Introduction
Lung cancer is still the leading cause of cancer
death in both sexes throughout the world. More people die of lung
cancer than of colon, breast, and prostate cancers combined, i.e.,
more than 1.2 million deaths each year (1). Non-small cell lung cancer (NSCLC)
accounts for approximately 85–90% of all lung cancers. The poor
prognosis of NSCLC is mainly due to late diagnosis, in as much as
only 20 to 30% of patients are eligible for surgical resection.
Despite the development of new chemotherapeutic drugs and
multimodal treatment strategies, the survival rate of NSCLC remains
unchanged and poor. Identification of new prognostic markers for
the characterization of lung-cancer biology may be helpful, as they
could serve as a basis for predicting response to radiation and
chemotherapy at a molecular level.
The alterations in epigenomes such as DNA
methylation and histone modifications play pivotal roles in
carcinogenesis (2–4). It has been reported that DNA
methylation level and global histone modification patterns may be
possible predictors of cancer recurrence and prognosis in a large
variety of cancer entities (5,6).
Post-translational modification of histone tails alters the
physical state of chromatin and has an essential role in both
transcriptional repression and activation during embryonic
development, lineage specification, terminal differentiation and
tumorigenesis as well (7,8). One such repressive modification, the
trimethylation of lysine 27 on histone H3 (H3K27me3), seemed to be
an epigenetic label mediating gene silencing; and a mark for de
novo DNA methylation in cancer cells by recruitment of DNA
methyltransferase (DNMTs) (9–11),
contributing to tumor progression through suppression of a certain
gene expression (12). In fact,
many recent studies have revealed that H3K27me3 may be involved in
the characterization of various types of human cancers, including
breast cancer (5,13), hepatocellular carcinoma (14), prostate cancer (15), Hodgekin’s lymphoma (16), esophageal cancer (17) and nasopharyngeal carcinoma
(18). Reports of H3K27me3 levels
in different cancer samples are somewhat contradictory. It is
demonstrated that low H3K27me3 levels predicted poor outcome in
breast, ovarian and pancreatic cancers (5) while high levels predicted poor
outcome in hepatocellular carcinoma (14) and esophageal squamous cell
carcinoma (17).
Moreover, H3K27 methylation is catalyzed by the SET
domain of its specific methyltransferase, enhancer of zeste homolog
2 (EZH2), and requires the presence of 2 additional proteins, i.e.,
suppressor of zeste 12 (SUZ12) and embryonic ectoderm development
(EED) (19). These proteins,
together with the histone binding proteins retinoblastoma binding
protein 4 (RBBP4) and RBBP7, comprise the core components of the
polycomb repressive complex 2 (PRC2). Overexpression of EZH2 was
also found in a variety of cancers, including lung cancer (20,21),
breast cancer (5,22–26),
melanoma (27), colorectal
(20) and pancreatic
adenocarcinoma (28) and ovarian
carcinoma (29), and turned out to
be closely associated with high proliferation rate and aggressive
tumor subgroups, resulting in worse clinical outcome thereafter.
Although many reports on the role of H3K27me3 in carcinogenesis are
available, its carcinogenic role in NSCLC and how it interacts with
EZH2 and DNA methylation remain unclear.
In the present study, we investigated the prognostic
value of immunostaining for H3K27me3 and its relationship with EZH2
expression in NSCLC patients. We examined the correlation between
these parameters and the level of DNA methylation at CCGG sites,
detected by histo-endonuclease-linked detection of methylation
sites of DNA (HELMET) (30). Since
there had been several reports to indicate that H3K27me3 and EZH2
were involved in the early stage of various cancers, we focused on
stage I NSCLCs in this study.
Materials and methods
Patients and tissue preparation
Five normal lung tissue and 42 NSCLC patients (22
adenocarcinomas and 20 squamous cell carcinomas) with early-stage
(stage I) were included in our study. Patients underwent radical
resection of primary tumor (lobectomy or pneumonectomy) and
systematic lymph-adenectomy at the First Department of Surgery,
Nagasaki University Hospital (Nagasaki, Japan), between 2000 and
2006. The patients’ clinicopathological data are shown in Table I. None of the patients had received
chemo- or radiotherapy before tissue collection. The
histopathologic features of the tumor specimens were classified in
accordance with the WHO criteria. The TNM staging was determined
according to the latest National Comprehensive Cancer Network
(NCCN, Version 2, 2013) guidelines for NSCLC. The study protocol
was approved by the Human Ethics Review Committee of Nagasaki
University School of Medicine, and a signed informed consent was
obtained from each patient. Each specimen was fixed overnight in
10% buffered formalin at room temperature (RT) and embedded in
paraffin. Serial sections were cut at a thickness of 4 μm
and placed onto 3-aminopropyltriethoxysilane-coated glass slides.
Some sections were stained with hematoxylin and eosin in a routine
manner for histological examination.
 | Table I.Clinicopathologic parameters. |
Table I.
Clinicopathologic parameters.
| No. of cases (%)
|
|---|
| Parameters | Adenocarcinoma | Squamous cell
carcinoma |
|---|
| Median age,
years | 68.50 | 69.75 |
| Age (y.o.) | | |
| ≤69 | 12 (54.5) | 12 (60) |
| >69 | 10 (45.5) | 8 (40) |
| Gender | | |
| Male | 10 (45.5) | 17 (85) |
| Female | 12 (54.5) | 3 (15) |
| Serum CEA
(ng/ml) | | |
| ≤5 | 22 (100) | 17 (85) |
| >5 | 0 | 3 (15) |
| P-factor | | |
| Positive | 1 (4.5) | 0 |
| Negative | 21 (95.5) | 20 (100) |
| LV-factor | | |
| Positive | 18 (81.8) | 13 (65) |
| Negative | 4 (18.2) | 7 (35) |
| V-factor | | |
| Positive | 8 (36.4) | 10 (50) |
| Negative | 14 (63.6) | 10 (50) |
| T-stage | | |
| 1a | 16 (72.7) | 13 (65) |
| 1b | 5 (22.7) | 7 (35) |
| 2a | 1 (4.6) | |
| Nodal status | | |
| N0 | 22 (100) | 20 (100) |
|
Differentiation | | |
| Well | 13 (59.1) | 3 (15) |
| Moderate | 6 (27.3) | 9 (45) |
| Poor | 3 (13.6) | 8 (40) |
| Relapse | | |
| Yes | 2 (9.1) | 8 (40) |
| No | 20 (90.9) | 12 (60) |
| Smoking status | | |
| Smoker | 7 (31.8) | 17 (85) |
| Non-smoker | 15 (68.2) | 3 (15) |
| Postoperative
metastasis | | |
| Yes | 3 (13.6) | 7 (35) |
| No | 19 (86.4) | 13 (65) |
| Median follow-up
(months) | 75.2 | 52.9 |
Chemicals and biochemicals
Bovine serum albumin (BSA) (essentially fatty acid
and globulin-free), Trizma base, 2-mercaptoethanol,
3-aminopropyltriethoxysilane, Triton X-100, and Brij-35 were from
Sigma Chemical Co. (St. Louis, MO, USA). Sodium dodecyl sulfate
(SDS)-polyacrylamide gel electrophoresis (PAGE) reagents and the
molecular marker set were purchased from Daiichi Pure Chemicals
(Tokyo, Japan). Polyvinylidene fluoride membrane (PVDF) was
purchased from Millipore (MA, USA). Lima bean trypsin inhibitor was
purchased from Worthington Biochemical (Lakewood, NJ, USA); the
protein assay kit and Coomassie Brilliant Blue were purchased from
Bio-Rad Laboratories (Hercules, CA, USA); and
3,3′-diaminobenzi-dine-4HCl (DAB) was purchased from Dojin Chemical
Co. (Kumamoto, Japan). Biotin-16-dUTP, digoxigenin-11-dUTP,
Rhodamine anti-Dig and terminal deoxynucleotidyl transferase (TdT)
were from Roche Diagnostics (Mannheim, Germany). Dideoxy ATP
(ddATP) and dideoxy TTP (ddTTP) were from Jena Bioscience (Jena,
Germany). HpaII and MspI were from Takara Bio Inc.
(Shiga, Japan). 4′,6-diamidino-2-phenyl-indole, dihydrochloride
(DAPI) was from Invitrogen Corporation (Carlsbad, CA, USA).
Permount was from Fisher Scientific Inc. (Bridgewater, NJ, USA).
All other reagents used in this study were from Wako Pure Chemicals
(Osaka, Japan) and were of analytical grade.
Immunohistochemistry for H3K27me3, EZH2,
PCNA and simultaneous localization of EZH2 and PCNA
Immunohistochemistry was performed with the indirect
enzyme-labeled antibody method, as described previously (31–33).
Antibodies used in IHC are listed in Table II. For detection of H3K27me3, EZH2
and PCNA, paraffin-embedded sections were deparaffinized with
toluene and rehydrated in graded alcohols. After autoclaved for 15
min at 120°C in 10 mM citrate buffer (pH 6.0) for antigen
retrieval, endogenous peroxidase was inactivated with 0.3% hydrogen
peroxide in methanol for 15 min. The sections were then
pre-incubated with 500 μg/ml normal goat IgG dissolved in 1%
BSA in PBS (pH 7.4) for 1 h, reacted with primary antibodies for 16
h, washed with 0.075% Brij 35 in PBS, and then incubated with
HRP-conjugated goat anti-rabbit IgG (H3K27me3/EZH2) or
HRP-conjugated goat anti-mouse IgG (PCNA) in 1% BSA in PBS for 1 h.
After washing with 0.075% Brij 35 in PBS, the sites of HRP were
visualized with DAB and H2O2 in the presence
of nickel and cobalt ions (34).
As a negative control, some sections were reacted with normal
rabbit IgG or normal mouse IgG instead of the specific antibodies.
For simultaneous staining of EZH2 and PCNA, the sections were
incubated with Alexa 546 anti-rabbit IgG and Alexa 488 anti-mouse
IgG (both 1:500) in darkness for 1 h, then washed with 0.075% Brij
35 in PBS in darkness and finally observed with 0.5 μg/ml
DAPI for 1 min. The stained slides were analyzed under a laser
scanning microscope (LSM 5 PASCAL; Carl Zeiss Inc., Germany).
 | Table II.List of antibodies used in
immunohistochemistry. |
Table II.
List of antibodies used in
immunohistochemistry.
| Antibody | Working
dilution/concentration | Manufacturer |
|---|
| Polyclonal, rabbit
anti-human H3K27me3 | 1:200 | Cell Signaling
Technology, MA, USA |
| Monoclonal, rabbit
anti-human EZH2 | 1:400 | Cell Signaling
Technology |
| Monoclonal, mouse
anti-human PCNA (clone: PC10) | 1:400 | DakoCytomation,
Glostrup, Denmark |
| HRP-conjugated goat
anti-rabbit/mouse IgG | 1:200 | Millipore Co., CA,
USA |
| Normal goat
IgG | 1:20 | Sigma Chemical Co.,
MO, USA |
| Alexa 488
anti-mouse/Alexa546 anti-rabbit | 1:500 | Invitrogen Co., CA,
USA |
| FITC-labeled goat
anti-biotin | 1:100 | Vector
Laboratories, CA, USA |
| Rhodamine-labeled
sheep anti-digoxigenin | 1:100 | Roche Diagnostics,
Mannheim, Germany |
Quantitative evaluation
Staining results were examined by two observers
masked to patients’ clinical information. Another reading by a
third observer was needed to reach a consensus when there was a
significant discrepancy between initial readings. At least 5
high-power fields and more than 2,000 cells were calculated in each
case with a light microscope (Zeiss 2021-85; Carl Zeiss Inc.) at
×400 magnification. Immunostaining results were evaluated by using
a semi-quantitative scoring system according to the method
described in the study by Ellinger et al (15). That is, the number of positive
cancerous cells was estimated as follows (0, no positive cells; 1,
0> and ≤25% positive cells; 2, >25 and ≤50% positive cells;
3, >50 and ≤75% positive cells; and 4, >75 and ≤100% positive
cells). These scores were multiplied with an intensity scale (0,
negative; 1, weak; 2, moderate; and 3, intensive), and the final
score ranged from 0–12.
Western blot analysis of H3K27me3
Western blot analysis was carried out as detailed
previously (35). In brief, human
lung cancer specimens and normal lung tissue were homogenized, and
the lysates were centrifuged. Soluble proteins were separated on
10% SDS-PAGE gel (Daiichi Pure Chemical, Tokyo, Japan) with equal
amounts (10 μg) of protein per lane. Separated proteins were
electrophoretically transferred onto polyvinylidene difluoride
(PVDF) membranes (Millipore), blocked with 10% non-fat milk in TBS
(20 mM Tris buffer, pH 7.6, and 150 mM NaCl) for 1 h and then
incubated overnight at 4°C with rabbit polyclonal anti-H3 (Cell
Signaling Technology, MA, USA) and H3K27me3 antibody. As a
secondary antibody, HRP-goat anti-rabbit IgG was reacted for 1 h
and then the bands were visualized with DAB, Ni, Co and
H2O2.
In situ evaluation of DNA
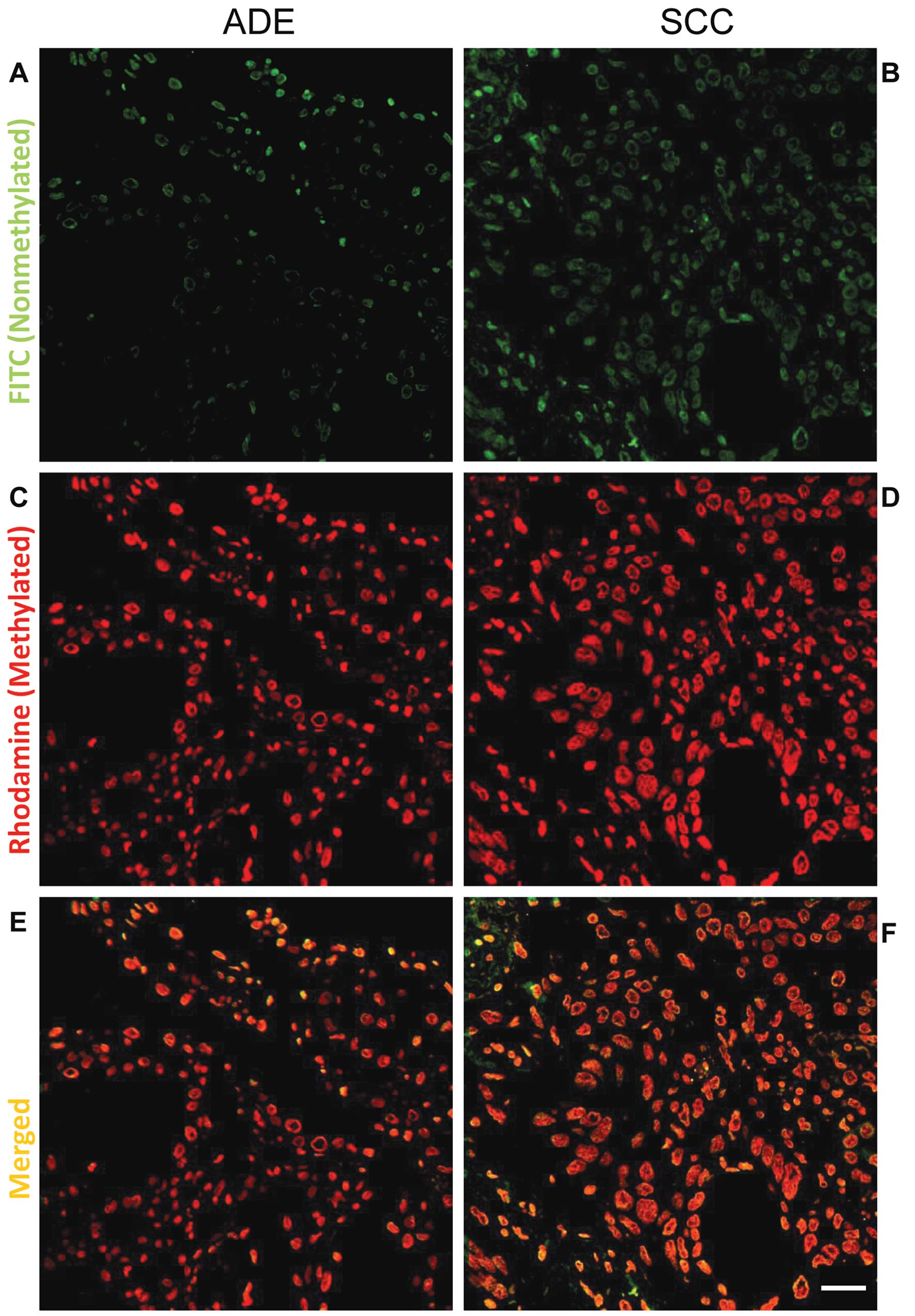
methylation
To evaluate the DNA methylation level of
pathological slides of NSCLC at CCGG sites,
histo-endonuclease-linked detection of methylation sites of DNA
(HELMET) was performed (30).
Paraffin sections were dewaxed and digested with 10 μg/ml of
proteinase K in PBS at 37°C for 15 min. Then the sections were
incubated with TdT buffer (25 mM Tris-HCl buffer, pH 6.6,
containing 0.2 M potassium cacodylate and 0.25 mg/ml BSA) alone at
RT for 30 min. After incubation, the slides were reacted with 800
U/ml of TdT dissolved in TdT buffer containing 20 μM ddATP,
20 μM ddTTP, 1.5 mM CoCl2 and 0.1 mM
dithiothreitol at 37°C for 2 h. After washing with PBS, the
sections were fixed with freshly-prepared 4% PFA in PBS for 5 min
and then rinsed with PBS. The non-methylated CCGG sites were
digested at 37°C for 2 h by 100 U/ml HpaII dissolved in 10
mM Tris-HCl buffer (pH 7.5), containing 10 mM MgCl2 and
1 mM dithiothreitol. The HpaII-cut sites were labeled with
biotin-16-dUTP by TdT reaction for 90 min. Then, the 3′-OH ends
were blocked with a mixture of dideoxynucleotides by TdT, as
described above, for 2 h. After fixation with 4% PFA in PBS, the
methylated CCGG sites were digested at 37°C for 2 h by 100 U/ml
MspI dissolved in Tris-HCl buffer (pH 7.9), containing 10 mM
MgCl2, 0.5 mM dithiothreitol, 66 mM potassium acetate,
and 0.1% BSA. The MspI-cut sites were then labeled with
digoxigenin-11-dUTP by TdT reaction for 90 min. Finally, the
sections were incubated with a mixture of 500 μg/ml normal
goat IgG and normal sheep IgG in 5% BSA/PBS for 1 h, and then
visualized by FITC-labeled goat anti-biotin and rhodamine-labeled
sheep anti-digoxigenin. The nuclei were stained with 0.5
μg/ml DAPI for 1 min.
Statistical analysis
The X-tile software program (Version 3.6.1; Yale
University School of Medicine, New Haven, CT, USA) as described
previously (36) was used to
determine the best threshold value of H3K27me3 for classifying
samples into groups of high and low expression. The SPSS 18.0
statistical software package (SPSS Inc, Chicago, IL, USA) was
employed for all analyses. The association between tested markers
and different clinicopathological characteristics of the patients,
including age, gender, tissue type, tumor differentiation,
P-factor, LV-factor, V-factor, smoking status, relapse,
postoperative metastasis and CEA level were evaluated by Pearson’s
χ2 or Fisher’s exact test as appropriate. The
Kaplan-Meier method with log-rank test was used for estimating
probability of overall survival. The Cox proportional hazard model
was used to evaluate the association between various markers and
patient’s survival. Univariate and multivariate analyses were
determined by Cox regression. A p-value <0.05 was considered
statistically significant.
Results
Clinicopathological data of patients
As shown in Table
I, the diagnosis of the 5 normal lung specimens was identically
pneumothorax. Three females and 2 males were included, with an
average age of 67.6 years. The cancer patient population included
27 males and 15 females and had a mean age of 69 years. By
histological classification, 20 cases were squamous cell carcinoma
and 22 were adenocarcinoma. Of the 22 cases of adenocarcinoma, 13
patients had well-differentiated tumor, 6 had moderately- and 3 had
poorly-differentiated tumor. In the 20 cases of squamous cell
carcinoma, the well-, moderately- and poorly-differentiated numbers
were 3, 9 and 8, respectively. All cases were TNM stage I and lymph
node negative. Postoperative follow-up data were available in all
cases, and the median follow-up duration in adenocarcinoma and
squamous cell carcinoma groups were 75.2 months and 52.9 months,
respectively.
Trimethylation level of histone H3 at
lysine 27 in normal lung and NSCLC tissues
Either in normal lung or NSCLC tissues, H3K27me3 was
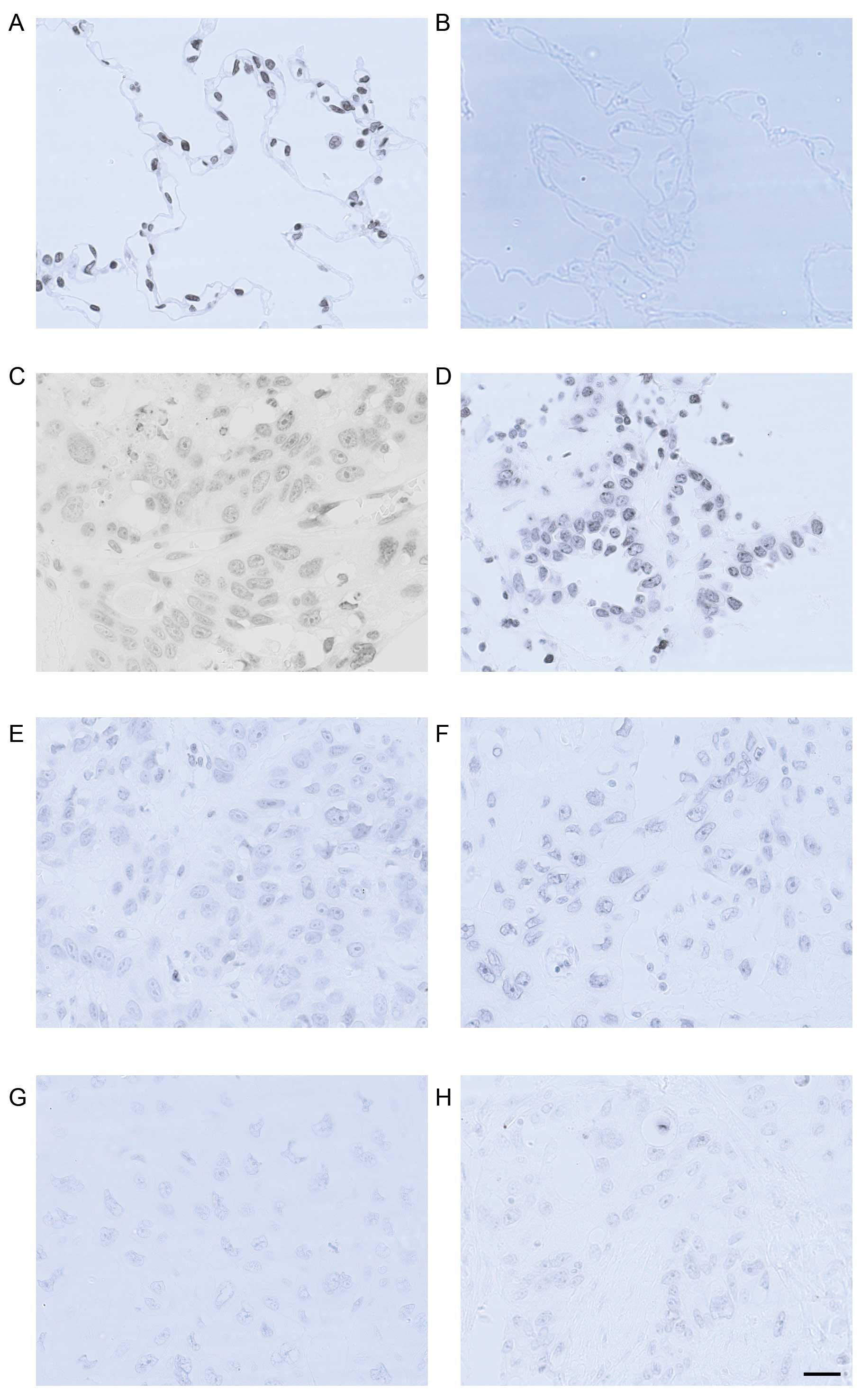
localized predominantly in the nuclei (Fig. 1). Specificity of the antibody for
H3K27me3 was determined by western blot analysis (Fig. 2H). Calculated staining score of
immunopositive cells ranged from 0 to 12 in all tested tissues.
According to the X-tile plots (Fig.
2D–F), we categorized the samples into low (IHC score ≤3) and
high (IHC score >3) expression subgroups based on a cut-point
determined by X-tile software related to survival status ((Fig. 2F, P<0.05). As shown in Fig. 2A, high expression of H3K27me3 was
observed in all 5 (100%) normal lung tissues, whereas in cancer
tissues, high methylation level of H3K27 was found in 18 (81.8%)
adenocarcinomas and 6 (30%) squamous cell carcinomas (P<0.01).
The staining score of H3K27me3 was significantly higher in normal
lung tissue compared to those of adenocarcinoma and squamous cell
carcinoma (11.2±0.8, 7.55±0.77 and 3.55±0.61, respectively,
P<0.05, Figs. 1 and 2A). In addition, a positive relationship
between tumor differentiation and H3K27me3 expression was found
(Figs. 1, 2B and C). In both cancer subtypes, higher
positive staining score was correlated with better cellular
differentiation (P<0.01). In the adenocarcinoma subgroup,
staining scores for well-, moderately- and poorly-differentiated
NSCLC samples were 8.69±1.00, 7.00±1.24 and 3.67±1.20, respectively
(P<0.01). In the squamous cell carcinoma subgroup, their
staining scores were 8.00, 2.78±0.70 and 2.75±0.82, respectively
(P<0.01).
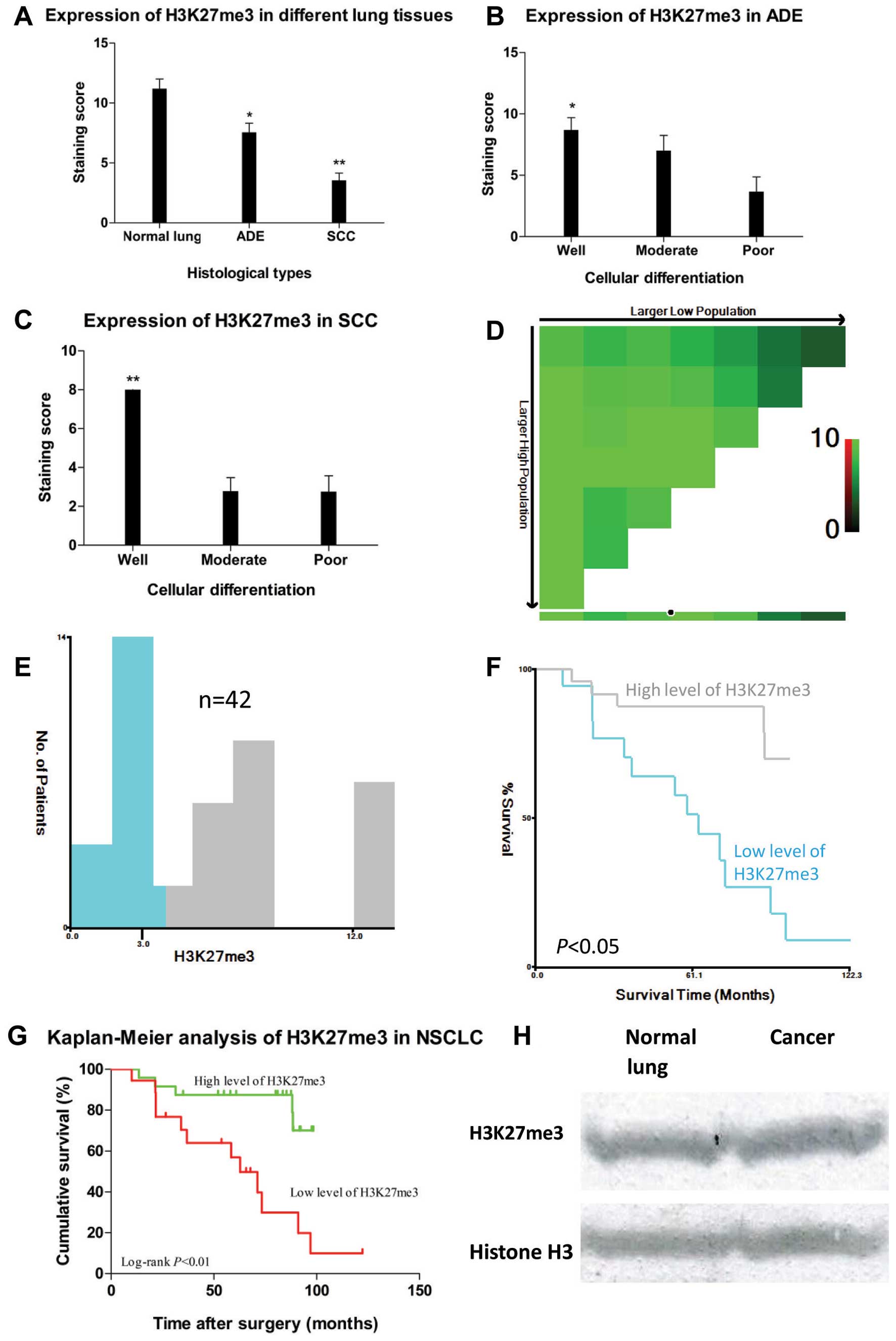 | Figure 2.(A) H3K27me3 expression differed in
different lung tissues, *P<0.05 and
**P<0.01 compared to normal lung; error bars, SD of
mean. (B) In ADE, H3K27me3 expression reduced markedly as
differentiation decreased, *P<0.05 compared to poor
differentiation. (C) In SCC, H3K27me3 expression reduced markedly
as differentiation decreased, **P<0.01 compared to
moderate or poor differentiation. (D) X-tile plots of H3K27me3
expression for optimal cut-point (3, P<0.05), which is
demarcated by the circle (black with white border). (E) The
cut-point was used to separate low H3K27me3 expression (blue) from
high H3K27me3 expression (gray) in the expression frequency
histogram of the whole sample set. (F) Kaplan-Meier curve for
testing the survival of sample subsets defined by H3K27me3
expression below 3 (green line) and above 3 (gray line). (G)
Kaplan-Meier curves of OS in different levels of H3K27me3
expression in NSCLC patients. High expression of H3K27me3 (green
line) was associated with better prognostic outcome and longer
disease-free survival time while low expression of H3K27me3 (red
line) with worse prognosis and shorter DFS/OS, P<0.05. (H) To
determine the specificity of H3K27me3 antibody, western blot
analysis (10 μg protein/lane) were performed to detect
H3K27me3 expression in both normal lung tissue (upper left panel)
and lung adenocarcinoma (upper right panel, well differentiation),
and histone H3 expression was used as internal control (lower
panel). |
Correlation of H3K27me3 expression with
clinicopathological parameters
To determine the correlation of H3K27me3 expression
and clinicopathological parameters, and to determine its prognostic
impact, χ2 analyses together with univariate,
multivariate and Kaplan-Meier survival analyses of cumulative
survival analysis were performed. Correlation analyses (Table III) revealed that H3K27me3
expression was significantly associated with non-SCC histology (P=
0.001), better cellular differentiation (P=0.002), non-smoking
status (P= 0.019), low EZH2 expression (P= 0.038), high methylation
level at CCGG sites (P= 0.049) and tumor-specific survival after
resection of primary tumors (P<0.01 by log-rank test, Fig. 2G). Kaplan-Meier survival analysis
revealed that patients with higher H3K27me3 expression in tumors
showed longer disease-free survival in contrast to those with low
expression. Univariate analysis revealed associations between poor
prognosis in NSCLC patients and several factors, including low
H3K27me3 expression (P= 0.002), histologic type (non-ADE, P=
0.011), smoking status (smoker, P= 0.002), relapse (P<0.01), and
postoperative metastasis (P<0.01, Table IV). Using a Cox proportional hazard
regression analysis, we found that expression of H3K27me together
with postoperative metastasis were independent predictors
associated with prognostic outcome (Table IV).
 | Table III.Association of H3K27me3, EZH2
expression and DNA methylation level at CCGG sites with
clinicopathologic parameters in NSCLC patients. |
Table III.
Association of H3K27me3, EZH2
expression and DNA methylation level at CCGG sites with
clinicopathologic parameters in NSCLC patients.
| H3K27me3
| EZH2
| DNA methylation
|
|---|
| Variables | All cases | H | L | P-value | All cases | H | L | P-value | All cases | H | L | P-value |
|---|
| Agea (y.o.) | | | | 0.857 | | | | 0.653 | | | | 0.245 |
| ≤69 | 18 | 10 | 8 | | 18 | 7 | 11 | | 24 | 9 | 15 | |
| >69 | 24 | 14 | 10 | | 24 | 11 | 13 | | 18 | 10 | 8 | |
| Gender | | | | 0.114 | | | | 0.026 | | | | 0.152 |
| Male | 27 | 13 | 14 | | 27 | 15 | 12 | | 27 | 10 | 17 | |
| Female | 15 | 11 | 4 | | 15 | 3 | 12 | | 15 | 9 | 6 | |
| Tissue type | | | | 0.001 | | | | 0.000 | | | | 0.059 |
| ADE | 22 | 18 | 4 | | 22 | 3 | 19 | | 22 | 13 | 9 | |
| SCC | 20 | 6 | 14 | | 20 | 15 | 5 | | 20 | 6 | 14 | |
|
Differentiation | | | | 0.002 | | | | 0.067 | | | | 0.261 |
| Well | 16 | 14 | 2 | | 16 | 4 | 12 | | 16 | 9 | 7 | |
|
Moderate/poor | 26 | 10 | 16 | | 26 | 14 | 12 | | 26 | 10 | 16 | |
| P-factorb | | | | 1.000 | | | | 1.000 | | | | 0.452 |
| Yes | 1 | 1 | 0 | | 1 | 0 | 1 | | 1 | 1 | 0 | |
| No | 41 | 23 | 18 | | 41 | 18 | 23 | | 41 | 18 | 23 | |
| LV-factorb | | | | 1.000 | | | | 1.000 | | | | 0.180 |
| Yes | 31 | 18 | 13 | | 31 | 13 | 18 | | 31 | 12 | 19 | |
| No | 11 | 6 | 5 | | 11 | 5 | 6 | | 11 | 7 | 4 | |
| V-factor | | | | 0.150 | | | | 0.038 | | | | 0.049 |
| Yes | 18 | 8 | 10 | | 18 | 11 | 7 | | 18 | 5 | 13 | |
| No | 24 | 16 | 8 | | 24 | 7 | 17 | | 24 | 14 | 10 | |
| Smoking status | | | | 0.019 | | | | 0.019 | | | | 0.016 |
| Smoker | 24 | 10 | 14 | | 24 | 14 | 10 | | 24 | 7 | 17 | |
| Non-smoker | 18 | 14 | 4 | | 18 | 4 | 14 | | 18 | 12 | 6 | |
| Relapseb | | | | 0.010 | | | | 0.720 | | | | 1.000 |
| Yes | 10 | 2 | 8 | | 10 | 5 | 5 | | 10 | 4 | 6 | |
| No | 32 | 22 | 10 | | 32 | 13 | 19 | | 32 | 15 | 17 | |
| Postoperative
metastasisb | | | | 0.281 | | | | 0.281 | | | | 0.305 |
| Yes | 10 | 4 | 6 | | 10 | 6 | 4 | | 10 | 3 | 7 | |
| No | 32 | 20 | 12 | | 22 | 12 | 20 | | 32 | 16 | 16 | |
| CEAb (ng/ml) | | | | 0.567 | | | | 0.071 | | | | 0.239 |
| ≤5 | 39 | 23 | 16 | | 39 | 15 | 24 | | 39 | 19 | 20 | |
| >5 | 3 | 1 | 2 | | 3 | 3 | 0 | | 3 | 0 | 3 | |
| EZH2c | | | | 0.038 | | | | | | | | 0.049 |
| ≤3.7 | 24 | 17 | 7 | | | | | | 24 | 14 | 10 | |
| >3.7 | 18 | 7 | 11 | | | | | | 18 | 5 | 13 | |
| H3K27me3d | | | | | | | | 0.038 | | | | 0.049 |
| ≤3 | | | | | 18 | 11 | 7 | | 18 | 5 | 13 | |
| >3 | | | | | 24 | 7 | 17 | | 24 | 14 | 10 | |
| DNA
methylationc | | | | 0.049 | | | | 0.049 | | | | |
| ≤2.64 | 23 | 10 | 13 | | 23 | 13 | 10 | | | | | |
| >2.64 | 19 | 14 | 5 | | 19 | 5 | 14 | | | | | |
| PCNAb | | | | 0.214 | | | | 0.029 | | | | 0.384 |
| ≤10% | 6 | 5 | 1 | | 6 | 0 | 6 | | 6 | 4 | 2 | |
| >10% | 36 | 19 | 17 | | 36 | 18 | 18 | | 36 | 15 | 21 | |
 | Table IV.Univariate and multivariate analyses
of factors associated with OS. |
Table IV.
Univariate and multivariate analyses
of factors associated with OS.
| Variables | Hazard ratio (95%
confidential interval, CI) | P-value |
|---|
| Univariate
analysis | | |
| Age (≤69 vs.
>69) | 1.248
(0.480–3.249) | 0.649 |
| Gender (male vs.
female) | 0.461
(0.161–1.319) | 0.149 |
| Tissue type (ADE
vs. SCC) | 0.276
(0.102–0.744) | 0.011 |
| Differentiation
(well vs. moderate/poor) | 0.338
(0.109–1.048) | 0.060 |
| P-factor (yes vs.
no) | 21.193
(0.000–1.031E7) | 0.648 |
| LV-factor (yes
vs. no) | 0.921
(0.323–2.622) | 0.877 |
| V-factor (yes vs.
no) | 0.573
(0.227–1.448) | 0.239 |
| Smoking status
(smoker vs. non-smoker) | 0.131
(0.036–0.472) | 0.002 |
| Relapse (yes vs.
no) | 0.134
(0.050–0.359) | 0.000 |
| Postoperative
metastasis (yes vs. no) | 0.115
(0.041–0.322) | 0.000 |
| Serum CEA level
(ng/ml) (≤5 vs. >5) | 0.043
(0.000–174.939) | 0.458 |
| H3K27me3 (high
vs. low) | 0.187
(0.066–0.531) | 0.002 |
| EZH2 (high vs.
low) | 1.975
(0.775–5.031) | 0.154 |
| DNA methylation
(high vs. low) | 0.441
(0.165–1.176) | 0.102 |
| PCNA (high vs.
low) | 0.755
(0.286–1.991) | 0.569 |
| Multivariate
analysis | | |
| Postoperative
metastasis (yes vs. no) | 0.115
(0.041–0.322) | 0.000 |
| H3K27me3 (high
vs. low) | 0.205
(0.068–0.614) | 0.005 |
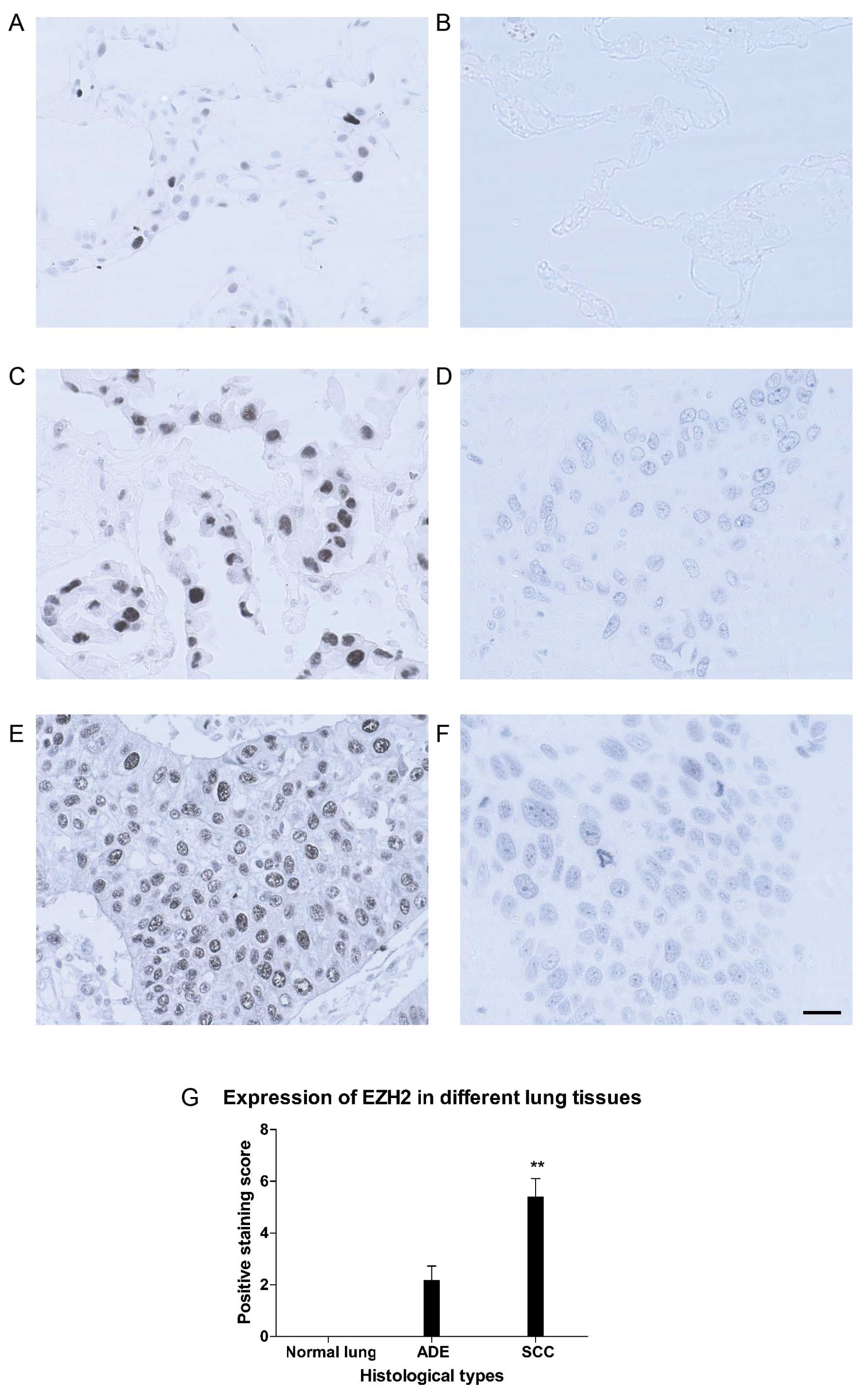
Expression of EZH2 in lung tissues and
its correlation with clinicopathological parameters
As shown in Fig. 3
(right panel), EZH2 was localized predominantly in nuclei, although
occasionally it could be seen in the cytoplasm as background. In
contrast with H3K27me3, no EZH2 staining was found in any of the 5
normal lung samples, and the staining for EZH2 in SCC was generally
more intense than that of ADE (5.40±0.71 vs. 2.18±0.54, P<0.01,
Fig. 3B, D, F and G). Of 22
adenocarcinomas, 3 cases (13.6%) were defined as high expression.
While in squamous cell carcinoma group, 15 out of 20 cases (75%)
were considered as high expression (Fig. 3G). χ2 analyses
demonstrated that when related to clinicopathological data
(Table III), expression of EZH2 in
NSCLC patients was found to be significantly associated with male
gender (P= 0.026), non-ADE histology (P<0.01), smoking status
(P=0.019), low H3K27me3 expression (P=0.038), invasion into veins
(V-factor, P=0.038), low methylation level at CCGG sites (P=0.049)
and high PCNA percentage (P= 0.029). It should be noted that the
expression of EZH2 was correlated with lower level of H3K27
methylation while with enhanced PCNA expression, indicating its
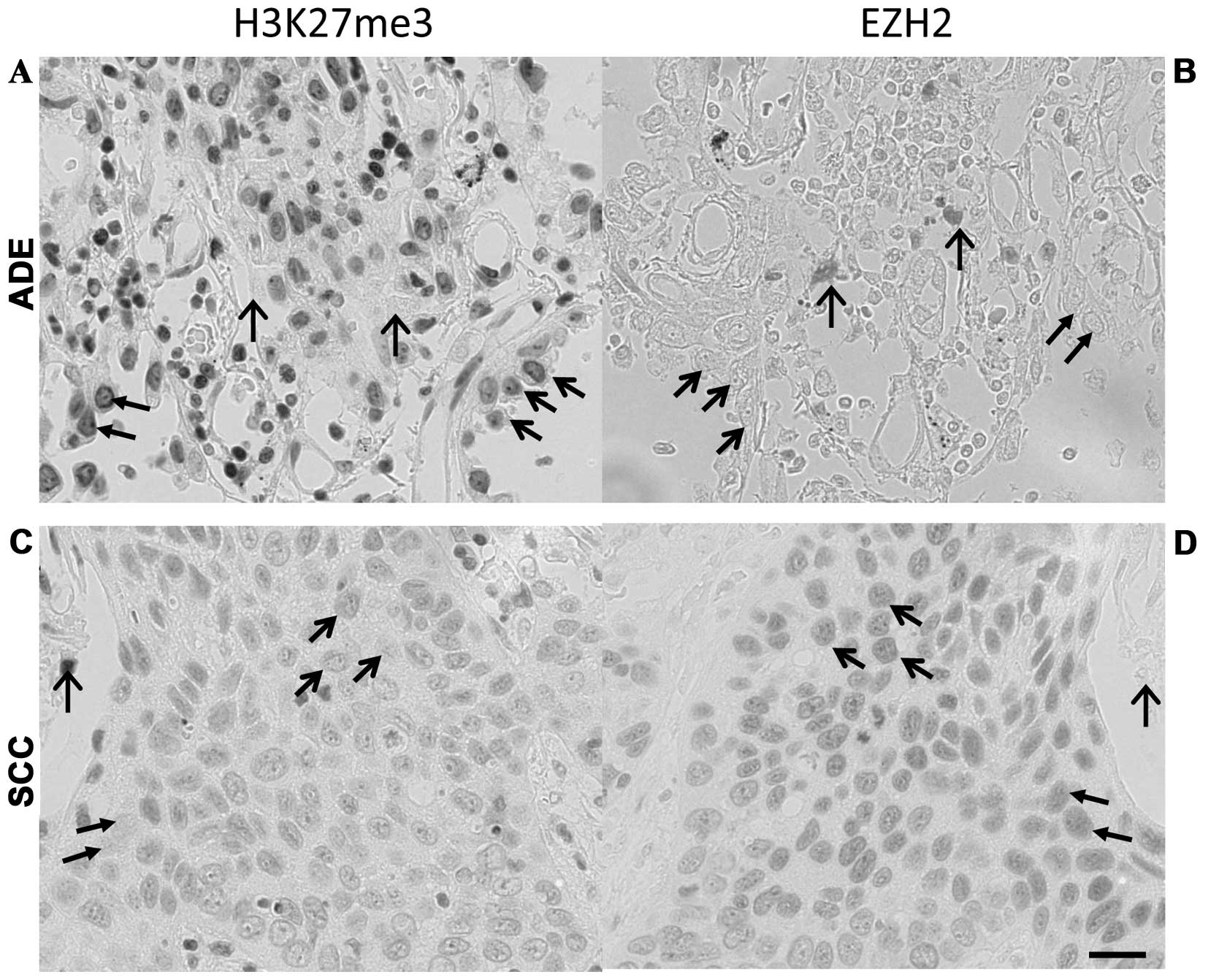
positive correlation with cell proliferating activity (Table III, Figs. 3A–F and 4). The reciprocal expression pattern of
EZH2 and H3K27me3 in NSCLC was also confirmed immunohistochemically
in mirror sections. As shown in Fig.
5, in lung adenocarcinoma (upper panel), the cells heavily
stained for H3K27me3 were essentially negative or weak for EZH2
staining. In lung squamous cell carcinoma (lower panel), the
expression of H3K27me3 seemed generally weaker than that of
adenocarcinoma, however, identical cells with negative or weak
staining of H3K27me3 were found with high level expression of EZH2.
Both correlation analysis (Table
III) and immunofluorescence double staining for EZH2 and PCNA
(Fig. 4) confirmed that PCNA index
was positively correlated with EZH2 expression. In contrast,
neither univariate nor multivariate analysis indicated that EZH2
was an independent prognostic factor for surgically treated NSCLC
patients enrolled in this study (P=0.720).
DNA methylation level at CCGG sites and
its relation with clinicopathological parameters
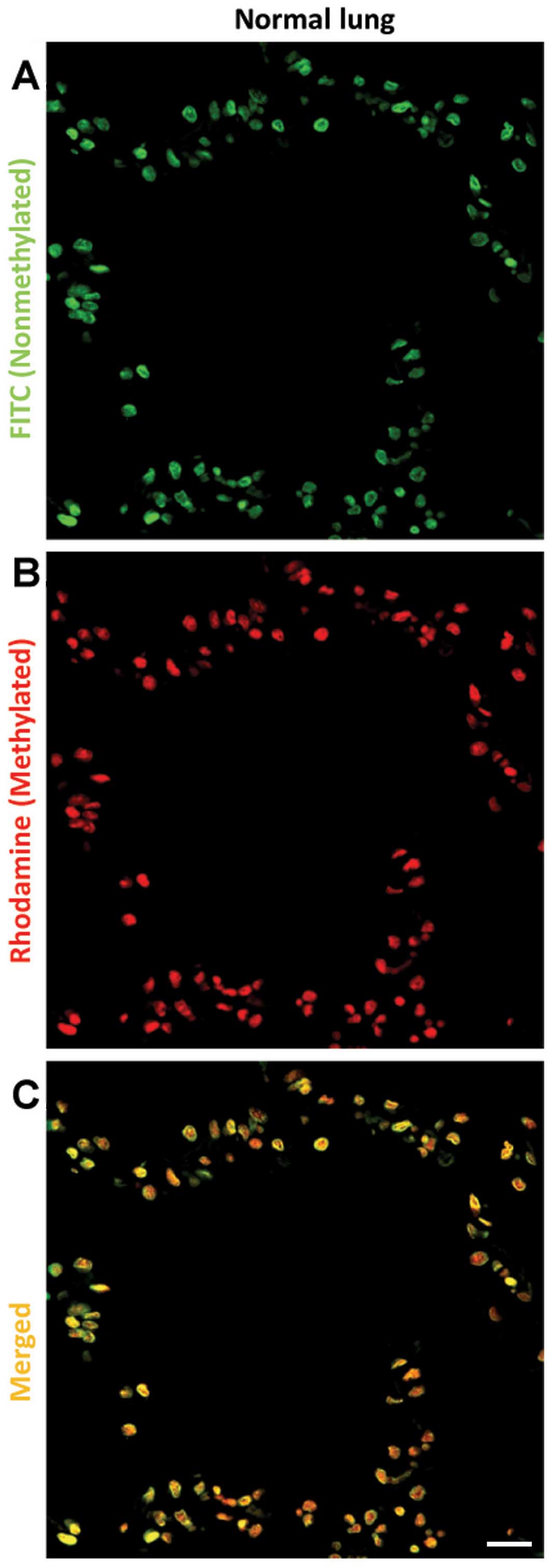
As shown in Figs. 6
and 7, CCGG sites in cancer cells
were generally hypermethylated, compared to that of normal cells.
Although no correlation was found between DNA methylation level at
CCGG sites and DFS/OS, we found that higher DNA methylation level
at CCGG sites in NSCLC patients was significantly associated with
less pulmonary vein invasion (V-factor, P= 0.049), non-smoking
status (P= 0.016), low EZH2 expression (P= 0.049) and high H3K27me3
expression (P= 0.049) (Table III).
Although the expression of H3K27me3 was correlated with
differentiation status, and DNA methylation level at CCGG sites was
correlated with the methylation level of H3K27 positively, no
correlation between DNA methylation and differentiation level was
found (P= 0.261).
Discussion
In the present study, we investigated the prognostic
value of H3K27me3 and EZH2 expression in human lung cancer
immunohistochemically, and found that, in comparison to normal lung
tissue, the level of H3K27me3 was significantly downregulated in
both ADE and SCC tissues of early-staged NSCLC. Furthermore, in
each cancer subtype, the level of H3K27me3 was antiparallel with
the cellular differentiation level, and the decrease in H3K27me3
was significantly correlated with tumor relapse and shorter overall
survival, strongly demonstrating that the level of H3K27me3 is a
new epigenetic marker in lung cancers. In contrast, although EZH2
is known to catalyze trimethylation of H3K27, the expression of
EZH2 in it was not significantly correlated with lung
carcinogenesis.
With regard to the prognostic impact of H3K27me3 in
various human cancers, it was reported that overexpression of
H3K27me3 was linked to more malignant behavior and worse prognosis
in patients with prostate (15),
esophageal (17), nasopharyngeal
(18) and hepatocellular
carcinomas (14). On the other
hand, in breast, ovarian and pancreatic cancers (5) and renal cell carcinoma (37), reduced expression of H3K27me3 was
associated with worse prognosis. In human NSCLCs, however, we have
found that a lower level of H3K27me3 was associated with higher
tumor invasiveness and/or poorer disease-free survival (DFS) in
this study. These contradictory findings may in part reflect that
H3K27me3 marks different genes for silencing in different cell
types. In the present study, we observed that low expression of
H3K27me3 was a strong and independent predictor of poor cellular
differentiation and shortened cancer-specific survival, as
evidenced by univariate and multivariate analyses (Table IV). When the Cox regression model
was constructed for the entire series, low H3K27me3 expression
remained an independent predictive factor of recurrence and/or
cancer death in NSCLC patients. To our knowledge, our data
presented here demonstrated for the first time a direct association
of expression of H3K27me3 with clinical outcome with NSCLCs. As
H3K27me3 serves as an epigenetic mark mediating silencing and
represses target gene expression, loss of it may result in
reactivation of these silenced genes, such as some oncogenes, and
therefore contributing to tumorigenesis or cancer progression.
EZH2 is involved in PRC2-directed gene silencing
through the formation of H3K27me3 as an epigenetic marker. It was
reported that EZH2 was found to be overexpressed at both mRNA and
protein levels in NSCLC and bladder cancer, and correlated with
invasiveness, increased proliferation and poor outcome (20,21).
In colorectal cancer, EZH2 overexpression indicated a good
prognosis, in contrast to the poor prognosis associated with EZH2
overexpression in NSCLCs (20). In
the current study, we found that the higher expression of EZH2 in
NSCLC patients was significantly associated with various
clinicopathological parameters including PCNA labeling index, which
was consistent with the findings of Takawa et al (20), while we failed to confirm the
significant correlation with poor prognosis (P=0.154). Further
study with a greater number of specimens is needed.
In addition, we found that the expression of
H3K27me3 was reversely correlated with that of EZH2 in NSCLC
(Table III and Fig. 5). Similar findings have been
recently reported by Holm et al (38) in breast cancer. Previous studies
indicated that the polycomb group protein EZH2 directly methylated
DNA in either normal (39) or
cancerous (11) tissues, and might
result in the inhibition of target gene expression through its
methylation, also overexpression of EZH2 promoted formation of a
different PRC complex, the PRC4, and exhibited differential histone
substrate specificities (40).
Loss of H3K27me3 in tumor might reflect the formation of the new
PRC complex to modify other histone residues. Since EZH2
methyltransferase activity requires its association with other PRC
components, it has been proposed that overexpression of EZH2 may
result in the disruption of the integrity of the PRC complexes or
may form new PRC complexes (41,42),
and thus the methyltransferase activity toward H3K27 may be
changed.
It is clear that histone methylation marks do not
act alone, but in a coordinated manner with other epigenetic
modifications (4). DNA methylation
principally occurs at cytosine residues located in dinucleotide CpG
sites (43). CpG dinucleotides are
statistically under-represented in the genome but are found to be
concentrated in CG-rich regions termed CpG islands that frequently
coincide with promoter or gene regulatory regions (3). Global hypomethylation appears to be
an early event for colon and breast cancer as well as chronic
lymphacytic leukemia (CLL) (44).
In the present study, we evaluated the DNA methylation level at
CCGG sites by means of HELMET, indicating that cancerous tissues
were relatively hypermethylated in DNA at CCGG sites in contrast to
that of normal lung tissue. This result is consistent with previous
finding, that gene hypermethylation is an early event in the
process of tumorigenesis of lung cancer (45). We also found that higher DNA
methylation level at CCGG sites were statistically correlated with
less chance of venous invasion, non-smoking status, lower
expression of EZH2 and higher expression of H3K27me3 as well,
although no significance of correlation with overall survival was
found. This finding might still suggest that hypermethylation at
CCGG sites contributes to better prognosis in NSCLC patients,
because those factors were more or less affecting the outcome of
NSCLC patients. On the other hand, Lin et al (46) have reported that hypermethylation
in CpG was correlated with poor prognosis in NSCLC, contrary to our
findings. Although the reason for this discrepancy is not known,
the genes that were hypermethylated in the studies might be
different. Since DNA methylation in cancer was mainly targeted to
polycomb-regulated genes and a very high percentage of the
methylated genes was pre-marked with trimethylated H3K27 in
addition to other polycomb components (10), further study is needed to clarify
which gene is involved and whether or not methylation of H3K27 also
plays a role in the interaction with this hypermethylation.
Some limitations of this research should be noted.
First of all, the dataset was small and as a result the statistical
power would be somewhat limited. Further study with larger number
of samples is necessary to validate the present results. Although
the number of specimens used here was limited, we did achieve some
significant indications, showing that high expression of H3K27me3
was correlated with longer disease-free survival. Secondly, all the
cases involved in this study were patients of TNM stage I. Although
chemotherapy or radiotherapy would be used after progression had
been proved, the contribution of other therapies to overall
survival had not been taken into account in the survival analysis,
for surgery was considered the key therapy for patients at this
stage, and thus would probably lead to some bias.
In conclusion, our study indicated that high
expression of H3K27me3 was associated with clinicopathological
parameters and correlated with better outcome in NSCLC patients as
well. Thus H3K27me3 can be used as a good marker, enabling us to
predict the prognosis of NSCLC patients, and to carry out a more
intensive follow-up according to H3K27me3 expression status in
resected specimens.
Abbreviations:
|
NSCLC
|
non-small cell lung cancer;
|
|
H3K27me3
|
trimethylated histone H3 at lysine
27;
|
|
EZH2
|
enhancer of zeste homolog 2;
|
|
PCNA
|
proliferating cell nuclear
antigen;
|
|
OS
|
overall survival;
|
|
DNMTs
|
DNA methyltransferase;
|
|
SUZ12
|
suppressor of zeste 12;
|
|
EED
|
embryonic ectoderm development;
|
|
RBBP
|
retinoblastoma binding protein;
|
|
PRC2
|
polycomb repressive complex 2;
|
|
HELMET
|
histo endonuclease-linked detection of
methylation sites of DNA
|
Acknowledgements
This study was supported in part by a
Grant-in-Aid for Scientific Research from the Japan Society for the
Promotion of Science (no. 18390060 to T.K.).
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Lund AH and van Lohuizen M: Epigenetics
and cancer. Genes Dev. 18:2315–2335. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Taniguchi H, Yamamoto H, Akutsu N, et al:
Transcriptional silencing of hedgehog-interacting protein by CpG
hypermethylation and chromatic structure in human gastrointestinal
cancer. J Pathol. 213:131–139. 2007. View Article : Google Scholar
|
|
4.
|
Zhang C, Li H, Zhou G, et al:
Transcriptional silencing of the TMS1/ASC tumour suppressor gene by
an epigenetic mechanism in hepatocellular carcinoma cells. J
Pathol. 212:134–142. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Wei Y, Xia W, Zhang Z, et al: Loss of
trimethylation at lysine 27 of histone H3 is a predictor of poor
outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog.
47:701–706. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Mitani Y, Oue N, Hamai Y, et al: Histone
H3 acetylation is associated with reduced p21(WAF1/CIP1) expression
by gastric carcinoma. J Pathol. 205:65–73. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Prystowsky MB, Adomako A, Smith RV, et al:
The histone deacetylase inhibitor LBH589 inhibits expression of
mitotic genes causing G2/M arrest and cell death in head and neck
squamous cell carcinoma cell lines. J Pathol. 218:467–477. 2009.
View Article : Google Scholar
|
|
9.
|
Ohm JE, McGarvey KM, Yu X, et al: A stem
cell-like chromatin pattern may predispose tumor suppressor genes
to DNA hypermethylation and heritable silencing. Nat Genet.
39:237–242. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Schlesinger Y, Straussman R, Keshet I, et
al: Polycomb-mediated methylation on Lys27 of histone H3 pre-marks
genes for de novo methylation in cancer. Nat Genet. 39:232–236.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Vire E, Brenner C, Deplus R, et al: The
Polycomb group protein EZH2 directly controls DNA methylation.
Nature. 439:871–874. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Karlic R, Chung HR, Lasserre J, Vlahovicek
K and Vingron M: Histone modification levels are predictive for
gene expression. Proc Natl Acad Sci USA. 107:2926–2931. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Yoo KH and Hennighausen L: EZH2
methyltransferase and H3K27 methylation in breast cancer. Int J
Biol Sci. 8:59–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Cai MY, Hou JH, Rao HL, et al: High
expression of H3K27me3 in human hepatocellular carcinomas
correlates closely with vascular invasion and predicts worse
prognosis in patients. Mol Med. 17:12–20. 2011.
|
|
15.
|
Ellinger J, Kahl P, von der Gathen J, et
al: Global histone H3K27 methylation levels are different in
localized and metastatic prostate cancer. Cancer Invest. 30:92–97.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Anderton JA, Bose S, Vockerodt M, et al:
The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus
and over-expressed in Hodgkin’s lymphoma. Oncogene. 30:2037–2043.
2011.PubMed/NCBI
|
|
17.
|
Tzao C, Tung HJ, Jin JS, et al: Prognostic
significance of global histone modifications in resected squamous
cell carcinoma of the esophagus. Mod Pathol. 22:252–260. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Cai MY, Tong ZT, Zhu W, et al: H3K27me3
protein is a promising predictive biomarker of patients’ survival
and chemoradioresistance in human nasopharyngeal carcinoma. Mol
Med. 17:1137–1145. 2011.PubMed/NCBI
|
|
19.
|
Chase A and Cross NC: Aberrations of EZH2
in cancer. Clin Cancer Res. 17:2613–2618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Takawa M, Masuda K, Kunizaki M, et al:
Validation of the histone methyltransferase EZH2 as a therapeutic
target for various types of human cancer and as a prognostic
marker. Cancer Sci. 102:1298–1305. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Huqun, Ishikawa R, Zhang J, et al:
Enhancer of zeste homolog 2 is a novel prognostic biomarker in
nonsmall cell lung cancer. Cancer. 118:1599–1606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Gong Y, Huo L, Liu P, et al: Polycomb
group protein EZH2 is frequently expressed in inflammatory breast
cancer and is predictive of worse clinical outcome. Cancer.
117:5476–5484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Kleer CG, Cao Q, Varambally S, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Chang CJ, Yang JY, Xia W, et al: EZH2
promotes expansion of breast tumor initiating cells through
activation of RAF1-beta-catenin signaling. Cancer Cell. 19:86–100.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Kunju LP, Cookingham C, Toy KA, Chen W,
Sabel MS and Kleer CG: EZH2 and ALDH-1 mark breast epithelium at
risk for breast cancer development. Mod Pathol. 24:786–793. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Alford SH, Toy K, Merajver SD and Kleer
CG: Increased risk for distant metastasis in patients with familial
early-stage breast cancer and high EZH2 expression. Breast Cancer
Res Treat. 132:429–437. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Bachmann IM, Halvorsen OJ, Collett K, et
al: EZH2 expression is associated with high proliferation rate and
aggressive tumor subgroups in cutaneous melanoma and cancers of the
endometrium, prostate, and breast. J Clin Oncol. 24:268–273. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Fujii S, Fukamachi K, Tsuda H, Ito K, Ito
Y and Ochiai A: RAS oncogenic signal upregulates EZH2 in pancreatic
cancer. Biochem Biophys Res Commun. 417:1074–1079. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Guo J, Cai J, Yu L, Tang H, Chen C and
Wang Z: EZH2 regulates expression of p57 and contributes to
progression of ovarian cancer in vitro and in vivo. Cancer Sci.
102:530–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Koji T, Kondo S, Hishikawa Y, An S and
Sato Y: In situ detection of methylated DNA by histo
endonuclease-linked detection of methylated DNA sites: a new
principle of analysis of DNA methylation. Histochem Cell Biol.
130:917–925. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
An S, Hishikawa Y and Koji T: Induction of
cell death in rat small intestine by ischemia reperfusion:
differential roles of Fas/Fas ligand and Bcl-2/Bax systems
depending upon cell types. Histochem Cell Biol. 123:249–261. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Shirendeb U, Hishikawa Y, Moriyama S, et
al: Human papillomavirus infection and its possible correlation
with p63 expression in cervical cancer in Japan, Mongolia, and
Myanmar. Acta Histochem Cytochem. 42:181–190. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Song N, Liu J, An S, Nishino T, Hishikawa
Y and Koji T: Immunohistochemical analysis of histone H3
modifications in germ cells during mouse spermatogenesis. Acta
Histochem Cytochem. 44:183–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Adams JC: Heavy metal intensification of
DAB-based HRP reaction product. J Histochem Cytochem. 29:7751981.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Yamayoshi T, Nagayasu T, Matsumoto K, Abo
T, Hishikawa Y and Koji T: Expression of keratinocyte growth
factor/fibroblast growth factor-7 and its receptor in human lung
cancer: correlation with tumour proliferative activity and patient
prognosis. J Pathol. 204:110–118. 2004. View Article : Google Scholar
|
|
36.
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: a new bioinformatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Rogenhofer S, Kahl P, Mertens C, et al:
Global histone H3 lysine 27 (H3K27) methylation levels and their
prognostic relevance in renal cell carcinoma. BJU Int. 109:459–465.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Holm K, Grabau D, Lovgren K, et al: Global
H3K27 trimethylation and EZH2 abundance in breast tumor subtypes.
Mol Oncol. 6:494–506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Chen H, Tu SW and Hsieh JT:
Down-regulation of human DAB2IP gene expression mediated by
polycomb Ezh2 complex and histone deacetylase in prostate cancer. J
Biol Chem. 280:22437–22444. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Kuzmichev A, Margueron R, Vaquero A, et
al: Composition and histone substrates of polycomb repressive group
complexes change during cellular differentiation. Proc Natl Acad
Sci USA. 102:1859–1864. 2005. View Article : Google Scholar
|
|
41.
|
Cao R and Zhang Y: The functions of
E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr
Opin Genet Dev. 14:155–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Kuzmichev A, Jenuwein T, Tempst P and
Reinberg D: Different EZH2-containing complexes target methylation
of histone H1 or nucleosomal histone H3. Mol Cell. 14:183–193.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Molloy PL and Watt F: DNA methylation and
specific protein-DNA interactions. Philos Trans R Soc Lond B Biol
Sci. 326:267–275. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Ross JP, Rand KN and Molloy PL:
Hypomethylation of repeated DNA sequences in cancer. Epigenomics.
2:245–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Digel W and Lubbert M: DNA methylation
disturbances as novel therapeutic target in lung cancer:
preclinical and clinical results. Crit Rev Oncol Hematol. 55:1–11.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT
and Wang YC: Alteration of DNA methyltransferases contributes to
5′CpG methylation and poor prognosis in lung cancer. Lung Cancer.
55:205–213. 2007.
|