Introduction
According to 2013 the National Comprehensive Cancer
Network (NCCN) Guidelines, gastric adenocarcinoma is spreading
globally, especially in Asian countries, such as Japan and China.
Gastric carcinoma is the fourth most common malignancy and the
second leading cause of death worldwide, with estimated 900,000 new
cases per year and a 20% 5-year survival rate (1,2). The
oncogenesis and development of gastric cancer is still vague. The
surrounding tumor microenvironment has begun to be recognized as an
active participant in the development and progression of cancer.
Chemokines, secreted by cells in the tumor microenvironment, can
regulate fundamental biological processes in both tumor and stromal
cells, including anti-angiogenesis, activation of host specific
immunity and autocrine stimulation of cell growth (3–5). It
has been reported that there is a difference of chemokine receptors
and function between tumor cells and normal cells (4,5),
which gives rise to the hypothesis that dysregulation of chemokines
may take part in the development of the malignancy.
CXCL14 (also named BRAK), one of the CXC chemokines,
was first reported by Hromas et al (6). Its gene locates in 5p31.1 chromosome
and is constituted by 77 amino acids (7). Physiologically, CXCL14 tends to play
a homeostatic role which was constitutively expressed in cerebrum,
small intestine, kidney, and epithelium, but not in lymphoid tissue
(8–10). In common inflammation, CXCL14 was
inclined to decrease (11,12). However, the opposite result of
increased CXCL14 was seen in infiltrating inflammatory cells around
tumor (9,13). As a member of ELR (Glu-Leu-Arg
motif immediately prior) negative chemokine family, CXCL14 contains
multiple functions of anti-angiogenesis, chemotaxis to natural
killer cells, B-cells, macrophages, monocytes, and immature
dendritic cells (3,8,11,14,15).
Beginning with breast cancer, several studies showed
its low or absence of expression of CXCL14 in renal carcinoma, lung
cancer, head and neck squamous cell carcinoma, and cervical cancer
(6,8,16,17).
However, CXCL14 has been found at high levels in part of prostate
and pancreas cancer (18,19). The expression of CXCL14 and its
clinical significance in gastric cancer is poorly understood. In
the present study, we explored the expression and clinical
significance of CXCL14 in gastric cancer tissues. Further
investigation showed downregulation of CXCL14 in gastric cancer
tissues was caused by unusual methylation in its promoter region.
Our findings suggest that the anticancer function of CXCL14
provides a new approach in gastric cancer diagnosis and
treatment.
Materials and methods
Patients and specimens
All the gastric adenocarcinoma patients in the study
cohort, diagnosed by endoscopic biopsy, were admitted for surgical
treatment in the First Affiliated Hospital of Wenzhou Medical
University (Zhejiang Province, China) from December 2008 to April
2009. None received radiation therapy or chemotherapy before
operation and received strict chemotherapy after surgery according
to the NCCN gastric cancer guidelines. The histopathological
diagnosis of gastric adenocarcinoma was confirmed by the Pathology
Department according to the criteria of the World Health
Organization after the operation. The patient characteristics and
clinicopathological features are listed in Table I. Paired specimens of gastric
adenocarcinoma tissues and corresponding normal gastric tissues
(obtained from negative resection margin) were obtained from the
above patients. Each sample was divided into several fractions.
Some were snap-frozen in liquid nitrogen within 30 min of the
resection and stored for RNA/DNA extraction. The remaining part of
the sample was formalin-fixed and paraffin-embedded for
immunohistochemistry. Informed written consent was obtained from
each patient and the study was approved by the Human Research
Ethics Committee at the First Affiliated Hospital of Wenzhou
Medical University.
 | Table I.Clinicopathological features and
CXCL14 mRNA (2−ΔΔCt) of 60 patients. |
Table I.
Clinicopathological features and
CXCL14 mRNA (2−ΔΔCt) of 60 patients.
| Clinicopathologic
variables | n | mRNA
(2−ΔΔCt) | P-value |
|---|
| Gender | | | |
| Male | 46 | 0.598
(0.025–1.425) | 0.332 |
| Female | 14 | 0.474
(0.010–1.475) |
| Age (years) | | | |
| <60 | 22 | 0.592
(0.170–1.226) | 0.745 |
| ≥60 | 38 | 0.556
(0.010–1.475) |
| Tumor location | | | |
| Upper third | 13 | 0.774 (0.
072–1.299) | 0.352 |
| Middle third | 15 | 0.561
(0.010–1.475) |
| Lower third | 32 | 0.516
(0.085–1.349) |
| Tumor size
(cm) | | | |
| <5 | 37 | 0.600
(0.025–1.475) | 0.509 |
| ≥5 | 23 | 0.528
(0.010–1.349) |
|
Differentiation | | | |
| Well or
moderately | 11 | 0.770 (0.170–1.
349) | 0.172b |
| Poorly or
none | 49 | 0.473
(0.010–1.475) |
| Primary
tumora | | | |
| T1 | 6 | 0.439
(0.325–0.581) |
<0.001b |
| T2 | 6 | 0.255
(0.072–0.523) |
| T3 | 25 | 0.390
(0.010–1.097) |
| T4 | 23 | 0.879
(0.025–1.475) |
| Regional lymph
nodesa | | | |
| N0 | 8 | 0.551 (0.
072–1.226) | 0.121 |
| N1 | 7 | 0.247
(0.085–0.541) |
| N2 | 29 | 0.583
(0.025–1.349) |
| N3 | 16 | 0.694
(0.010–1.475) |
| Anatomic
stagea | | | |
| Stage I | 6 | 0.334
(0.072–0.581) | 0.142b |
| Stage II | 8 | 0.576
(0.170–1.226) |
| Stage III | 31 | 0.678
(0.010–1.349) |
| Stage IV | 15 | 0.475
(0.085–1.475) |
| Carcinoembryonic
antigen (CEA; μg/l) | | | |
| ≤5 | 49 | 0.575
(0.025–1.475) | 0.799 |
| >5 | 11 | 0.540
(0.010–1.226) |
| Carbohydrate
antigen 19-9 (CA19-9; U/ml) | | | |
| ≤37 | 43 | 0.522
(0.072–1.350) | 0.640b |
| >37 | 17 | 0.688
(0.010–1.475) |
Tumor cell lines and treatment
Gastric cancer cell lines, AGS obtained from the
American Type Culture Collection (ATCC®, Manassas, VA,
USA), SGC7901, BGC823 and MGC803 were purchased from the Type
Culture Collection of Chinese Academy of Sciences (Shanghai
Institute of Biochemistry and Cell Biology®, Chinese
Academy of Sciences, Shanghai, China), cultured in Ham’s F-12K
medium (Gibco Life Technologies®, Shanghai, China)
supplemented with 10% fetal bovine serum at 37°C under 5%
CO2. Cells, harvest from 25 cm2 culture
flasks, were seeded in 6-well plate with low density
(1×106 cells per well). Twenty-four hour adhesion and 16
h serum starvation was performed before drug treatment. The cells
were treated by 5-Aza-2′-deoxycytidine (Sigma®, St.
Louis, MO, USA) for 5 days with fresh medium containing the drug
changed every 24 h. The final concentration of drug in the well was
0, 5, 10, 15, 25 μmol/l, respectively. CXCL14 mRNA, DNA and
protein were respectively isolated for further research.
RNA isolation and purification
Total RNA were extracted from gastric adenocarcinoma
tissues, corresponding normal gastric tissues and cancer cell lines
using TRIzol reagent (Invitrogen Life Technologies®,
Grand Island, NY, USA) following the supplier’s instructions with
some modifications. Briefly, the extracted RNA precipitated in
isopropanol were incubated at −20°C overnight to enhance
precipitation efficiency of low-molecular-weight RNA. Quantified by
the ultraviolet spectrophotometer (Beckman Coulter®,
Miami, FL, USA), RNA was purified with DNase I (Thermo
Scientific®, Waltham, MA, USA) and re-extracted using
phenol/chloroform according to the manufacturer’s instructions. The
pure RNAs were dissolved in diethylpyrocarbonate (DEPC)-treated
water and stored at −80°C. The concentration and purity of total
RNA were qualified by the ultraviolet spectrophotometer at 260 and
280 nm. Only the RNA samples with ratio of >2.0 A260/A280>1.8
were used for the experiment.
Reverse transcription PCR and real-time
PCR
First-strand complementary DNA was reverse
transcribed with ReverTra Ace® qPCR RT kit (Toyobo®,
Tokyo, Japan) from 1 μg of total RNA and preserved at −20°C
until use. We extended the reverse transcription time to 60 min in
comparison with specification. CXCL14 RNA expression was assessed
by real-time PCR with RNA-direct™ SYBR® Green Real-time
PCR Master Mix (Toyobo) and specific primers (forward,
5′-AGCCAAAGTACCCGCACTG-3′; and reverse, 5′-AGACCCTGCGCTTCTCGTTC-3′;
156 bp). hGAPDH was chosen as internal control gene (forward,
5′-CAGGGCTGCTTTTAACTCTGGTAA-3′; and reverse,
5′-GGGTGGAATCATATTGGAACATGT-3′; 101 bp). In StepOne™ Real-Time PCR
system (Applied Biosystems®, Grand Island, NY, USA), PCR
cycles involved at 95°C for 5 min; then followed by 40
amplification cycles of 94°C for 30 sec, 57°C for 30 sec, 72°C for
30 sec. Melting curves were generated for each real-time PCR to
verify the specificity of each PCR reaction. Duplication was
performed in real-time PCR for accuracy judgement.
Western blot analysis
Protein, obtained from samples by cell lysis buffer
(Beyotime®, Shanghai, China) was quantified to 50
μg per lane by Enhanced BCA Protein Assay kit (Beyotime).
After separating in 12% PAGE gels and transferring to
nitrocellulose membrane, the membrane was blocked with 5% defatted
milk in tris-buffered-saline with Tween (TBS-T) for 2 h. The same
protein lysates were incubated with CXCL14 antibody (12 kDa, 0.2
μg/ml, Abcam®, Cambridge, MA, USA) and β-actin
antibody (42 kDa, dilution 1:1,000, Beyotime) at 4°C overnight,
respectively. Followed by washing, the related
horseradish-peroxidase (HRP) conjugated secondary antibodies
(Beyotime) were incubated for 1 h at room temperature (25°C).
BeyoECL Plus (Beyotime) was used for detection of the final
chemiluminescence reaction.
Immunohistochemistry
Paired formalin-fixed and paraffin-embedded tissue
blocks (n=60) from gastric adenocarcinoma and normal resection
margin were cut into 5 μm sections and adhered to 0.1%
poly-L-Lysine treated glass slides (Maixin-Bio®, Fuzhou,
Fujian Province, China); dewaxed in oven at 61°C 1 h and a series
of xylene; rehydrated using graded ethanol (100, 90, 80 and 70%);
followed by distilled water. Antigen retrieval was carried out by
high-pressure antigen retrieval for 2 min in citrate antigen
retrieval solution (pH 6.0, Maixin-Bio). After cooling to room
temperature, slides were washed three times by 0.01 mol/l phosphate
buffer solution (PBS, pH 7.4). 0.3% hydrogen peroxide was used to
block endogenous peroxidase activity for 10 min and the primary
antibodies (5 μg/ml, Abcam) were incubated in room
temperature for 3 h and 30-min incubation with HRP-conjugated
secondary goat anti-rabbit antibodies (Maixin-Bio). The color
reaction was developed using the DAB kit (Zhongshan Golden Bridge
Bio®, Beijing, China) for 5 min according to the
manufacturer’s protocol. Specimens were counterstained with
hematoxylin, rinsed in PBS, dehydrated through graded ethanol (80,
90 and 100%) and by dimethylbenzene.
For a semiquantitative analysis of CXCL14 protein
expression: −, was graded for no expression; +, was graded for
<25% expression; ++, was graded for 25–50%; +++, was graded for
>50–75%; and ++++, was graded for >75% expression.
Bisulfite modification and BSP
We obtained the genomic DNAs from paired specimens
of tumor/normal gastric tissues and gastric cancer cell lines
before/after 5-Aza-2′-deoxycytidine treatment using TIANamp Genomic
DNA kit (Tiangen Biotech®, Beijing, China) according to
their protocol. Before modification with bisulfite, quantified DNA
was measured by ultraviolet spectrophotometer. Extracted DNA (0.5
μg) from tissues and cells was used for bisulfite
modification with EZ DNA Methylation-Gold kit (Zymo
Research®, Irvine, CA, USA) according to the
manufacturer’s protocol. Modified DNA was amplified immediately
because of the unstable situation of CT conversion. The
bisulfite-sequencing PCR primers (16) (forward,
5′-GTTGTGGTATGGGTGTGTAAG-3′; and reverse,
5′-CRCCAAAAACCTCATACTAACC-3′) and Taq Hot Start™
(Takara®, Otsu, Shiga, Japan) were necessary for
amplifying CpG islands in the promoter region with the expected
product of 505 bp in length. For follow-on research, the PCR
reaction solution in T100 thermal cycler (Bio-Rad®,
Hercules, CA, USA) was geometrically amplified to 50 μl.
After initial denaturation step at 95°C for 5 min, the PCR profile
was limited to 35 cycles for mutation at 94°C for 30 sec and 60°C
for 1 min and 72°C for 1 min; final extension was performed at 72°C
for 5 min. The PCR products following electrophoresis were purified
from 1.5% agarose gel by TIANgel Midi Purification kit (Tiangen
Biotech®, Beijing, China) per supplier’s specification.
For the veracity of DNA sequencing, PCR products were cloned into
pMD®19-T Simple Vector (Takara) and transformed into
competent cells of Escherichia coli DH5α. Using
LB-Agar-Power medium with ampicillin (100 μg/ml), five
monoclones per sample were picked for sequencing by 3730xl DNA
Analyzer (Applied Biosystems). Quality control for DNA methylation
data was performed using BiQ (software tool for DNA methylation
analysis; http://biq-analyzer.bioinf.mpi-inf.mpg.de/).
Statistical analysis
The Ct value (threshold cycle) was defined as the
fractional cycle number at which the fluorescence passed the fixed
threshold. ΔCt represented the expression difference between target
RNA and internal control gene. ΔΔCt represented the difference in
value between ΔCt of tumor tissues or the cells in experimental
group and ΔCt of normal tissues or control group. The normalized
mRNA expression level in a cancer specimen is 2−ΔΔCt.
For the paired normal tissue sample or control group, ΔΔCt equal to
0 and 2−ΔΔCt equal to 1. Because most mRNA level fit the
gaussian distribution, one-sample t-test, independent sample t-test
and Kruskal-Wallis H test are used to evaluate the differences of
the miRNA expression between different groups. The difference of
DNA methylation in paired specimens were analysed by χ2
test. In Kaplan-Meier survival curve, we define high expression as
the fold change >1 and low expression as <1, probability of
survival was compared by log-rank test. The quantified RNA data and
other clinicopathological features were selected by stepwise
regression selection as covariates to access the effects to
survival time in Cox proportional hazard regression model. Level of
significance was defined as P<0.05. All analysis were carried
out by SPSS version 16.0 for Windows.
Results
Downregulation of CXCL14 expression in
human gastric adenocarcinoma
To explore the expression of CXCL14 in gastric
cancer, we firstly design a sensitive and specific real-time PCR to
access the mRNA level. Electrophoresis indicated that a single band
of CXCL14 or hGAPDH at the appropriate position (156 bp for CXCL14
and 101 bp for hGAPDH) and no PCR product was obtained from the
‘minus-RT’ control in which reverse transcriptase was omitted from
the reactions (Fig. 1A). The
melting-curves of CXCL14 and hGAPDH were sharply defined curves
with a narrow peak, indicating that pure, homogeneous PCR products
were produced (Fig. 1B). The
combination of melting curves and gel electrophoresis confirmed the
PCR specificity.
 | Figure 1.Expression of CXCL14 in human gastric
cancer and gastric tissues on mRNA or protein levels. (A) CXCL14
mRNA expression by improved protocol; L1, blank control; L2, RNA
control; L3-8, tumor 1, normal 1, tumor 2, normal 2, tumor 3,
normal 3. (B) Melting curve; *hGAPDH;
**CXCL14. (C) CXCL14 protein levels by western blotting;
L1-8, tumor 1, normal 1, tumor 2, normal 2, tumor 3, normal 3,
tumor 4, normal 4. (D) Expression and localization of CXCL14
protein in gastric tissues by immunohistochemistry; magnification
×200 and ×400, respectively. (E) CXCL14 mRNA levels in normal
gastric tissues (n=60) and gastric cancer (n=60);
*P<0.001. (F) Semi-quantitative analysis of CXCL14
immunoreactivity; n=60; *P<0.001. |
Using the real-time PCR method described above, we
further endeavored to determine the expression patterns in paired
samples consisting of tumor and normal gastric tissues (n=60
respectively). As shown in Fig.
1E, the expression level of CXCL14 in tumor samples was lower
than that in the controls (P<0.001). The mean of relative
expression of CXCL14 (2−ΔΔCt) was 0.569 in tumor
samples, with that in non-tumor control samples set at 1.000.
The specificity of the antibody against CXCL-14 was
examined by western blotting (Fig.
1C). However, the difference of chemiluminescence reaction
between paired specimens was probably eliminated by the normal
gastric epithelial cells in tumor tissues. Immunohistochemical
analysis also indicated that CXCL14 protein levels in gastric
adenocarcinoma tissues presented a loss of CXCL14 expression when
compared with paired normal gastric tissues (Fig. 1D). Normal tissues (n=30) were
strongly positive stained by CXCL14 antibody, whereas tumor tissues
(n=30) were low or absent of CXCL14 expression. Semi-quantitative
analysis (Table II) indicated that
approximately half the cancer samples had downregulated protein
expression >25%, the remaining cancer samples declined more
severely by ≥50% (mean, 60%). On the contrary, most normal tissues
were filled with brown staining (mean, 90%, P<0.001) under the
microscope (Fig. 1F).
 | Table II.Semi-quantitative analysis of CXCL14
immunoreactivity in cancer and normal tissues. |
Table II.
Semi-quantitative analysis of CXCL14
immunoreactivity in cancer and normal tissues.
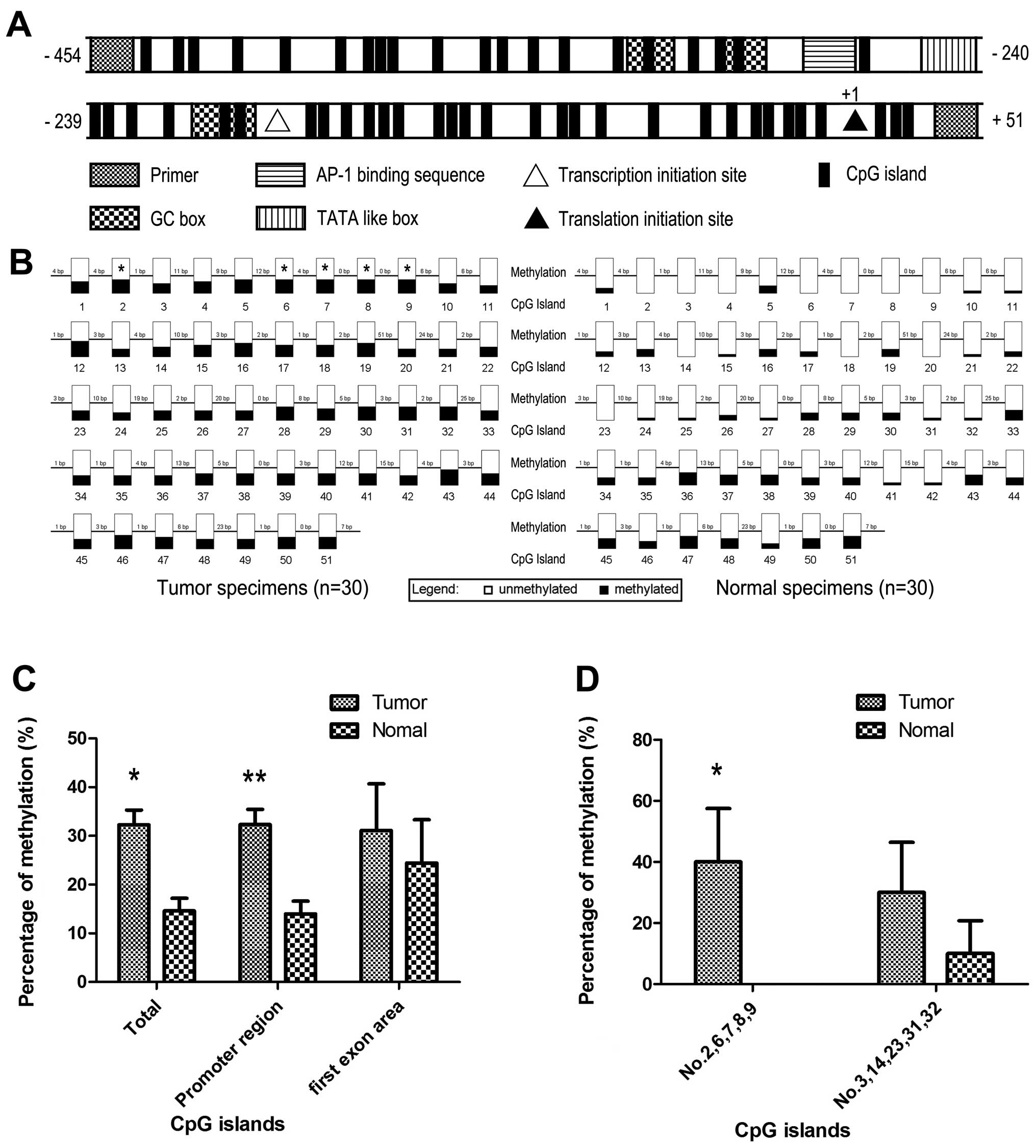
Abnormal hypermethylation of CXCL14
promoter region in tumor tissues
Following NCBI and Komori et al (20) conclusion, there was an atypical
TATA-like TATTAA sequence, an AP-1 binding sequence, 4 GC boxes and
51 CpG islands included in this CXCL14 bisulfite-sequencing PCR
product. It has been reported that transcription initiation site
(downstream 60 bp of TATTAA sequence), translation initiation site
(ATG) and the first exon area also exist (20) (Fig.
2A). They are likely to be the composition of methylation
patterns. Considering the downregulation of CXCL14 in gastric
cancer and the many cis-function elements contained in CXCL14
genome, we further analyzed the methylation state of CXCL14 in
promoter and first exon area.
Bisulfite sequencing of tumor and normal tissues
revealed that 14.58% (223/1530) of the 51 CpG sites in normal
specimens were methylated, in contrast of 32.29% (494/1530) in
tumor (Fig. 2B and C). The
methylation status of combined CpG islands showed statistical
significance (P<0.001). Further investigation indicated that
CXCL14 promoter region revealed statistical difference between
samples obtained from normal (13.96%, 201/1440) and tumor tissues
(32.36%, 466/1440, P<0.001), whereas no methylation difference
was found between normal and cancer samples in first exon area
(P=0.498) (Fig. 2C). Furthermore,
we found that each CpG island had different effect on methylation
status in cancer. Hypermethylation of nos. 2, 6, 7, 8 and 9 CpG
island, as group 1, had statistical difference between normal and
tumor tissues (P<0.001) and nos. 3, 14, 23, 31 and 32 CpG
island, as group 2, showed a potential difference existed (P=0.053)
(Fig. 2D). Each CpG island
inner-group showed the same methylation state in hypermethylated
CpG islands.
Demethylation restored the expression of
CXCL14 in gastric cancer cell lines
To further verify the above phenomenon, AGS, BGC823,
MGC803 and SGC7901 gastric cancer cell lines were treated with
5-Aza-2′-deoxycytidine to recover the demethylation state of CXCL14
gene CpG islands. Fig. 3A
illustrates that with 5-Aza-2′-deoxycytidine treatment, AGS cells
were restored to upregulate CXCL14 mRNA level (P=0.019) compared
with control group (0 μmol/l). The BSP verified the
demethylating efficacy and reversible methylation affected CXCL14
expression. As shown in Fig. 3B,
the rate of methylated CpG islands in the CXCL14 promoter region
was reduced from 85.62% (655/765) to 12.55% (96/765) (P<0.001)
but no statistical difference was revealed with concentration
gradients (5, 10, 15 and 25 μmol/l, P=0.825). The same
results were explored in BGC823 and SGC7901, it seems that the
restoration of CXCL14 expression was associated with the presence
of hypomethylating agents, not dose-dependently (P<0.001, both)
(Fig. 3C and D). However, no
statistical difference was shown between study cohort and control
cohort in MGC803 (P=0.353).
Correlation between CXCL14 mRNA
expression and clinical analysis
The depth of penetration was associated with CXCL14
mRNA relative level (especially for T4, P<0.001). However, there
was no correlation between CXCL14 relative expression and general
clinicopathological features of gastric cancer, such as age,
gender, tumor location, differentiation, lymph node metastasis, TNM
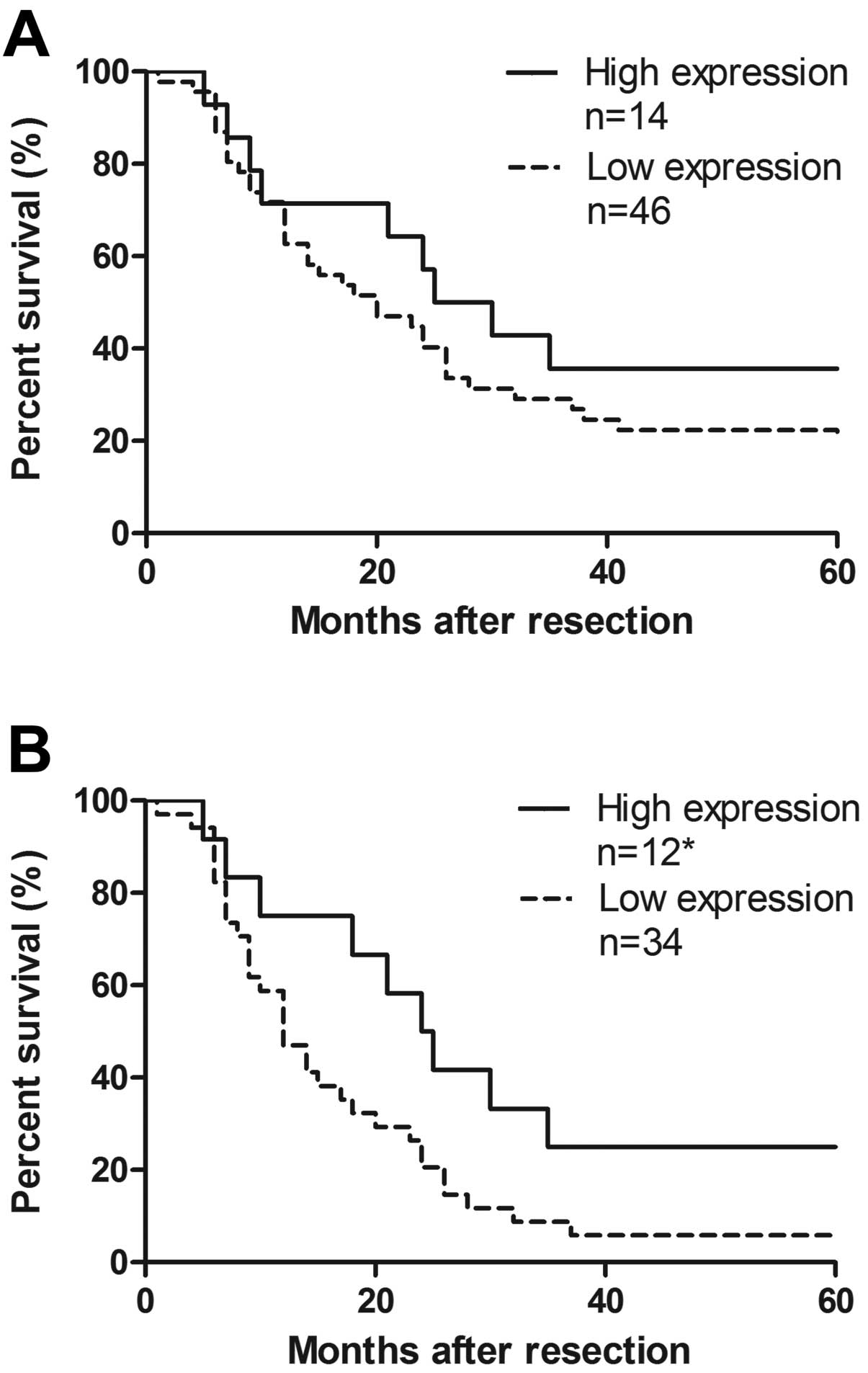
classification, and tumor markers (Table I). We defined high CXCL14 relative
expression as the fold change >1 and low expression as <1.
The overall survival rate illustrated that the cohort with higher
expression of CXCL14 showed no improved survival compared to that
with lower level (P=0.270, Fig.
4A). Respective median survival time of stage III/IV in the two
groups showed significant difference, to 12 or 24 months,
respectively (P=0.046, Fig. 4B).
Analysis of Cox proportional hazard model showed most
clinicopathological features are excluded, but combined TNM
classification, lymphatic invasion and CXCL14 mRNA expression has
an effect on survival time (P<0.001). The risk ratio of CXCL-14
reached 0.394 (95% CI, 0.195, 0.793; P=0.009) as TNM was 2.952 (95%
CI, 1.549, 4.335; P<0.001) and lymphatic invasion was 2.133 (95%
CI, 1.344, 3.386; P=0.001). Therefore, CXCL14 might be an
independent positive factor in prognosis, as the death risk of low
expression group was 2.538-fold higher than the high one.
Discussion
The development of neoplasm is dependent on the
balance of tumor progression and inhibition genes. As a
multifunctional chemokine, CXCL14 might use distinct signal
transduction pathways to take part in the inhibition and
development of neoplasm. In tissues of prostate and pancreas
cancers, CXCL14 showed higher expression compared to normal control
(18,19). However, in our present study, we
reported for the first time the downregulation of CXCL14 expression
in gastric cancer. Survival analysis showed CXCL14 levels were
positively correlated with survival time in stage III/IV and
invasive depth although CXCL14 expressions showed no correlation
with clinicopathological features of gender, age, tumor location,
size, differentiation, lymph node metastasis, anatomic stage or
common tumor markers. Consistent with our results, several studies
also showed the decreased or absent expression of CXCL14 in breast
cancers, renal carcinomas, lung cancers, head and neck squamous
cell carcinomas and cervical cancer (6,8,16,17,21).
It is well reported that CXCL14 can chemoattract
several classes of immune cells including monocyte-divided
macrophagocyte, immature dendritic cells (iDCs), natural killer
(NK) cells and B cells (8,11,22).
Due to lack of CXCL14 expression in solid tumors, few immune cells
were assembled in cancer tissues resulting in local immune response
deficiencies, including attenuated immune surveillance, immune
evasion, weakened antigen presentation and disordered immune
internal environment (11).
Besides immunological anticancer mechanism, CXCL14 could also
suppress tumorous vasculature by inhibiting the chemotaxis of
vascular smooth muscle cells and the formation of microvascular
system (3,7,23).
CXCL14 also directly affects the proliferation, invasion and
migration of tumor cells (13,16,23).
Mechanisms involved in CXCL14 function in the development of
gastric cancer still needs to be further explored.
Literature over the years have investigated several
CXCL14 related up-stream genes to state the different expression
between normal and tumor tissues. It has been reported that the
Ras/Raf/MEK/ERK/MAPK signal pathway, RhoA, ROCK signal pathway,
calcium/calmodulin signal pathway, reactive oxygen species
imbalance and transcription factor SP-1 could influence the CXCL14
expression (24–27). DNA abnormal hypermethylation in CpG
islands of promoters, related to gene silencing, is now recognized
as a common feature for malignant tumors (28). In our study, methylation in CXCL14
promoter was analyzed by BSP method. Data indicated that
methylation in CXCL14 promoter exists in gastric cancer and normal
tissues, but the level of methylation of the latter is far below
the former. No statistical difference existed in the first exon
area. When checking 48 CpG islands in promoter, nos. 2, 6, 7, 8 and
9 CpG showed significant hypermethylation in tumor tissues
(P<0.001). In addition, nos. 3, 14, 23, 31 and 32 have a
potential difference (P=0.053). These islands might act as a
potential diagnostic marker for gastric cancer. Which islands are
involved in CXCL14 silencing in gastric cancer requires further
investigation.
In conclusion, as a new member of CXC subfamily of
chemokines, CXCL14 has a role in the development and progression of
gastric cancer. Hypermethylation in promoter region causes the low
expression of CXCL14 in gastric adenocarcinoma tissues. The level
of CXCL14 expression provides a valuable adjuvant parameter in
predicting the prognosis of gastric cancer patients and thus a
potential therapeutic target.
Acknowledgements
This study was supported by National
Natural Science Foundation of China (81001343), the Zhejiang
Provincial Natural Science Foundation of China (Y2100660 and
Y2100909) and the Wenzhou Science and Technology Bureau
(H20100028).
References
|
1.
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar
|
|
2.
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006.
|
|
3.
|
Shellenberger TD, Wang M, Gujrati M, et
al: BRAK/CXCL14 is a potent inhibitor of angiogenesis and a
chemotactic factor for immature dendritic cells. Cancer Res.
64:8262–8270. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Balkwill F: The significance of cancer
cell expression of the chemokine receptor CXCR4. Semin Cancer Biol.
14:171–179. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Zlotnik A, Burkhardt AM and Homey B:
Homeostatic chemokine receptors and organ-specific metastasis. Nat
Rev Immunol. 11:597–606. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Hromas R, Broxmeyer HE, Kim C, et al:
Cloning of BRAK, a novel divergent CXC chemokine preferentially
expressed in normal versus malignant cells. Biochem Biophys Res
Commun. 255:703–706. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Hara T and Nakayama Y: CXCL14 and insulin
action. Vitam Horm. 80:107–123. 2009. View Article : Google Scholar
|
|
8.
|
Sleeman MA, Fraser JK, Murison JG, et al:
B cell- and monocyte-activating chemokine (BMAC), a novel non-ELR
alpha-chemokine. Int Immunol. 12:677–689. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Frederick MJ, Henderson Y, Xu X, et al: In
vivo expression of the novel CXC chemokine BRAK in normal and
cancerous human tissue. Am J Pathol. 156:1937–1950. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Meuter S and Moser B: Constitutive
expression of CXCL14 in healthy human and murine epithelial
tissues. Cytokine. 44:248–255. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Kurth I, Willimann K, Schaerli P, Hunziker
T, Clark-Lewis I and Moser B: Monocyte selectivity and tissue
localization suggests a role for breast and kidney-expressed
chemokine (BRAK) in macrophage development. J Exp Med. 194:855–861.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Maerki C, Meuter S, Liebi M, et al: Potent
and broad-spectrum antimicrobial activity of CXCL14 suggests an
immediate role in skin infections. J Immunol. 182:507–514. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Shurin GV, Ferris RL, Tourkova IL, et al:
Loss of new chemokine CXCL14 in tumor tissue is associated with low
infiltration by dendritic cells (DC), while restoration of human
CXCL14 expression in tumor cells causes attraction of DC both in
vitro and in vivo. J Immunol. 174:5490–5498. 2005. View Article : Google Scholar
|
|
14.
|
Starnes T, Rasila KK, Robertson MJ, et al:
The chemokine CXCL14 (BRAK) stimulates activated NK cell migration:
implications for the downregulation of CXCL14 in malignancy. Exp
Hematol. 34:1101–1105. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Juremalm M and Nilsson G: Chemokine
receptor expression by mast cells. Chem Immunol Allergy.
87:130–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Tessema M, Klinge DM, Yingling CM, Do K,
Van Neste L and Belinsky SA: Re-expression of CXCL14, a common
target for epigenetic silencing in lung cancer, induces tumor
necrosis. Oncogene. 29:5159–5170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Balkwill FR: The chemokine system and
cancer. J Pathol. 226:148–157. 2012. View Article : Google Scholar
|
|
18.
|
Augsten M, Hagglof C, Olsson E, et al:
CXCL14 is an autocrine growth factor for fibroblasts and acts as a
multi-modal stimulator of prostate tumor growth. Proc Natl Acad Sci
USA. 106:3414–3419. 2009. View Article : Google Scholar
|
|
19.
|
Wente MN, Mayer C, Gaida MM, et al: CXCL14
expression and potential function in pancreatic cancer. Cancer
Lett. 259:209–217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Komori R, Ozawa S, Kato Y, Shinji H,
Kimoto S and Hata R: Functional characterization of proximal
promoter of gene for human BRAK/CXCL14, a tumor-suppressing
chemokine. Biomed Res. 31:123–131. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Park CR, You DJ, Kim DK, et al: CXCL14
enhances proliferation and migration of NCI-H460 human lung cancer
cells overexpressing the glycoproteins containing heparan sulfate
or sialic acid. J Cell Biochem. 114:1084–1096. 2013. View Article : Google Scholar
|
|
22.
|
Mokhtar NM, Cheng CW, Cook E, Bielby H,
Smith SK and Charnock-Jones DS: Progestin regulates chemokine
(C-X-C motif) ligand 14 transcript level in human endometrium. Mol
Hum Reprod. 16:170–177. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Izukuri K, Suzuki K, Yajima N, et al:
Chemokine CXCL14/BRAK transgenic mice suppress growth of carcinoma
cell transplants. [corrected]. Transgenic Res. 19:1109–1117.
2010.PubMed/NCBI
|
|
24.
|
Miyamoto C, Maehata Y, Ozawa S, et al:
Fasudil suppresses fibrosarcoma growth by stimulating secretion of
the chemokine CXCL14/BRAK. J Pharmacol Sci. 120:241–249. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Ikoma T, Ozawa S, Suzuki K, et al:
Calcium-calmodulin signaling induced by epithelial cell
differentiation upregulates BRAK/CXCL14 expression via the binding
of SP1 to the BRAK promoter region. Biochem Biophys Res Commun.
420:217–222. 2012. View Article : Google Scholar
|
|
26.
|
Ozawa S, Kato Y, Ito S, et al: Restoration
of BRAK/CXCL14 gene expression by gefitinib is associated with
antitumor efficacy of the drug in head and neck squamous cell
carcinoma. Cancer Sci. 100:2202–2209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Pelicano H, Lu W, Zhou Y, et al:
Mitochondrial dysfunction and reactive oxygen species imbalance
promote breast cancer cell motility through a CXCL14-mediated
mechanism. Cancer Res. 69:2375–2383. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Issa JP: CpG island methylator phenotype
in cancer. Nat Rev Cancer. 4:988–993. 2004. View Article : Google Scholar : PubMed/NCBI
|