Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer and third leading cause of cancer-related deaths
worldwide and its prognosis is poor despite advances in diagnostic
and therapeutic modalities (1,2). A
minority of patients with HCC are diagnosed during early stages
when the disease is amenable to potentially curative treatments
such as surgery and topical therapy (for example, radiofrequency
ablation) (3). The prognosis is
poor for patients diagnosed at an advanced stage or with recurrent
lesions after surgery or topical therapies, owing to underlying
liver damage and lack of effective treatment options (1,4).
Therefore, investigations that clarify the mechanisms of
carcinogenesis and tumor progression in HCC are urgently needed to
discover targets for therapy and prognostic markers for recurrence
and progression of HCC.
Evidence indicates that the pathogenesis of HCC
occurs in a stepwise process driven by oncogene activation and
inactivation of tumor suppressor genes (TSGs) (5,6).
Epigenetic alterations such as promoter hypermethylation can lead
to the transcriptional silencing of TSGs (7,8). We
identified several tumor-related genes frequently silenced through
promoter hypermethylation in HCC, suggesting that expression status
and promoter hypermethylation of these genes can serve as a
biomarker for early detection of HCC (9–11).
The differentially expressed in normal and
neoplastic cells (DENN) domain proteins regulate Rab GTPases and
represent a newly recognized class of membrane trafficking proteins
(12–16). The Rab family comprises 70 members
in humans and represents the largest family of small GTPases
(17). Rab proteins cycle between
inactive GDP-bound and active GTP-bound states. In the active
state, they recruit effectors that control multiple aspects of
membrane trafficking (13,18,19).
The DENN domain present in members of the connecdenn family of
proteins, interacts directly with Rab35 and functions as a guanine
nucleotide exchange factor (GEF) for this GTPase (20). GEFs activate Rabs by mediating the
exchange of GDP for GTP. The human genome encodes eight DENND (DENN
domain) proteins that form eight families based on homology and
domain structure as follows: DENND1A-1C, DENND2A-2D, DENND3,
DENND4A-4C, DENND5A/5B, DENND6A/6B, MTMR5/13 and DENN/MADD. The
DENN domain is located toward the N-terminus, except for the DENND2
family, where it is located toward the C-terminus (21–24).
There is no significant sequence similarity to other proteins
outside of the DENN domain.
Little is known about the functions and expression
patterns of DENND family proteins in malignant tumors, although
they play important roles in intracellular signaling pathways by
integrating the activity of Rab pathways. DENND2D, which is
located on chromosome 1p13.3, encodes a 53-kDa protein, which
suppresses the proliferation and tumorigenicity of non-small cell
lung cancer cells (23,25); however, the role of DENND family
proteins in gastroenterological cancers has not been reported.
Accordingly, we focused on DENND2D and investigated the regulation
of its expression in an attempt to identify a TSG regulated by
silencing through promoter hypermethylation. Our present study also
focused on developing novel epigenetic biomarkers for HCC.
Materials and methods
Sample collection
Nine HCC cell lines (Hep3B, HepG2, HLE, HLF, HuH1,
HuH2, HuH7, PLC/PRF/5 HepG2 and SK-Hep1) were obtained from the
American Type Culture Collection (Manassas, VA, USA) and cultured
in RPMI-1640 medium supplemented with 10% fetal bovine serum at
37°C in an atmosphere containing 5% CO2. Primary HCC
tissues and corresponding non-cancer tissues were collected from 92
patients (75 men, 17 women; mean age, 63.2±9.9 years; range, 34–84
years) who underwent liver resection for HCC at Nagoya University
Hospital between January, 1998 and July, 2008. Mean duration of
patient follow-up was 48.3±36.9 months (range, 0.8–147 months).
Specimens were classified histologically using the 7th edition of
the Union for International Cancer Control (UICC) classification
(26). Written informed consent
for surgery and use of clinical data, as required by the
institutional review board, was obtained from all patients. Tissue
samples were immediately flash frozen in liquid nitrogen and stored
at −80°C. Microscopic evaluations were performed to ensure that
tumor samples contained >80% tumor cells, whereas non-cancer
liver tissue samples did not contain regenerative or dysplastic
nodules.
Reverse transcription-polymerase chain
reaction (RT-PCR) and quantitative real-time RT-PCR (qPCR)
The levels of DENND2D mRNA expression were analyzed
using RT-PCR and qRT-PCR. Total RNAs (10 μg) isolated from
each of the HCC cell lines listed above, 92 primary HCC tissues and
corresponding non-cancer tissues were used as templates to generate
complementary DNAs (cDNAs). PCR primers for DENND2D were as
follows: sense (S) (5′-CACTGCTCTACCCCTTCAGC-3′ in exon 7) and
anti-sense (AS) (5′-TTTTTCATCACCAACCGACA-3′ in exons 9 and 10),
which amplify a 204-base-pair (bp) product. RT-PCR amplification
was performed as follows: 40 cycles at 94°C for 30 sec, 60°C for 30
sec and 72°C for 30 sec after an initial denaturation step at 94°C
for 5 min. To confirm that equal amounts of cDNA were used as
templates, RT-PCR of β-actin was performed. Each RT-PCR product was
loaded directly onto 2% agarose gels, electrophoresed, stained with
ethidium bromide and visualized with ultraviolet light. The qPCR
reactions were performed using a SYBR® Green PCR Core
Reagents kit (Life Technologies, Carlsbad, CA, USA) under the
following conditions: 1 cycle at 50°C for 2 min, 1 cycle at 95°C
for 10 min and 45 cycles at 95°C for 15 sec and 60°C for 30 sec.
Real-time detection of the SYBR-Green emission was conducted with
an ABI PRISM® 7000 Sequence Detection System (Life
Technologies). The primers were those described above. For
standardization, GAPDH (TaqMan®, GAPDH Control Reagents,
Life Technologies) was amplified in each sample. Nine HCC cell
lines and 92 pairs of clinical samples and negative control
reactions without templates were analyzed. Reactions were performed
in triplicate. The amount of amplified DENND2D DNA in each
sample was normalized to that of GAPDH. The expression of
DENND2D mRNA was defined as downregulated in tumor tissues
when its level was less than one-third that of the corresponding
non-cancer tissues.
Analysis of the promoter region of
DENND2D
The nucleotide sequence of the DENND2D
promoter region was analyzed to determine the presence or absence
of CpG islands defined as: ≥200-bp region of DNA with a high GC
content (>50%) and an Observed CpG/Expected CpG ratio ≥0.6
(27). We used CpG Island Searcher
software (http://cpgislands.usc.edu/) to
determine the locations of CpG islands (28).
Methylation-specific PCR (MSP)
DNA samples from HCC cell lines, HCC tissues and
corresponding non-cancer tissues were treated with bisulfite.
Briefly, 2 μg of DNA was denatured with NaOH, reacted with
sodium bisulfite and purified using the Wizard® PCR
Preps DNA Purification System resin (Promega, Madison, WI, USA),
treated again with NaOH, precipitated with ethanol and resuspended
in water. The sequences of the unmethylated primer pairs that
amplify a 102-bp product were derived from the DENND2D
promoter region upstream of exon 1 are: S
(5′-GATATGTGTTTTTGTGGATT-3′) and AS (5′-ACACATCCAAAACTAAAC-3′).
Primer sequences derived from the DENND2D promoter region
used to detect methylated DNA amplify a 193-bp product and are: S
(5′-AGGTGGCGTCGTTTAGTTTC-3′) and AS (5′-GCGAATCCGACACTTTCACT-3′).
DNA was amplified as follows: 45 cycles of 94°C for 30 sec 58°C for
30 sec and 72°C for 30 sec after an initial denaturation step at
94°C for 5 min. Each PCR product was loaded directly on 2% agarose
gels, electrophoresed, stained with ethidium bromide and visualized
with ultraviolet light.
Bisulfite sequence analysis
Genomic bisulfite-treated DNAs from HCC cell lines
were sequenced to verify the MSP results. The sequences of the
primer pair used to generate a fragment for sequencing was derived
from the DENND2D promoter region: S
(5′-GGAGGTTAAGGATAGGGG-3′) and AS (5′-ACACTAACCCCCATAACC-3′), which
amplify a 133-bp product. DNA was amplified as follows: 50 cycles
at 94°C for 30 sec, 63°C for 30 sec and 72°C for 30 sec following
an initial denaturation step at 94°C for 5 min. PCR products were
purified directly using the QIAquick PCR Purification kit (Qiagen,
Hilden, Germany). Purified DNA fragments were subcloned into the TA
cloning vector (Life Technologies). Each DNA sample was mixed with
3 μl of specific primer (M13) and 4 μl of Cycle
Sequence Mix (BigDye® Terminator v1. 1 Cycle Sequencing
kit, Life Technologies, Grand Island, NY, USA). Sequences were
analyzed using an Applied Biosystems ABI PRISM 310 DNA Analyzer and
sequence electropherograms were generated using ABI Sequence
Analysis 3.0 software (Life Technologies).
5-Aza-2′-deoxycytidine (5-aza-dC)
treatment
To assess the relation of promoter hypermethylation
to DENND2D expression, HCC cells (1.5×106) were
treated with 5-aza-dC (Sigma-Aldrich, St. Louis, MO, USA) to
inhibit DNA methylation and were cultured for 6 days with medium
changes on days 1, 3 and 5. RNA was extracted and RT-PCR was
performed as described above.
Immunohistochemistry (IHC)
We used IHC to investigate DENND2D localization in
30 representative sections of well preserved HCC tissues.
Formalin-fixed, paraffin-embedded tissues were treated with 3%
H2O2 to inhibit endogenous peroxidase,
followed by epitope retrieval using five incubations in 10 mM
citrate buffer at 95°C, 5 min each. The samples were incubated with
Histofine SAB-PO(R) (Nichirei, Tokyo, Japan) for 5 min to limit
non-specific reactivity and were then incubated for 1 h at room
temperature with a rabbit antibody against DENND2D (PA5-24032,
Thermo Fisher Scientific Inc., Rockford, IL, USA) diluted 1:100 in
Antibody Diluent (Dako). Sections were developed for 2 min using
liquid 3, 3′-diaminobenzidine as the substrate (Nichirei). Staining
properties were determined using surrounding hepatic veins as
internal controls and staining patterns were compared between HCCs
and the corresponding non-cancer tissues. To avoid subjectivity,
specimens were randomized and coded before analysis by two
independent observers blinded to the status of the samples. Each
observer evaluated all specimens at least twice within a given time
interval to minimize intraobserver variation.
Statistical analysis
The relative mRNA expression levels
(DENND2D/GAPDH) between HCC and non-cancer tissues were
analyzed using the Mann-Whitney U test. The χ2 test was
used to analyze the association between the expression and
methylation status of DENND2D and clinicopathological
parameters. Overall and disease-free survival rates were calculated
using the Kaplan-Meier method and the difference in survival curves
was analyzed using the log-rank test. We performed multivariable
regression analysis to detect prognostic factors using the Cox
proportional hazards model and variables with a P<0.05 were
entered into the final model. All statistical analysis was
performed using JMP® 10 software (SAS Institute Inc,
Cary, NC, USA). P<0.05 was considered statistically
significant.
Results
DENND2D mRNA expression in HCC cell lines
and tumor tissues
The levels of DENND2D mRNA detected using qPCR were
reduced significantly relative to median value of normal liver
controls in all HCC cell lines except HuH1 (Fig. 1A). In particular, the
DENND2D mRNA levels were 20-times lower in Hep3B, HLE, HuH2
and PLC-PRF5 cells. When we compared the levels of DENND2D
mRNA in non-cancer tissues of HCC patients with (n=35) or without
(n=57) cirrhosis, no significant difference was observed,
suggesting that the expression of DENND2D mRNA in non-cancer
liver tissue was not affected by liver fibrosis (Fig. 1B). The expression level of
DENND2D mRNA in 72 (78%) of 92 patients was lower in HCC
tissues than in the corresponding normal tissues. Further, the mean
expression of DENND2D mRNA was 2-fold lower in HCC tissues
than in corresponding normal tissues (P<0.001, Fig. 1C).
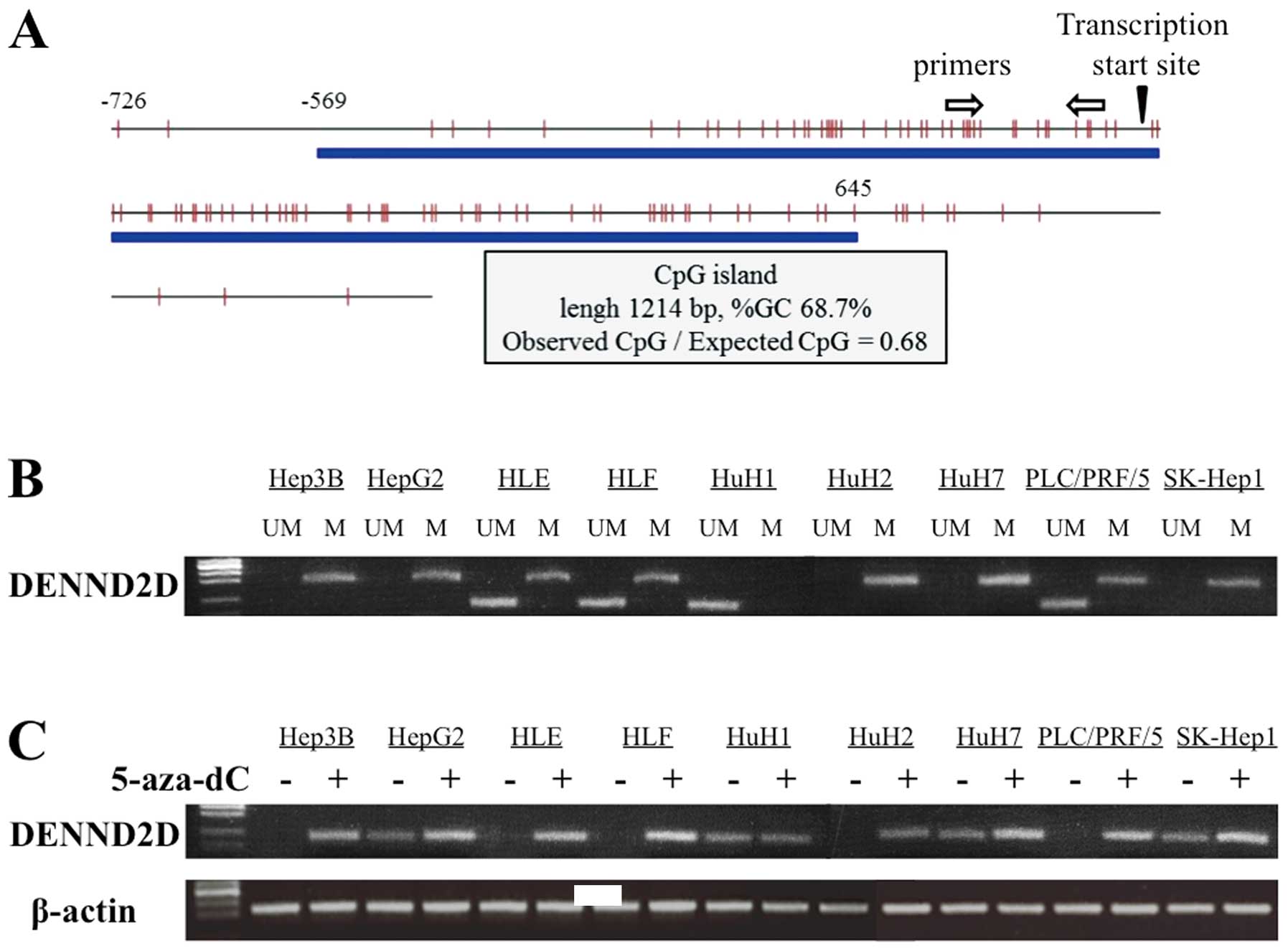
Identification of a CpG island in the
DENND2D promoter
A CpG island was identified at the DENND2D
promoter region using the CpG Island Searcher. The properties of
the CpG island are: 1214 bp, 68.7% GC and 0.68 Observed
CpG/Expected CpG ratio (Fig. 2A).
Therefore, we hypothesized that hypermethylation of the CpG islands
regulates the expression of DENND2D in HCC tissue.
MSP analysis of HCC cell lines
MSP was conducted to verify the hypothesis described
above. We first determined the methylation status of DENND2D
in nine HCC cell lines. Bands consistent with methylated DNA were
detected in all HCC cell lines except HuH1. PCR using unmethylated
primers, amplified bands from HLE, HLF, HuH1 and PLC/PRF/5
DENND2D DNA (Fig. 2B). We
conclude that methylation of the DENND2D promoter was
complete in Hep3B, HepG2, HuH2, HuH7 and SK-Hep1; partial in HLE,
HLF and PLC/PRF/5 cells and undetectable in HuH1 cells.
Transcription of DENND2D in cells treated
with 5-aza-dC
To determine whether promoter hypermethylation leads
to the suppression of DENND2D transcription, we analyzed HCC
cell lines before and after treatment with the DNA methylation
inhibitor, 5-aza-dC. Using semi-quantitative RT-PCR, reactivation
or an increase in DENND2D expression was detected in all HCC
cell lines except HuH1, consistent with the results of the MSP
analysis (Fig. 2C).
Bisulfite sequence analysis
To confirm the results of the MSP experiments, we
directly sequenced the DENND2D promoter in HepG2 (complete
methylation) and HuH1 (undetectable methylation) cells and found
that all CpGs in the HepG2 fragment were CG, while TG was present
at the corresponding position in HuH1 cells (Fig. 3), thus confirming the MSP data.
Prognostic value of DENND2D expression in
92 patients with HCC
Downregulation of DENND2D mRNA was detected
in tumor samples from 34 of 92 (37.0%) patients with HCC. Median
disease specific survival (32.6 versus 112 months, P=0.020,
Fig. 4A) and relapse-free survival
(10.3 versus 23.0 months, P<0.001, Fig. 4B) were significantly shorter in
patients with downregulation of DENND2D mRNA. Tumor size
≥3.0 cm and vascular invasion, but not DENND2D expression,
were identified as independent prognostic factors for disease
specific survival using multivariate analysis (Table I). In contrast, univariate analysis
for recurrence-free survival showed that serosal infiltration,
vascular invasion and downregulation of DENND2D mRNA were
significantly prognostic of adverse outcomes. Multivariate analysis
identified downregulation of DENND2D mRNA as an independent
prognostic factor for recurrence-free survival (hazard ratio 2.86,
P=0.002, Table II). Downregulation
of DENND2D mRNA was not significantly associated with other
clinicopathological parameters.
 | Table I.Prognostic factors for
disease-specific survival in 92 patients with hepatocellular
carcinoma. |
Table I.
Prognostic factors for
disease-specific survival in 92 patients with hepatocellular
carcinoma.
| Variable | n | Univariate | Multivariate |
|---|
|
|
|---|
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥65) | 47 | 1.61 | 0.87–3.03 | 0.130 | | | |
| Gender (male) | 77 | 1.18 | 0.55–2.92 | 0.695 | | | |
| Background liver
(cirrhosis) | 36 | 1.44 | 0.77–2.67 | 0.250 | | | |
| Pugh-Child’s
classification (B) | 7 | 1.28 | 0.31–3.57 | 0.692 | | | |
| AFP (>20
ng/ml) | 47 | 1.90 | 1.03–3.57 | 0.042a | 1.70 | 0.87–3.36 | 0.121 |
| PIVKA II (>40
mAU/ml) | 54 | 2.25 | 1.18–4.51 | 0.013a | 1.36 | 0.65–2.99 | 0.415 |
| Tumor multiplicity
(multiple) | 24 | 1.97 | 1.00–3.69 | 0.049a | 2.26 | 1.08–4.62 | 0.032a |
| Tumor size (≥3.0
cm) | 67 | 2.41 | 1.16–5.67 | 0.017a | 1.18 | 0.49–3.07 | 0.712 |
| Tumor
differentiation (well) | 28 | 0.51 | 0.23–1.03 | 0.060 | | | |
| Growth type
(invasive growth) | 17 | 1.16 | 0.51–2.35 | 0.705 | | | |
| Serosal
infiltration | 25 | 2.33 | 1.16–4.49 | 0.018a | 1.05 | 0.46–2.34 | 0.898 |
| Formation of
capsule | 68 | 0.98 | 0.51–2.00 | 0.952 | | | |
| Infiltration to
capsule | 54 | 1.07 | 0.58–2.03 | 0.822 | | | |
| Septum
formation | 34 | 0.91 | 0.49–1.75 | 0.776 | | | |
| Vascular
invasion | 23 | 3.33 | 1.73–6.23 | <0.001a | 2.35 | 1.08–5.11 | 0.032a |
| Margin status
(positive) | 24 | 2.23 | 1.17–4.15 | 0.016a | 1.89 | 0.96–3.60 | 0.064 |
| Hypermethylation of
DENND2D | 69 | 1.22 | 0.61–2.73 | 0.588 | | | |
| Downregulation of
DENND2D mRNA | 34 | 2.08 | 1.10–3.89 | 0.025a | 1.79 | 0.89–3.59 | 0.101 |
 | Table II.Prognostic factors for
recurrence-free survival in 92 patients with hepatocellular
carcinoma. |
Table II.
Prognostic factors for
recurrence-free survival in 92 patients with hepatocellular
carcinoma.
| Variable | n | Univariate | Multivariate |
|---|
|
|
|---|
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥65) | 47 | 1.72 | 0.96–3.08 | 0.068 | | | |
| Gender (male) | 77 | 1.41 | 0.64–3.73 | 0.418 | | | |
| Background liver
(cirrhosis) | 36 | 1.68 | 0.86–3.10 | 0.124 | | | |
| Pugh-Child’s
classification (B) | 7 | 0.93 | 0.28–2.32 | 0.889 | | | |
| AFP (>20
ng/ml) | 47 | 1.58 | 0.89–2.82 | 0.116 | | | |
| PIVKA II (>40
mAU/ml) | 54 | 1.10 | 0.61–2.03 | 0.759 | | | |
| Tumor multiplicity
(multiple) | 24 | 1.06 | 0.54–1.96 | 0.861 | | | |
| Tumor size (≥3.0
cm) | 67 | 1.93 | 0.95–4.47 | 0.073 | | | |
| Tumor
differentiation (well) | 28 | 1.05 | 0.51–2.02 | 0.879 | | | |
| Growth type
(invasive growth) | 17 | 1.33 | 0.57–2.75 | 0.483 | | | |
| Serosal
infiltration | 25 | 2.62 | 1.35–5.00 | 0.005a | 2.06 | 1.04–4.02 | 0.039a |
| Formation of
capsule | 68 | 0.57 | 0.29–1.22 | 0.139 | | | |
| Infiltration to
capsule | 54 | 0.84 | 0.47–1.55 | 0.577 | | | |
| Septum
formation | 34 | 0.68 | 0.38–1.26 | 0.214 | | | |
| Vascular
invasion | 23 | 2.41 | 1.27–4.39 | 0.008a | 2.12 | 1.08–4.01 | 0.029a |
| Margin status
(positive) | 24 | 1.79 | 0.97–3.23 | 0.064 | | | |
| Hypermethylation of
DENND2D | 69 | 1.11 | 0.59–2.24 | 0.749 | | | |
| Downregulation of
DENND2D mRNA | 34 | 2.86 | 1.52–5.35 | 0.001a | 2.86 | 1.49–5.44 | 0.002a |
Methylation status of DENND2D in 92
clinical HCC samples
MSP analysis revealed that 69 (75.0%) out of 92 HCC
tissue samples and only 4 (4.3%) of 92 corresponding non-cancer
tissues showed hypermethylation of the DENND2D promoter.
There was no significant association between hypermethylation of
DENND2D in HCC tissues and overall or recurrence-free
survival (Tables I and II). Analysis of the associations between
the methylation status of DENND2D and clinicopathological
factors, including demographics, background liver status,
pathological findings and expression level of DENND2D mRNA
showed that promoter hypermethylation of DENND2D in HCCs was
significantly associated with tumor size ≥3 cm, serosal
infiltration and downregulation of DENND2D (P=0.048, 0.049 and
0.004, respectively, Table
III).
 | Table III.Association between methylation
status of DENND2D and clinicopathological parameters in 92
HCC patients. |
Table III.
Association between methylation
status of DENND2D and clinicopathological parameters in 92
HCC patients.
| Clinicopathological
parameters | Methylation
positive in tumor tissue (n) | Methylation
negative in tumor tissue (n) | P-value |
|---|
| Age | | | |
| <65 year | 31 | 15 | 0.090 |
| ≥65 year | 38 | 8 | |
| Gender | | | |
| Male | 59 | 16 | 0.101 |
| Female | 10 | 7 | |
| Background
liver | | | |
| Normal liver | 4 | 5 | 0.080 |
| Chronic
hepatitis | 36 | 12 | |
| Cirrhosis | 29 | 6 | |
| Pugh-Child’s
classification | | | |
| A | 64 | 21 | 0.823 |
| B | 5 | 2 | |
| Hepatitis
virus | | | |
| Absent | 9 | 5 | 0.617 |
| HBV | 19 | 6 | |
| HCV | 41 | 12 | |
| AFP (ng/ml) | | | |
| ≤20 | 34 | 13 | 0.547 |
| >20 | 35 | 10 | |
| PIVKA II
(mAU/ml) | | | |
| ≤40 | 32 | 7 | 0.175 |
| >40 | 37 | 16 | |
| Tumor
multiplicity | | | |
| Solitary | 53 | 16 | 0.493 |
| Multiple | 16 | 7 | |
| Tumor size | | | |
| <3.0 cm | 23 | 3 | 0.048a |
| ≥3.0 cm | 46 | 20 | |
|
Differentiation | | | |
| Well | 21 | 6 | 0.689 |
| Moderate to
poor | 48 | 17 | |
| Growth type | | | |
| Expansive
growth | 58 | 18 | 0.533 |
| Invasive
growth | 11 | 5 | |
| Serosal
infiltration | | | |
| Absent | 54 | 13 | 0.049a |
| Present | 15 | 10 | |
| Formation of
capsule | | | |
| Absent | 19 | 7 | 0.790 |
| Present | 50 | 16 | |
| Infiltration to
capsule | | | |
| Absent | 29 | 11 | 0.628 |
| Present | 40 | 12 | |
| Septum
formation | | | |
| Absent | 26 | 8 | 0.803 |
| Present | 43 | 15 | |
| Vascular
invasion | | | |
| Absent | 53 | 17 | 0.779 |
| Present | 16 | 6 | |
| Margin status | | | |
| Negative | 51 | 17 | 1.000 |
| Positive | 18 | 6 | |
| UICC pathological
stage | | | |
| I, II | 44 | 10 | 0.089 |
| III, IV | 25 | 13 | |
| Downregulation of
DENND2D | | | |
| Absent | 38 | 20 | 0.004a |
| Present | 31 | 3 | |
IHC
The expression of DENND2D was determined using IHC
in 30 cases showing relative overexpression, underexpression, or
equivalent DENND2D mRNA expression in HCC tissues compared
with the corresponding non-cancer tissues. Two representative cases
with the lowest expression level of DENND2D mRNA in HCC
tissues showed downregulation of DENND2D in the membrane and
cytoplasm of tumor cells compared with adjacent non-cancer tissues
(Fig. 5A and B). In contrast,
equivalent expression of DENND2D protein in tumor and normal cells
was detected in the case without reduced DENND2D mRNA
expression in HCC tissues (Fig.
5C). The MSP analysis of these cases is shown in Fig. 5D.
Discussion
Research conducted over the past decade has
demonstrated that certain TSGs are epigenetically inactivated in
HCC, indicating that this is one of the major molecular alterations
that occurs during hepatocarcinogenesis (7,29).
Moreover, there is growing evidence that a major mechanism of
epigenetic silencing in cancers involves hypermethylation of the
TSG promoter (10,30,31).
Promoter hypermethylation can be used as a sensitive marker for TSG
identification, cancer diagnosis and predicting prognosis. In the
present study, DENND2D was identified as another candidate
TSG that is epigenetically inactivated in HCC.
We show here that the expression level of
DENND2D mRNA was independent of fibrosis of the liver,
because there was no significant difference in DENND2D mRNA
levels between patients with and without cirrhosis. In contrast,
the level of DENND2D mRNA was reduced in 8/9 HCC cell lines
and in 72/92 surgically resected specimens and the mean expression
level was significantly lower in cancer tissues than in
corresponding normal tissues. This result indicates that DENND2D
plays an important role in hepatocarcinogenesis but not in the
fibrous response of the liver. Further, the expression pattern of
DENND2D was consistent with that of its mRNA according to the
results of qPCR and IHC analyses. A striking discovery was that a
significant reduction in DENND2D mRNA level in HCC tissues
correlated with remarkably earlier recurrence (hazard ratio 2.86,
P=0.002, Table I) and subsequent
adverse prognosis. These findings suggest that DENND2D acts
as a TSG and are consistent with the results of a study on DENND2D
expression in lung cancer (25).
HCC frequently relapses after hepatectomy, becoming multicentric
metastatic within the liver. Downregulation of DENND2D
expression in HCC tissues could therefore be a biomarker of HCC
recurrence.
To understand the regulation of DENND2D
transcription, we performed methylation analysis of DENND2D
after we identified a CpG island within its promoter. The
DENND2D promoter was hypermethylated in 8/9 HCC cell lines
and DENND2D transcription could be reactivated in cells
treated with an inhibitor of methylation. Hypermethylation was
frequently (75%) detected in HCC tissues and significantly
associated with substantial (≥3-times) reduction of DENND2D
mRNA levels. Therefore, we consider promoter hypermethylation as a
potent regulatory factor of DENND2D transcription in HCC.
Further, hypermethylation of the DENND2D promoter in HCC
tissues significantly associated with tumor size and serosal
infiltration, indicating the importance of DENND2D in
tumor-cell growth and migration. Therefore, hypermethylation of
DENND2D should provide an important new biomarker of HCC
progression.
The DENND2 family is the only example where the DENN
domain is located within the C-terminal region (23,32).
Each DENND2 protein acts as a GEF for Rab9a/b. DENND2D is composed
of only a DENN domain (33);
therefore, the DENN domain mediates GEF activity. Rab9 functions in
retrograde trafficking of the mannose phosphate receptor from late
endosomes to the trans-Golgi network and depletion of DENND2A, but
not other DENND2 family members, disrupting trafficking (34,35).
This suggests that DENND2 proteins activate other Rab9 functions,
such as biogenesis of lysosome-related organelles (36).
This study is limited by its lack of sufficient
functional analysis of DENND2D, which tempers the conclusion that
it acts as a tumor suppressor for HCC. Further studies will be
required to clarify the molecular mechanisms underlying the
biological activities of DENND2D in HCC.
In conclusion, we propose DENND2D as a
candidate tumor suppressor gene that is inactivated by promoter
hypermethylation in HCC and shows promise as a novel biomarker of
early recurrence of HCC.
References
|
1.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar
|
|
3.
|
Della C and Colombo M: Surveillance for
hepatocellular carcinoma. Semin Oncol. 39:384–398. 2012. View Article : Google Scholar
|
|
4.
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Yu MC and Yuan JM: Environmental factors
and risk for hepatocellular carcinoma. Gastroenterology. 127:72–78.
2004. View Article : Google Scholar
|
|
6.
|
Ponder BA: Cancer genetics. Nature.
411:336–341. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Cheng YY, Jin H, Liu X, Siu JM, Wong YP,
Ng EK, Yu J, Leung WK, Sung JJ and Chan FK: Fibulin 1 is
downregulated through promoter hypermethylation in gastric cancer.
Br J Cancer. 99:2083–2087. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar
|
|
9.
|
Kanda M, Nomoto S, Okamura Y, Hayashi M,
Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S and Nakao A:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Kanda M, Nomoto S, Okamura Y, Nishikawa Y,
Sugimoto H, Kanazumi N, Takeda S and Nakao A: Detection of
metallothionein 1G as a methylated tumor suppressor gene in human
hepatocellular carcinoma using a novel method of double combination
array analysis. Int J Oncol. 35:477–483. 2009.
|
|
11.
|
Kanda M, Nomoto S, Nishikawa Y, Sugimoto
H, Kanazumi N, Takeda S and Nakao A: Correlations of the expression
of vascular endothelial growth factor B and its isoforms in
hepatocellular carcinoma with clinico-pathological parameters. J
Surg Oncol. 98:190–196. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Stenmark H: Rab GTPases as coordinators of
vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Pawson T and Nash P: Assembly of cell
regulatory systems through protein interaction domains. Science.
300:445–452. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Marat AL and McPherson PS: The connecdenn
family, Rab35 guanine nucleotide exchange factors interfacing with
the clathrin machinery. J Biol Chem. 285:10627–10637. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Sa=to M, Sato K, Liou W, Pant S, Harada A
and Grant BD: Regulation of endocytic recycling by C. elegans Rab35
and its regulator RME-4, a coated-pit protein. EMBO J.
27:1183–1196. 2008.PubMed/NCBI
|
|
16.
|
Levivier E, Goud B, Souchet M, Calmels TP,
Mornon JP and Callebaut I: uDENN, DENN, and dDENN: indissociable
domains in Rab and MAP kinases signaling pathways. Biochem Biophys
Res Commun. 287:688–695. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Allaire PD, Marat AL, Dall’Armi C, Di
Paolo G, McPherson PS and Ritter B: The Connecdenn DENN domain: a
GEF for Rab35 mediating cargo-specific exit from early endosomes.
Mol Cell. 37:370–382. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Knödler A, Feng S, Zhang J, Zhang X, Das
A, Peränen J and Guo W: Coordination of Rab8 and Rab11 in primary
ciliogenesis. Proc Natl Acad Sci USA. 107:6346–6351.
2010.PubMed/NCBI
|
|
19.
|
Denef N, Chen Y, Weeks SD, Barcelo G and
Schüpbach T: Crag regulates epithelial architecture and polarized
deposition of basement membrane proteins in Drosophila. Dev Cell.
14:354–364. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Rink J, Ghigo E, Kalaidzidis Y and Zerial
M: Rab conversion as a mechanism of progression from early to late
endosomes. Cell. 122:735–749. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Girard M, Allaire PD, McPherson PS and
Blondeau F: Non-stoichiometric relationship between clathrin heavy
and light chains revealed by quantitative comparative proteomics of
clathrin-coated vesicles from brain and liver. Mol Cell Proteomics.
4:1145–1154. 2005. View Article : Google Scholar
|
|
22.
|
Majidi M, Hubbs AE and Lichy JH:
Activation of extracellular signal-regulated kinase 2 by a novel
Abl-binding protein, ST5. J Biol Chem. 273:16608–16614. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Marat AL, Dokainish H and McPherson PS:
DENN domain proteins: regulators of Rab GTPases. J Biol Chem.
286:13791–13800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Postel EH, Weiss VH, Beneken J and Kirtane
A: Mutational analysis of NM23-H2/NDP kinase identifies the
structural domains critical to recognition of a c-myc regulatory
element. Proc Natl Acad Sci USA. 93:6892–6897. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Ling B, Zheng H, Fu G, Yuan J, Shi T, Chen
S, Liu Y, Liu Y, Cao Y, Zheng S, Guo S, Han N, Gao Y, Cheng S and
Zhang K: Suppression of non-small cell lung cancer proliferation
and tumorigenicity by DENND2D. Lung Cancer. 79:104–110. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
International Union Against Cancer: TNM
Classification of Malignant Tumors. 7th edition. Wiley-Blackwell;
New York, NY: 2009
|
|
27.
|
Takai D and Jones PA: Comprehensive
analysis of CpG islands in human chromosomes 21 and 22. Proc Natl
Acad Sci USA. 99:3740–3745. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Takai D and Jones PA: The CpG island
searcher: a new WWW resource. In Silico Biol. 3:235–240.
2003.PubMed/NCBI
|
|
29.
|
Mann CD, Neal CP, Garcea G, Manson MM,
Dennison AR and Berry DP: Prognostic molecular markers in
hepatocellular carcinoma: a systematic review. Eur J Cancer.
43:979–992. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Herath NI, Leggett BA and MacDonald GA:
Review of genetic and epigenetic alterations in
hepatocarcinogenesis. J Gastroenterol Hepatol. 21:15–21. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Tischoff I and Tannapfe A: DNA methylation
in hepatocellular carcinoma. World J Gastroenterol. 14:1741–1748.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Bloethner S, Mould A, Stark M and Hayward
NK: Identification of ARHGEF17, DENND2D, FGFR3, and RB1 mutations
in melanoma by inhibition of nonsense-mediated mRNA decay. Genes
Chromosomes Cancer. 47:1076–1085. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Yoshimura S, Gerondopoulos A, Linford A,
Rigden DJ and Barr FA: Family-wide characterization of the DENN
domain Rab GDP-GTP exchange factors. J Cell Biol. 191:367–381.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Carroll KS, Hanna J, Simon I, Krise J,
Barbero P and Pfeffer SR: Role of Rab9 GTPase in facilitating
receptor recruitment by TIP47. Science. 292:1373–1376. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Kloer DP, Rojas R, Ivan V, Moriyama K, van
Vlijmen T, Murthy N, Ghirlando R, van der Sluijs P, Hurley JH and
Bonifacino JS: Assembly of the biogenesis of lysosome-related
organelles complex-3 (BLOC-3) and its interaction with Rab9. J Biol
Chem. 285:7794–7804. 2010. View Article : Google Scholar : PubMed/NCBI
|