lymphomas
(32), prostate cancer (33) and pancreatic cancer (34) and it directly or indirectly
participates in carcinogenesis and tumorigenesis. Our studies
focused on the role of CD59 during tumorigenesis of NSCLC cells
(NCI-H157) upon silencing CD59.
In the present study, we first detected high
expression of CD59 in tissues of patients with lung cancer
(Fig. 1). CD59 expression in
non-small cell lung cancer tissues is much higher than in the
surrounding tissue based on IHC. This suggests that CD59 might be a
new biomarker for lung cancer progression.
RNA interference has emerged as a genetic tool for
silencing gene expression. Because siRNA can be integrated into the
host genome, long-term gene silencing can be achieved of siRNAs
(17,35). Retroviral infection of siCD59 to
NCI-H157 cells was able to significantly decrease CD59 mRNA and
protein levels compared to siCD59-C cells (Fig. 3). Knockdown of CD59 decreased cell
viability and increased cell damage and apoptosis of lung cancer
cells (Fig. 4). Furthermore, CD59
knockdown also reduced tumor growth in vivo (Fig. 6).
Loss of CD59 can induce complement-mediated
cytolysis and lead to apoptosis of tumor cells (5). Complement-mediated cytolysis can also
assist in apoptosis by inducing phagocytosis (36,37).
MAC-triggered cell death can occur through a caspase-dependent
pathway, specifically via caspase-3. Animal models of renal disease
also implicate that MAC can trigger apoptosis (38–40).
Korty et al demonstrated that CD59 can also increase calcium
flux to increase cytoplasmic calcium levels which induces
mitochondrial DNA damage and cytochrome c(41). Fas expression also initiates
apoptosis under certain conditions and Fas antigen and ICAM-1
molecule can have a synergistic effect (42). Bcl-2 has also emerged as a
proto-oncogene that blocks programmed cell death independently of
promoting cell division (43).
Bcl-2 can inhibit the synthesis and activation of caspase-3 to
inhibit apoptosis. Moreover, Bcl-2 can also be degraded by
caspase-3 a specific enzyme, which activates apoptosis. Apoptosis
can inhibit tumorigenesis by removing unwanted and damaged cells
(52).
Cell apoptosis typically occurs during tumor
development and regression. Therefore, an important mechanism to
prevent tumorigenesis is the induction of cell apoptosis that takes
place continuously in many tissues to remove unwanted, damaged or
aberrant cells. In this study, we investigated the effects of CD59
silencing to understand the biochemical mechanism underlying CD59
inactivation-induced apoptosis. We also studied the role of CD59 in
regulating the growth and apoptosis of NCI-H157 cells. We showed
that CD59 knockdown significantly decreased the growth and
increased apoptosis of NCI-H157 cells compared to siCD59-C cells
(Fig. 4), consistent with previous
observations (44,45). CD59-induced apoptosis might be
mediated by MAC. Further studies are needed to understand this
mechanism.
NCI-H157 cells treated by human complement (NHS, 8%,
v/v) were used to assess MAC-mediated cytolysis. We showed that
CD59 knockdown significantly inhibited the viability of NCI-H157
cells (Table I), decreased Bcl-2
expression and increased caspase-3 and Fas expression (Fig. 5). Caspases are executioners of
apoptosis and regulate Fas/TNF-R1, mitochondria dysfunction and
TNF-related apoptosis-inducing ligand (TRAIL). Caspase-3 activation
was observed upon Fas/TNF-R1 treatment (Fig. 5A). Overall, the present data
suggest that the loss of CD59 induces caspase-dependent apoptosis
in cultured cells.
To investigate the inhibitory role of siCD59 in
vivo, siCD59 infected cells were injected into athymic nude
mice. Knockdown of CD59 significantly decreased tumor weight and
growth and increased the survival rate of mice compared to siCD59-C
cells and NCI-H157 cells (Fig. 6),
which suggests that silencing CD59 gene expression could markedly
inhibit the growth of cancer in vivo. The silencing of CD59
expression in tumor tissue was confirmed by RT-PCR and western
blotting (Fig. 7A), which showed
significantly less CD59 expression in siCD59 group than in siCD59-C
group, or the NCI-H157 cell group. Decreased tumor growth could be
attributed to apoptosis, as indicated by Fas and Bcl-2 expression
in these tumor tissues (Fig. 7B).
The present data suggest that the CD59 loss induces
caspase-dependent apoptosis in vitro and in vivo.
Overall, our study illustrates that CD59 is
increased in human lung cancer and loss or inactivation of CD59 can
lead to apoptosis of NCI-H157 lung cancer cells. This may be
mediated by inducing Fas expression on the surface of NCI-H157
cells, leading to apoptosis through caspase-3 activation and
complement-induced cytolysis. This might provide insight into new
treatment for lung cancer patients by inhibiting CD59
expression.
In conclusion, the present study demonstrates that
CD59 is overexpressed in human lung cancer and retroviral-mediated
RNAi delivery is an efficient system for CD59 gene silencing of
human lung cancer. Suppression of CD59 expression enhanced
complement-mediated cytolysis of lung cancer cells, which may be
mediated by induction of Fas expression on the surface of NCI-H157
cells, leading to apoptosis through caspase-3 activation, CD59 may
serve as a candidate targeting gene in gene therapy for human
carcinomas such as lung, ovarian, prostate carcinoma and cervical
carcinoma.
Acknowledgements
This study was supported by Grants from the
Scientific Research and Technological Development Projects of
Guangxi, China (no. 0816004-8).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Fidias P and Novello S: Strategies for
prolonged therapy in patients with advanced non-small-cell lung
cancer. J Clin Oncol. 28:5116–5123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fuld AD, Dragnev KH and Rigas JR:
Pemetrexed in advanced non-small-cell lung cancer. Expert Opin
Pharmacother. 11:1387–1402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Whitehurst AW, Bodemann BO, Cardenas J, et
al: Synthetic lethal screen identification of chemosensitizer loci
in cancer cells. Nature. 446:815–819. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li B, Chu X, Gao M and Xu Y: The effects
of CD59 gene as a target gene on breast cancer cells. Cell Immunol.
272:61–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi XX, Zhang B, Zang JL, Wang GY and Gao
MH: CD59 silencing via retrovirus-mediated RNA interference
enhanced complement-mediated cell damage in ovary cancer. Cell Mol
Immunol. 6:61–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fonsatti E, Altomonte M, Coral S, et al:
Emerging role of protectin (CD59) in humoral immunotherapy of solid
malignancies. Clin Ter. 151:187–193. 2000.PubMed/NCBI
|
|
8
|
Chen S, Caragine T, Cheung NK and
Tomlinson S: CD59 expressed on a tumor cell surface modulates
decay-accelerating factor expression and enhances tumor growth in a
rat model of human neuroblastoma. Cancer Res. 60:3013–3018.
2000.PubMed/NCBI
|
|
9
|
Wickham SE, Hotze EM, Farrand AJ, et al:
Mapping the intermedilysin-human CD59 receptor interface reveals a
deep correspondence with the binding site on CD59 for complement
binding proteins C8alpha and C9. J Biol Chem. 286:20952–20962.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gelderman KA, Tomlinson S, Ross GD and
Gorter A: Complement function in mAb-mediated cancer immunotherapy.
Trends Immunol. 25:158–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fishelson Z, Donin N, Zell S, Schultz S
and Kirschfink M: Obstacles to cancer immunotherapy: expression of
membrane complement regulatory proteins (mCRPs) in tumors. Mol
Immunol. 40:109–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fonsatti E, Di Giacomo AM and Maio M:
Optimizing complement-activating antibody-based cancer
immunotherapy: a feasible strategy? J Transl Med. 2:212004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sioud M: Promises and challenges in
developing RNAi as a research tool and therapy. Methods Mol Biol.
703:173–187. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashihara E: RNA interference for cancer
therapies. Gan To Kagaku Ryoho. 37:2033–2041. 2010.PubMed/NCBI
|
|
15
|
Ashihara E, Kawata E and Maekawa T: Future
prospect of RNA interference for cancer therapies. Curr Drug
Targets. 11:345–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:550–553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sui G, Soohoo C, Affar el B, Gay F and Shi
Y, Forrester WC and Shi Y: A DNA vector-based RNAi technology to
suppress gene expression in mammalian cells. Proc Natl Acad Sci
USA. 99:5515–5520. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shafer SH and Williams CL: Non-small and
small cell lung carcinoma cell lines exhibit cell type-specific
sensitivity to edelfosine-induced cell death and different cell
line-specific responses to edelfosine treatment. Int J Oncol.
23:389–400. 2003.PubMed/NCBI
|
|
19
|
Geis N, Zell S, Rutz R, et al: Inhibition
of membrane complement inhibitor expression (CD46, CD55, CD59) by
siRNA sensitizes tumor cells to complement attack in vitro. Curr
Cancer Drug Targets. 10:922–931. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fleming TJ, O’HUigin C and Malek TR:
Characterization of two novel Ly-6 genes. Protein sequence and
potential structural similarity to alpha-bungarotoxin and other
neurotoxins. J Immunol. 150:5379–5390. 1993.
|
|
21
|
Sugita Y, Nakano Y, Oda E, Noda K, Tobe T,
Miura NH and Tomita M: Determination of carboxyl-terminal residue
and disulfide bonds of MACIF (CD59), a
glycosyl-phosphatidylinositol-anchored membrane protein. J Biochem.
114:473–477. 1993.
|
|
22
|
Treon SP, Shima Y, Grossbard ML, Preffer
FI, Belch AR, Pilarski LM and Anderson KC: Treatment of multiple
myeloma by antibody mediated immunotherapy and induction of myeloma
selective antigens. Ann Oncol. 11(Suppl 1): 107–111. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zaltzman AB, Van den Berg CW, Muzykantov
VR and Morgan BP: Enhanced complement susceptibility of
avidin-biotin-treated human erythrocytes is a consequence of
neutralization of the complement regulators CD59 and decay
accelerating factor. Biochem J. 307:651–656. 1995.
|
|
24
|
Hakulinen J and Meri S: Expression and
function of the complement membrane attack complex inhibitor
protectin (CD59) on human breast cancer cells. Lab Invest.
71:820–827. 1994.PubMed/NCBI
|
|
25
|
Madjd Z, Pinder SE, Paish C, Ellis IO,
Carmichael J and Durrant LG: Loss of CD59 expression in breast
tumours correlates with poor survival. J Pathol. 200:633–639. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thorsteinsson L, O’Dowd GM, Harrington PM
and Johnson PM: The complement regulatory proteins CD46 and CD59,
but not CD55, are highly expressed by glandular epithelium of human
breast and colorectal tumour tissues. APMIS. 106:869–878. 1998.
View Article : Google Scholar
|
|
27
|
Macor P, Mezzanzanica D, Cossetti C,
Alberti P, Figini M, Canevari S and Tedesco F: Complement activated
by chimeric anti-folate receptor antibodies is an efficient
effector system to control ovarian carcinoma. Cancer Res.
66:3876–3883. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koretz K, Brüderlein S, Henne C and Moller
P: Expression of CD59, a complement regulator protein and a second
ligand of the CD2 molecule, and CD46 in normal and neoplastic
colorectal epithelium. Br J Cancer. 68:926–931. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hosch SB, Scheunemann P, Lüth M, et al:
Expression of 17-1A antigen and complement resistance factors CD55
and CD59 on liver metastasis in colorectal cancer. J Gastrointest
Surg. 5:673–679. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bjørge L, Hakulinen J, Wahlström T, Matre
R and Meri S: Complement-regulatory proteins in ovarian
malignancies. Int J Cancer. 70:14–25. 1997.
|
|
31
|
Mäenpää A, Junnikkala S, Hakulinen J,
Timonen T and Meri S: Expression of complement membrane regulators
membrane cofactor protein (CD46), decay accelerating factor (CD55),
and protectin (CD59) in human malignant gliomas. Am J Pathol.
148:1139–1152. 1996.
|
|
32
|
Treon SP, Mitsiades C, Mitsiades N, Young
G, Doss D, Schlossman R and Anderson KC: Tumor cell expression of
CD59 is associated with resistance to CD20 serotherapy in patients
with B-cell malignancies. J Immunother. 24:263–271. 2001.
View Article : Google Scholar
|
|
33
|
Jarvis GA, Li J, Hakulinen J, Brady KA,
Nordling S, Dahiya R and Meri S: Expression and function of the
complement membrane attack complex inhibitor protectin (CD59) in
human prostate cancer. Int J Cancer. 71:1049–1055. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Crnogorac-Jurcevic T, Efthimiou E, Nielsen
T, et al: Expression profiling of microdissected pancreatic
adenocarcinomas. Oncogene. 21:4587–4594. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu CM, Liu DP, Dong WJ and Liang CC:
Retrovirus vector-mediated stable gene silencing in human cell.
Biochem Biophys Res Commun. 313:716–720. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Riedemann NC, Guo RF, Laudes IJ, et al:
C5a receptor and thymocyte apoptosis in sepsis. FASEB J.
16:887–888. 2002.PubMed/NCBI
|
|
37
|
Guo RF, Huber-Lang M, Wang X, et al:
Protective effects of anti-C5a in sepsis-induced thymocyte
apoptosis. J Clin Invest. 106:1271–1280. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Niculescu T, Weerth S, Soane L, et al:
Effects of membrane attack complex of complement on apoptosis in
experimental autoimmune encephalomyelitis. Ann NY Acad Sci.
1010:530–533. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nauta AJ, Daha MR, Tijsma O, van de Water
B, Tedesco F and Roos A: The membrane attack complex of complement
induces caspase activation and apoptosis. Eur J Immunol.
32:783–792. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hughes J, Nangaku M, Alpers CE, Shankland
SJ, Couser WG and Johnson RJ: C5b-9 membrane attack complex
mediates endothelial cell apoptosis in experimental
glomerulonephritis. Am J Physiol Renal Physiol. 278:F747–F7757.
2000.PubMed/NCBI
|
|
41
|
Korty PE, Brando C and Shevach EM: CD59
functions as a signal-transducing molecule for human T cell
activation. J Immunol. 146:4092–4098. 1991.PubMed/NCBI
|
|
42
|
Möller P, Henne C, Leithäuser F, et al:
Coregulation of the APO-1 antigen with intercellular adhesion
molecule-1 (CD54) in tonsillar B cells and coordinate expression in
follicular center B cells and in follicle center and mediastinal
B-cell lymphomas. Blood. 81:2067–2075. 1993.
|
|
43
|
Grobholz R, Zentgraf H, Köhrmann KU and
Bleyl U: Bax, Bcl-2, fas and Fas-L antigen expression in human
seminoma: correlation with the apoptotic index. APMIS. 110:724–732.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jha P, Sohn JH, Xu Q, Wang Y, Kaplan HJ,
Bora PS and Bora NS: Suppression of complement regulatory proteins
(CRPs) exacerbates experimental autoimmune anterior uveitis (EAAU).
J Immunol. 176:7221–7231. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Donev RM, Gray LC, Sivasankar B, Hughes
TR, van den Berg CW and Morgan BP: Modulation of CD59 expression by
restrictive silencer factor-derived peptides in cancer
immunotherapy for neuroblastoma. Cancer Res. 68:5979–5987. 2008.
View Article : Google Scholar : PubMed/NCBI
|