Introduction
The transcriptional potential of the cell nucleus is
determined by availability of transcription factors and by the
structural organization of genes in the context of chromatin. Even
in terminally differentiated cells, specific regions of chromatin
must be remodelled to allow transcription activation or repression
in response to intra- and/or extracellular signals. Linker histone
H1 binds DNA in between nucleosomes and regulates chromatin higher
order structures (1–5), thus also mediating differential gene
expression (6,7).
The H1 family is the most divergent among histone
proteins, with at least 11 different genes in humans, most of which
form a cluster on chromosome 6 (chromosome 13 in mouse, and
chromosome 17 in the rat) (8). In
general, it is possible to distinguish between two types of H1
histone genes: clustered- and single-genes. Moreover, this peculiar
distribution of H1 genes is conserved among human, mouse and rat.
Each H1 protein subtype has been also suggested to have specific
distribution and function in chromatin (7,9,10).
In comparison with the intermediate chromatin condensing activity
of H1.3, for example, other subtypes have been classified as weak
condensers (H1.1 and H1.2) and strong condensers (H1.0, H1.4, H1.5,
and H1.x) (10,11).
Functional differences among subtypes have been,
however, difficult to identify since they probably have redundant
activities in development (9,12).
It is clear that linker histones are highly mobile in chromatin and
that they interact with both cytosolic and nuclear proteins, thus
regulating a variety of cellular processes (10); they are also able to promote
epigenetic silencing of genes, by regulating both DNA methylation
and histone H3 methylation (13).
H1° is a linker histone subtype the expression of which has been
mostly correlated with terminal differentiation (14,15).
In developing rat brain, the concentration of H1°
mRNA decreases in vivo between the embryonal day 18 (E18)
and the postnatal day 10 (P10), with inverse correlation to protein
accumulation (16); the
concentration of H1° mRNA also decreases in isolated neurons,
between the second and the fifth day of culture in a serum-free
medium, while an active synthesis of the corresponding proteins can
be observed (17). The H1° gene is
transcribed at the same rate at any stage studied, suggesting that
it is regulated mainly at post-transcriptional level (17). Since post-transcriptional control
processes are mediated by several classes of RNA-binding proteins
(18–21), it was likely that developing rat
brain contained mRNA-binding factors involved in H1° mRNA binding
and regulation. We indeed already reported identification of a
variety of H1° mRNA-binding proteins probably involved in the
regulation of H1° mRNA metabolism (22–30).
A number of cell types can shed into the environment
microvesicles of different sizes (MVs) under both physiological and
pathological conditions (31–35).
MVs contain a wide array of biological molecules, such as proteins,
lipids, DNA, microRNAs and mRNAs, and have been suggested to act as
means of cell-to-cell communication (36). MVs can trigger in target cells
various events, including apoptosis (31,37),
and cell survival and proliferation (38,39).
They have also been shown to contain metalloproteinases able to
digest extracellular matrix components, thus probably contributing
to tissue invasion (40).
In the present study, we analyzed expression of the
H1° gene in murine oligodendroglioma cells in order to shed further
light on possible functions of this linker histone variant which
still remains incompletely understood.
Materials and methods
Experimental animals
Wistar rats (Harlan, Udine, Italy) were housed in
the animal house of STEBICEF Department, University of Palermo,
Palermo, Italy. Procedures involving animals were in agreement with
the European Community Council Directive 2010/63/EU and were
approved by the University licensed veterinary. The number of
animals used was minimized as much as possible.
Cell cultures and immunofluorescence
Astrocytes were isolated from brain cortices of
2-day old newborn rats, as previously described (41), and cultured in DME/Ham’s F-12
(2/1), supplemented with 10% heat-inactivated fetal calf serum
(Sigma-Aldrich, MO, USA), and 100,000 U penicillin, 100 mg
streptomycin and 250 μg amphotericin B (Sigma-Aldrich) per
liter.
G26/24 oligodendroglioma cells were cultured in
DMEMHam’s F-12 (2:1) medium supplemented with 10% fetal calf serum
(FCS), and 100,000 U penicillin, 100 mg streptomycin and 250
μg amphotericin B (Sigma-Aldrich) per liter, for the same
time. Cell cultures were maintained in humidified 5%
CO2/95% air, at 37°C.
Some cultures of both astrocytes and
oligodendroglioma cells were then progressively adapted to a medium
known as Maat-medium (MM) (41)
and maintained in culture for additional 3 days, as previously
described (42,43).
For immunofluorescence, cells were fixed in 96%
ethanol; then astrocytes were immunostained with rabbit anti-glial
fibrillary acidic protein antibodies (GFAP; Sigma-Aldrich), and
oligodendroglioma cells with goat anti-actin antibodies (Santa
Cruz, CA, USA). The secondary antibodies were rhodamine- or
fluorescein isothiocyanate-conjugated anti-rabbit- or anti-goat
immuno globulins (both from Sigma-Aldrich). Cells were finally
observed under a fluorescence microscope (Olympus BX-50).
Northern blot analysis
Northern blot analysis was performed as previously
described (16). Total RNA was
purified from astrocytes cultured in NIH and in MM, and from G26/24
oligodendroglioma cells, according to Chomczynski and Sacchi
(44), and were separated on 1.5%
agarose, 6% formaldehyde denaturing gels, transferred to nylon
membranes and hybridized to a 33P-labeled (Perkin-Elmer,
MA, USA) fragment from the plasmid pMH1° (EMBL ID: X70685), cut
with EcoRI.
Purification of total cell extracts
Cells were collected and homogenized in
homogenization buffer (0.32 M sucrose; 50 mM sodium phosphate
buffer, pH 6.5; 50 mM KCl, 0.5 mM spermine; 0.15 mM spermidine; 2
mM EDTA, and 0.15 mM EGTA), containing the protease inhibitors
aprotinin (2 μg/ml), antipain (2 μg/ml), leupeptin (2
μg/ml), pepstatin A (2 μg/ml), benzamidine (1.0 mM),
and phenylmethylsulfonyl fluoride (1.0 mM), all purchased from
Sigma-Aldrich. Protein concentration was determined according to
Bradford (45).
Preparation of microvesicles from the
cell culture medium
Vesicles were prepared from oligodendroglioma G26/24
and astrocyte subconfluent healthy cells grown in FCS-free medium,
as previously described (33,37,40).
After 24 h of culture, conditioned media were centrifuged at 2,000
× g for 10 min and then at 4,000 × g for 15 min. The supernatant
was centrifuged at 105,000 × g (Ti60 Rotor, Beckman) for 90 min at
4°C. Pelleted vesicles were suspended with phosphate-buffered
saline, pH 7.5 (PBS) and protein concentration in isolated vesicles
was determined using Qubit® Protein Assay Kit
(Invitrogen, OR, USA).
Western blot analysis
Proteins (15 μg of total cell extracts) were
separated by electrophoresis on denaturing 12.5% polyacrylamide
slab gels (SDS-PAGE) and transferred to PVDF membrane (Immobilon P,
Millipore, MA, USA), as previously described (30). Samples on the membrane were
visualized by staining with Ponceau Red for 5 min. Membranes were
immunostained with rabbit polyclonal anti-H1° antibodies (Santa
Cruz) and mouse monoclonal anti-Hsc70 antibodies (Santa Cruz). The
secondary antibodies were anti-mouse IgG (H+L), AP conjugate, and
anti-rabbit IgG (Fc), AP conjugate (Promega Corporation, WI,
USA).
Western blots were scanned by the ImageJ program and
the values obtained were used to calculate the relative
concentration of H1° in cell extracts and vesicles. The values
obtained by this analysis were normalized respect to the value
obtained with Hsc70 antibodies or by scanning the membrane stained
with Ponceau Red. The measurements obtained from at least 3
independent experiments were finally used to calculate the relative
concentrations of the analyzed proteins in the different
conditions, as well as standard deviations (SD).
Preparation of in vitro transcripts and
T1 RNase protection assay
33P-radiolabeled H1° RNA was prepared as
previously described (23), using
as a template the plasmid pMH1° (46), which contains the H1° insert (EMBL
ID: X70685). H1° RNA was mixed with total cell extracts (15
μg), prepared as described above. For the T1 protection
assay, we used a previously described method (23) except that cross-linking of RNA to
proteins was performed before incubation with T1 RNase (EC
3.1.27.3; Roche, Switzerland). RNA-protein complexes were analyzed
by SDS-PAGE. At the end of the run, the gel was directly exposed to
X-ray film for autoradiography. The gels were also stained with
Coomassie Brilliant Blue R-250 (Sigma-Aldrich), to confirm loading
of equal amounts of proteins per lane.
Results
Expression of H1° linker histone in
G26/24 oligodendroglioma cells and astrocytes
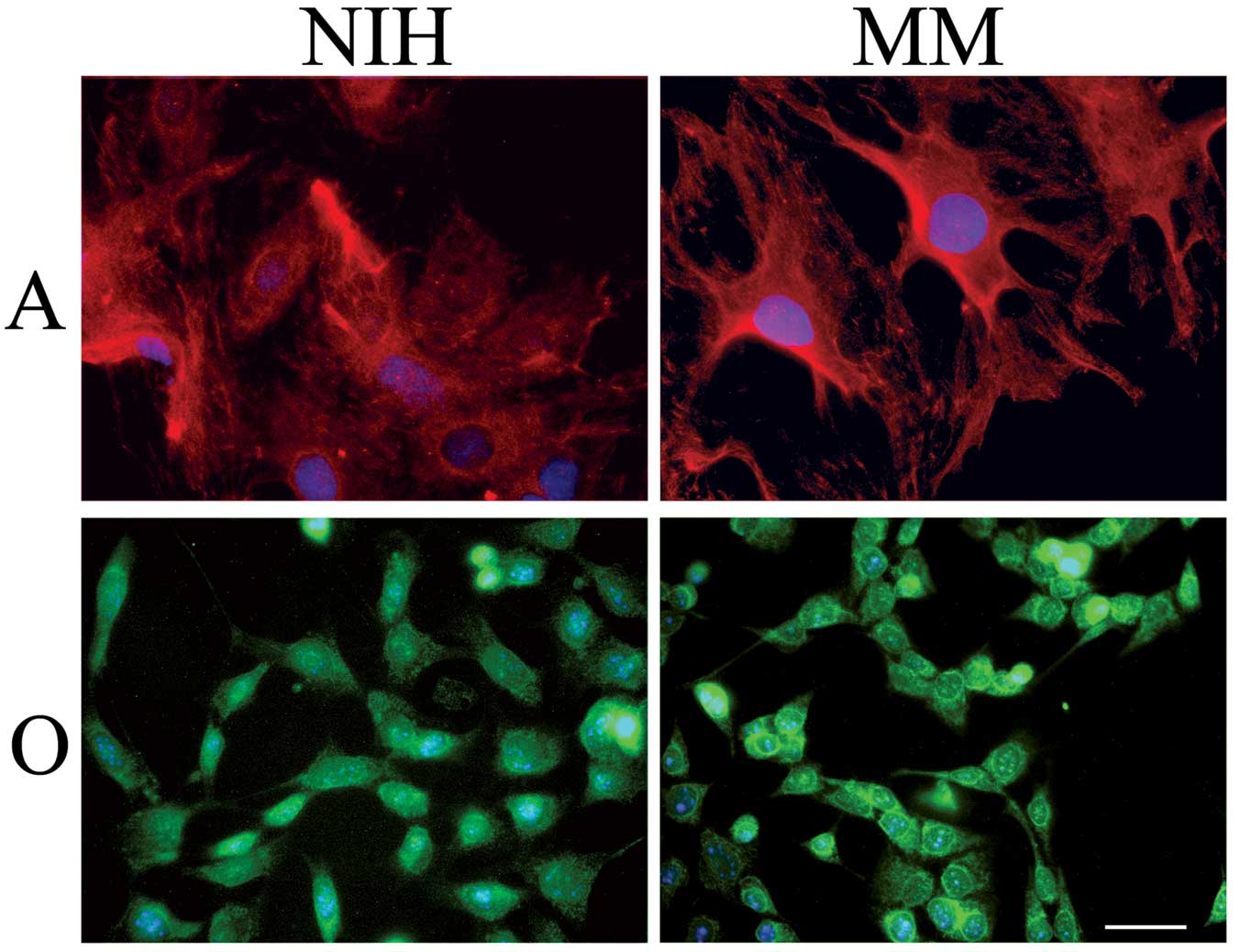
Astrocytes were cultured either in a serum-rich
(NIH)- or in a serum-free medium (MM) for 72 h. As shown in
Fig. 1, immunostaining of the
astrocyte-specific GFAP evidenced a higher number of star-like
brilliant cells when cells had been cultured in MM respect to cells
cultured in NIH. Pictures of this kind suggested that astrocytes
cultured in MM were a step forward, on the differentiation pathway,
in respect to cells cultured in NIH. In agreement with this
hypothesis, the linker histone H1°, a differentiation-specific
histone variant, was expressed at higher levels in astrocytes
cultured in MM (Fig. 2, lane 3)
than in astrocytes cultured in NIH-medium (Fig. 2, lane 2). Concentration of H1°
protein was even higher than in cortical fetal neurons, cultured in
MM (Fig. 2, lane 1), already
studied in the past (17). Once
the relationship between H1° protein expression and differentiation
was confirmed, we analyzed H1° expression in glial tumor cells. As
shown in Fig. 1, there is no
morphological difference between oligodendroglioma cells cultured
in NIH or in MM. We found, however, that these cells (Fig. 2, lane 4) accumulate the linker
histone H1° at levels comparable with those found in highly
differentiated astrocytes cultured in MM (Fig. 2, lane 3). Expression of H1° histone
in G26/24 cells did not change when cells were cultured in MM (data
not shown).
Since in neurons we had found that the increase of
H1° protein was accompanied by a decrease of the corresponding mRNA
levels (16,17), we also investigated, by northern
blot analysis, H1° RNA expression. As shown in Fig. 3, we found that the same correlation
exists also in astrocytes: H1° mRNA almost disappears in
differentiating astrocytes (Fig. 3,
lane 2) while it is abundant in astrocytes cultured in
NIH-medium (Fig. 3, lane 1).
Interestingly, in G26/24 oligodendroglioma cells both H1° protein
(Fig. 2, lane 4) and mRNA
(Fig. 3, lane 3) are expressed at
high levels.
H1° RNA-binding proteins in G26/24
oligodendroglioma cells and astrocytes
The fact that the concentration of H1° mRNA
decreased in astrocytes with an inverse correlation to H1° protein
accumulation suggested that in these brain cells, like in neurons
and whole rat brain, concentration of H1° histone was largely
regulated at the post-transcriptional level. Since this level of
gene expression control involves a variety of RNA-binding proteins,
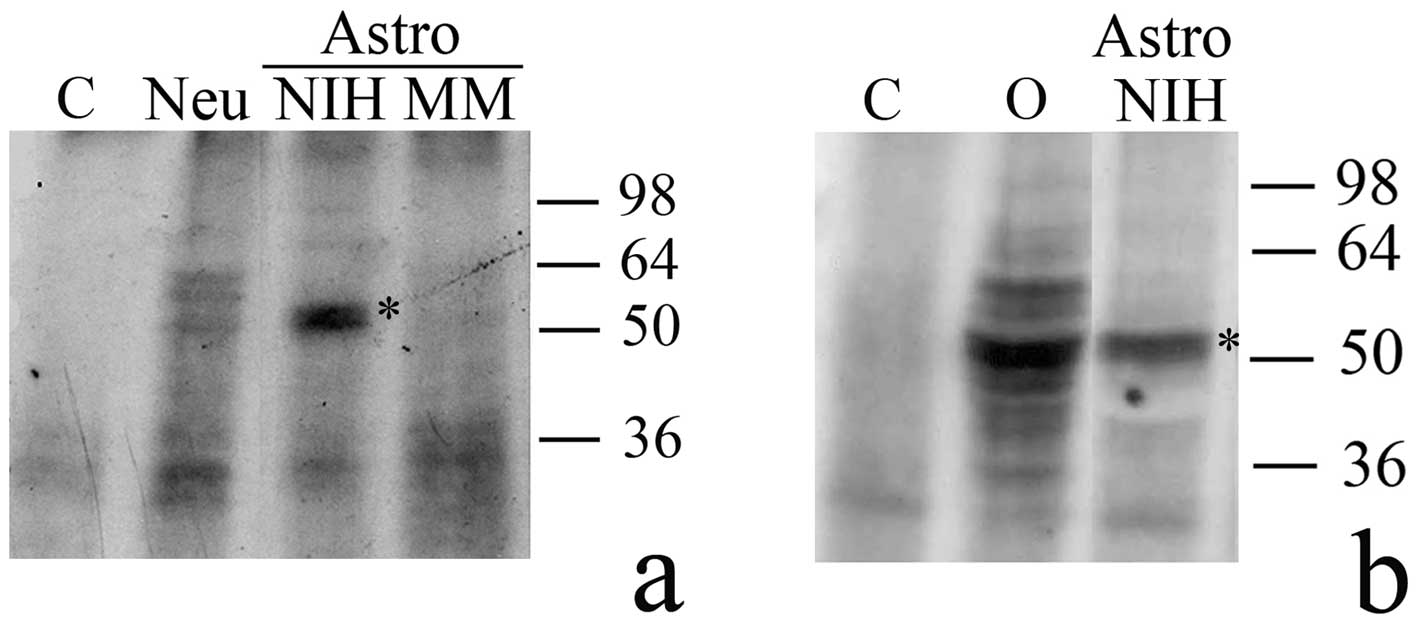
we tested astrocyte extracts for the presence of H1° RNA-binding
factors. As shown in Fig. 4, in
astrocytes cultured in the serum-rich medium (Astro, NIH) binding
factors are present which forms a major complex of about 50 kDa
with H1° RNA. No complex of the same apparent mass was evident in
either astrocytes (Fig. 4a, Astro,
MM) or neurons (Fig. 4a, Neu)
cultured in MM. A major signal of about 50 kDa, and several minor
ones, due to formation of H1° RNA-protein complexes, were also
visible when G26/24 cell extracts were analyzed (Fig. 4b, O).
H1° histone protein is shed by G26/24
cells through extracellular membrane vesicles
G26/24 cells were recently found to shed
extracellular membrane vesicles which can induce neuronal death
(31,37) and contain a variety of cell
proteins, among which extracellular matrix metalloproteases
(40). In the present study, we
also tested the presence of H1° histone in the vesicles. As shown
in Fig. 5a, H1° is clearly present
in the vesicles shed from G26/24 tumor cells but not in those shed
by astrocytes. This analysis also confirmed the already reported
presence of Hsc70 in vesicles shed from oligodendroglioma cells
(37). Moreover, in the present
study, we report that Hsc70 is also found in vesicles released from
astrocytes (Fig. 5a). The
statistical analysis performed on at least three different
experiments (Fig. 5b) suggested
that H1° is specifically enriched in vesicles: the relative
proportion of H1° in vesicles (V) shed from G26/24 (O), respect to
lysates (L) of the same cells, is indeed clearly higher in
comparison with the relative proportion of Hsc70 in the same
samples (Fig. 5c).
Discussion
The transcriptional potential of the cell nucleus is
determined by availability of transcription factors as well as by
the structural organization of genes in the context of chromatin.
Even in terminally differentiated cells, specific regions of
chromatin are remodelled to allow transcription activation or
repression in response to intra- and/or extracellular signals. One
of the mechanisms at the basis of chromatin dynamics is likely to
be synthesis and incorporation of replacement histone variants,
such as the core H3.3 histone and the linker H1° histone. In
developing brain, as well as in fetal neurons differentiating in
culture, H1° mRNA is progressively down-regulated in vivo at
the time of rat brain maturation (16,17),
with an inverse correlation to synthesis and accumulation of H1°
protein, although the transcriptional activity of H1° gene is
unaffected by terminal differentiation. This finding suggested that
H1° expression in the brain was mostly regulated at the
post-transcriptional level (17).
As shown in the present study, like in the whole
brain and in isolated neurons, the linker histone H1° is expressed
at higher levels in astrocytes cultured in a serum-free medium (MM)
in which they acquire a clearly differentiated star-like
appearance. In these cells, concentration of H1° protein is even
higher than in cortical fetal neurons, cultured in the same medium.
Moreover, like in neurons, H1° mRNA almost disappears with protein
accumulation. It is likely that H1° mRNA is destabilized and
degraded at higher rates concomitant with its increased engagement
with the translational apparatus.
How can the availability of H1° mRNA to the
ribosomes be controlled? We already knew that a variety of H1°
mRNA-binding factors exist in the rat brain (22–30).
Now we evidenced an RNA-protein covalent complex of about 50 kDa,
which disappears when astrocytes are cultured in differentiating
conditions. Since the T1 RNase assay is a functional test, we
cannot say whether the factor disappears during differentiation or
undergoes a post-translational modification that abolishes its
binding activity.
Unexpectedly, in glial tumor cells concentration of
both H1° mRNA and protein is very high and does not correlate with
a decrease of proliferation rate. Thus the still unknown mechanism
responsible for the inverse correlation between H1° mRNA and
protein concentrations does not work in oligodendroglioma cells.
Complexes with a size similar to the complex seen in
undifferentiated astrocytes do form, but they are probably not able
to block access to ribosomes. Most importantly, synthesis of a high
level of H1° histone protein does not correlate with a decrease of
proliferation rate.
Since we already knew that G26/24 cells actively
shed extracellular microvesicles (MVs), which contain a variety of
proteins (31,37,40),
we asked whether oligodendroglioma cells are able to unload H1°
into the extracellular environment. Here we report that indeed H1°
histone is present in MVs shed by oligodendroglioma cells but not
in those shed by astrocytes, even if both populations of MVs
contain, for example, Hsc70 chaperone. Although the role of
shedding in tumor cells is not yet completely understood, it could
be also involved in eliminating proteins from cells (such as the
H1° histone) that could be otherwise able to counteract
proliferation.
Acknowledgements
This study was supported by a special
grant of Merck Serono S.p.A. to G.S. and by the University of
Palermo (Università degli Studi di Palermo, Palermo, Italy; ex
60%). P.S. received a PhD studentship from the University of
Palermo.
References
|
1.
|
Allan J, Hartman PG, Crane-Robinson C and
Aviles FX: The structure of histone H1 and its location in
chromatin. Nature. 288:675–679. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Bates DL, Butler PJ, Pearson EC and Thomas
JO: Stability of the higher-order structure of chicken-erythrocyte
chromatin in solution. Eur J Biochem. 119:469–476. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Huang HC and Cole RD: The distribution of
H1 histone is nonuniform in chromatin and correlates with different
degrees of condensation. J Biol Chem. 259:14237–14242.
1984.PubMed/NCBI
|
|
4.
|
Hill DA: Influence of linker histone H1 on
chromatin remodeling. Biochem Cell Biol. 79:317–324. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Fan Y, Nikitina T, Zhao J, Fleury TJ,
Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL and Skoultchi
AI: Histone H1 depletion in mammals alters global chromatin
structure but causes specific changes in gene regulation. Cell.
123:1199–1212. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Crane-Robinson C: How do linker histones
mediate differential gene expression? Bioessays. 21:367–371. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Izzo A, Kamieniarz K and Schneider R: The
histone H1 family: specific members, specific functions? Biol Chem.
389:333–343. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Marzluff WF, Gongidi P, Woods RK, Jin J
and Maltais LJ: The human and mouse replication-dependent histone
genes. Genomics. 80:487–498. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Happel N and Doenecke D: Histone H1 and
its isoforms. Contribution to chromatin structure and function.
Gene. 431:1–12. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Kowalski A and Palyga J: Linker histone
subtypes and their allelic variants. Cell Biol Int. 36:981–996.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Clausell J, Happel N, Hale TK, Doenecke D
and Beato M: Histone H1 subtypes differentially modulate chromatin
condensation without preventing ATP-dependent remodeling by SWI/SNF
or NURF. PLoS One. 4:e00072432009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Fan Y, Sirotkin A, Russel RG, Ayala J and
Skoultchi AI: Individual somatic H1 subtypes are dispensable for
mouse development even in mice lacking the H1(0) replacement
subtypes. Mol Cell Biol. 21:7933–7943. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Yang SM, Byung JK, Norwood Toro L and
Skoultchi AI: H1 linker histone promotes epigenetic silencing by
regulating both DNA methylation and histone H3 methylation. Proc
Natl Acad Sci USA. 110:1708–1713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Zlatanova J and Doenecke D: Histone H1
zero: a major player in cell differentiation. FASEB J. 8:1260–1268.
1994.PubMed/NCBI
|
|
15.
|
Gabrilovich DI, Cheng P, Fan Y, Yu B,
Nikitina E, Sirotkin A, Shurin M, Oyama T, Adachi Y, Nadaf S,
Carbone DP and Skoultchi AI: H1° histone and differentiation of
dendritic cells. A molecular target for tumor-derived factors. J
Leukoc Biol. 72:285–296. 2002.
|
|
16.
|
Castiglia D, Cestelli A, Scaturro M,
Nastasi T and Di Liegro I: H1° and H3.3B mRNA levels in developing
rat brain. Neurochem Res. 19:1531–1537. 1994.
|
|
17.
|
Scaturro M, Cestelli A, Castiglia D,
Nastasi T and Di Liegro I: Post-transcriptional regulation of H1°
and H3.3 histone genes in differentiating rat cortical neurons.
Neurochem Res. 20:969–976. 1995.
|
|
18.
|
Burd CG and Dreyfuss G: Conserved
structures and diversity of functions of RNA-binding proteins.
Science. 265:615–621. 1994. View Article : Google Scholar
|
|
19.
|
Hentze MW: Translational regulation:
versatile mechanisms for metabolic and developmental control. Curr
Opin Cell Biol. 7:393–398. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Siomi H and Dreyfuss G: RNA-binding
proteins as regulators of gene expression. Curr Opin Genet Dev.
7:345–353. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Derrigo M, Cestelli A, Savettieri G and Di
Liegro I: RNA-protein interactions in the control of stability and
localization of messenger RNA (Review). Int J Mol Med. 5:111–123.
2000.PubMed/NCBI
|
|
22.
|
Castiglia D, Scaturro M, Nastasi T,
Cestelli A and Di Liegro I: PIPPin, a putative RNA-binding protein,
specifically expressed in the rat brain. Biochem Biophys Res
Commun. 218:390–394. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Scaturro M, Nastasi T, Raimondi L,
Bellafiore M, Cestelli A and Di Liegro I: H1° RNA-binding proteins
specifically expressed in the rat brain. J Biol Chem.
273:22788–22791. 1998.
|
|
24.
|
Nastasi T, Scaturro M, Bellafiore M,
Raimondi L, Beccari S, Cestelli A and Di Liegro I: PIPPin is a
brain-specific protein that contains a cold-shock domain and binds
specifically to H1° and H3.3 mRNAs. J Biol Chem. 274:24087–24093.
1999.PubMed/NCBI
|
|
25.
|
Raimondi L, D’Asaro M, Proia P, Nastasi T
and Di Liegro I: RNA-binding activity of PIPPin requires the entire
protein. J Cell Mol Med. 7:35–42. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Scaturro M, Sala A, Cutrona G, Raimondi L,
Cannino G, Fontana S, Pucci-Minafra I and Di Liegro I: Purification
by affinity chromatography of H1° RNA-binding proteins from rat
brain. Int J Mol Med. 11:509–513. 2003.
|
|
27.
|
Bono E, Compagno V, Proia P, Raimondi L,
Schiera G, Favaloro V, Campo V, Donatelli M and Di Liegro I:
Thyroid hormones induce sumoylation of the cold shock
domain-containing protein PIPPin in developing rat brain and in
cultured neurons. Endocrinology. 148:252–257. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Sala A, Scaturro M, Proia P, Schiera G,
Balistreri E, Aflalo-Rattenbach R and Di Liegro I: Cloning of a
rat-specific long PCP4/PEP19 isoform (LPI). Int J Mol Med.
19:501–509. 2007.PubMed/NCBI
|
|
29.
|
Saladino P, Di Liegro CM, Proia P, Sala A,
Schiera G, Lo Cicero A and Di Liegro I: RNA-binding activity of the
rat calmodulin-binding PEP-19 protein and of the long Pep-19
isoform. Int J Mol Med. 29:141–145. 2012.PubMed/NCBI
|
|
30.
|
Di Liegro CM, Schiera G, Proia P, Saladino
P and Di Liegro I: Identification in the rat brain of a set of
nuclear proteins interacting with H1° mRNA. Neuroscience.
229:71–76. 2013.PubMed/NCBI
|
|
31.
|
D’Agostino S, Salamone M, Di Liegro I and
Vittorelli ML: Membrane vesicles shed by oligodendroglioma cells
induce neuronal apoptosis. Int J Oncol. 29:1075–1085.
2006.PubMed/NCBI
|
|
32.
|
Schiera G, Proia P, Alberti C, Mineo M,
Savettieri G and Di Liegro I: Neurons produce FGF2 and VEGF and
secrete them at least in part by shedding extracellular vesicles. J
Cell Mol Med. 11:1384–1394. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Proia P, Schiera G, Mineo M, Ingrassia AM,
Santoro G, Savettieri G and Di Liegro I: Astrocytes shed
extracellular vesicles that contain fibroblast growth factor-2 and
vascular endothelial growth factor. Int J Mol Med. 2:63–67.
2008.PubMed/NCBI
|
|
34.
|
D’Asti E, Garnier D, Lee TH, Montermini L,
Meehan B and Rak J: Oncogenic extracellular vesicles in brain tumor
progression. Front Physiol. 3:2942012.PubMed/NCBI
|
|
35.
|
Corrado C, Raimondo S, Chiesi A, Ciccia F,
De Leo G and Alessandro R: Exosomes as intercellular signaling
organelles involved in health and disease: basic science and
clinical applications. Int J Mol Sci. 14:5338–5366. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Mathivanan S, Ji H and Simpson RJ:
Exosomes: extracellular organelles important in intercellular
communication. J Proteomics. 73:1907–1920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Lo Cicero A, Schiera G, Proia P, Saladino
P, Savettieri G, Di Liegro CM and Di Liegro I: Oligodendroglioma
cells shed microvesicles which contain TRAIL as well as molecular
chaperones and induce cell death in astrocytes. Int J Oncol.
39:1353–1357. 2011.PubMed/NCBI
|
|
38.
|
Deregibus MC, Cantaluppi V, Calogero R, Lo
Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B and Camussi G:
Endothelial progenitor cell derived microvesicles activate an
angiogenic program in endothelial cells by a horizontal transfer of
mRNA. Blood. 110:2440–2448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WTJ, Carter BS, Krichevsky AM
and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Lo Cicero A, Majkowska I, Nagase H, Di
Liegro I and Troeberg L: Microvesicles shed by oligodendroglioma
cells and rheumatoid synovial fibroblasts contain aggrecanase
activity. Matrix Biol. 31:229–233. 2012.PubMed/NCBI
|
|
41.
|
Cestelli A, Savettieri G, Ferraro D and
Vitale F: Formulation of a novel synthetic medium for selectively
culturing rat CNS neurons. Brain Res. 354:219–227. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Schiera G, Bono E, Raffa MP, Gallo A,
Pitarresi GL, Di Liegro I and Savettieri G: Synergistic effects of
neurons and astrocytes on differentiation of brain capillary
endothelial cells in culture. J Cell Mol Med. 7:165–170. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Schiera G, Sala S, Gallo A, Raffa MP,
Pitarresi GL, Savettieri G and Di Liegro I: Permeability properties
of a three-cell type in vitro model of Blood-Brain barrier. J Cell
Mol Med. 9:373–379. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Bradford MM: A rapid and sensitive method
for the quantification of microgram quantities of protein utilizing
the principle of protein dye binding. Anal Biochem. 72:248–254.
1978. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Castiglia D, Gristina R, Scaturro M and Di
Liegro I: Cloning and analysis of cDNA for rat histone H1°. Nucleic
Acids Res. 21:16741993.
|