Introduction
Epigenetic regulation in alteration of chromatin
structure is critical for many important cellular processes,
including gene transcription, recombination, DNA replication and
damage repair (1).
Post-translational modifications of the N-terminal tails of
histones such as acetylation, methylation, ubiquitilation and
phosphorylation are important mechanisms of epigenetic regulation.
Current evidence indicates that cancer can occur via
disproportionate histone modifications through altering gene
expressions including aberrant regulation of oncogenes and/or tumor
suppressors (2,3) and unbalanced histone modifications
affect genome integrity and/or chromosome segregation (4). In contrast to DNA mutations or
deletions, each histone modification process is reversible, thus
novel therapies that work by reversing epigenetic effects are being
increasingly explored (5).
Histone modification status in cells is dynamically
regulated by chromatin modifying enzymes that add and remove
covalent modifications to histone proteins. Histone acetylation or
methylation as the well-characterized epigenetic modifications is
controlled by histone acetyltransferases (HATs) or histone
methyltransferases (HMTs) and histone deacetylases (HDAC) or
histone demethylases (HDMs). HATs and HMTs can add acetyl and
methyl groups, respectively, whereas HDACs and HDMs can remove
acetyl or methyl, respectively (6,7).
Studies have suggested that epigenetic alterations may play a key
role in initiating events in forms of some cancers. Therefore,
efforts have been made to understand the role of global changes of
epigenetics in the initiation and propagation of various cancers
(8,9). For instance, global loss of histone
H4K16 acetylation (ac) and histone H4K20 tri-metylation (me3) as a
hallmark of several human cancers have been reported (10). In contrast to this, global changes
of other histone modifications seem to be more tumor-specific; for
example, low histone H3K4 dimethylation (me2) and H3K9ac levels
were shown in breast cancer cells, whereas lung cancer cells show
low H3K4me2 but high H3K9ac levels (11,12).
Furthermore, global changes of histone modifications also closely
associated with clinicopathological factors in some cancers. In
patients with renal clear cell carcinoma (RCC), total acetylation
levels of histone H3 were inversely correlated with pT-stage,
distant metastasis, Fuhrman grading and RCC progression, whereas
total histone H4ac deacetylation was correlated with pT-stage and
grading (13).
hMOF (hMYST1), a member of the MYST family of HATs,
is responsible for histone H4K16ac. Depletion of hMOF in cells
leads to genomic instability, reduced transcription of certain
genes, defective DNA damage repair and early embryonic lethality
(14–16). Biochemical purifications have
revealed that hMOF forms at least two distinct multi-protein
complexes, MSL and NSL, in mammalian cells. Although the functions
of MSL and NSL complexes in human cells are not entirely clear,
both complexes can acetylate histone H4K16, suggesting the
importance of acetylation of H4K16 in cells (17–19).
Abnormal gene expression of the hMOF and its corresponding
modification of H4K16 have been found in certain primary cancer
tissues. The expression patterns of hMOF in different
primary cancers varied. Frequent downregulation of hMOF expression
was found in breast cancer, medulloblastoma and RCC (20,21).
On the contrary, hMOF was overexpressed in non-small cell
lung carcinoma tissues (22,23).
In all occasions, hMOF protein expression tightly correlated with
acetylation of histone H4K16. Above observations strongly suggest
that histone acetyltransferase hMOF and its corresponding histone
of H4K16ac might be involved in certain tumorigenic pathways.
With aging and urbanization progresses, the number
of cancer patients has increased dramatically. Colorectal cancer
(CRC) has become one of the major causes of mortality and
morbidity, and is the third most common cancer in men and the
second most common cancer in women worldwide (24). RCC is one of the most common
genitourinary malignancies, accounting for 3% of all cancer
worldwide (25). However, gastric
cancer, another common tumor, is the second most common cause of
cancer deaths in the world (26).
In spite of recent studies indicating that global histone
modification, such as methylation of H3K4 and H3K27, correlate with
the outcomes in patients with above-mentioned cancer (27,28),
there have so far been no reports on HAT hMOF and its corresponding
modification in CRC and gastric cancer. Here we first examined the
hMOF mRNA and protein expression levels in primary CRC and gastric
cancer by quantitative PCR (qPCR) and western blotting (WB). In
addition, we recently reported the hMOF HAT is frequently
downregulated in ovarian cancer and RCC (29,21),
here we further analyzed the relationship between low-expression of
hMOF and clinicopathological features in RCC.
Materials and methods
Tissue collection
Human tumor tissues (CRC, gastric cancer and RCC)
and normal tissues were collected from patients with primary CRC
cases between July 2012 and March 2013, from patients with primary
stomach cancers between September 2008 and August 2011, and from
patients with primary RCC cases between May 2009 and May 2012.
Cancer patients underwent radical tumor surgery at the First
Hospital of Jilin University. Written informed consent was obtained
from all participants, and the study was approved by the
Institutional Ethics Board of School of Medicine, Jilin University.
Patient medical records including patient age and gender, tumor
staging, pathological diagnosis, and surgical records were
reviewed. Tumors were staged according to the 2010 TNM
classification system using the American Joint Committee on Cancer
(AJCC) stage grouping (30). None
of the patients received chemotherapy or radiotherapy before
surgery.
Antibodies
Anti-H4K16ac (H9164) polyclonal antibody was
purchased from Sigma. Anti-GAPDH and anti-hMOF were raised against
bacterially expressed proteins (Jilin University).
Reverse transcription PCR (RT-PCR)
Total RNA from tumor (CRC, gastric cancer, ovarian
cancer or RCC) or normal tissues were isolated using
TRIzol® LS Reagent (Invitrogen, CA, USA). Then, 1
μg of RNA from each sample was used as a template to produce
cDNA with PrimeScript 1st Strand cDNA Synthesis kit (Takara). hMOF
and GAPDH mRNA levels were analyzed by quantitative PCR with an Eco
Real-Time PCR System (Illumina). All PCR reactions were finished as
follows: initial denaturation step at 95°C for 30 sec, followed by
40 cycles of denaturation at 95°C for 5 sec, annealing at 60°C for
30 sec and extension at 72°C for 30 sec. Primer sets used for PCR
were as follows: GAPDH, 5′-ATCACTGCCACC CAGAAGAC-3′ (forward) and
5′-ATGAGGTCCACCACCCT GTT-3′ (reverse), yielding a 460-bp product;
hMOF, 5′-GGCT GGACGAGTGGGTAGACAA-3′ (forward) and 5′-TGGTG
ATCGCCTCATGCTCCTT-3′ (reverse), yielding a 227-bp product.
Western blotting (WB)
Cancer tissue (200 mg) or normal tissue samples were
homogenized with liquid nitrogen and solubilized in 200 μl
cold PBS containing 1.0% Nonidet P-40, 0.5% Na-deoxycholate, 0.1%
SDS, 0.05 mM PMSF and protease inhibitor cocktail. The homogenate
was swirled and kept on ice for 30 min. Whole-cell extracts were
sonicated (Scientz-IID, China) for 10 sec with 50% duty cycle and
centrifugation (13,000 × g for 30 min). The protein concentration
of the resulting supernatant was measured using the Bio-Rad Protein
Assay kit (500–0201). Equal amounts of protein from tissue
whole-cell lysates were mixed with 4X SDS-containing sample buffer
and boiled for 5 min at 95°C. Denatured proteins were then
separated by 12% SDS-PAGE. Specific proteins were detected by WB
using hMOF and GAPDH polyclonal antibodies.
Immunohistochemical staining (IHC)
Formalin-fixed and paraffin-embedded CRC tissue
blocks were supplied from The First Clinical Hospital of Jilin
University. IHC was performed essentially as described (31,32).
Anti-hMOF and acetylated H4K16 polyclonal antibodies (H9164) were
used at a 1:500 dilution.
Statistical analysis
The western blot images were scanned and quantified
by Quantity One Basic software (Bio-Rad). Differences in gene and
protein expression between tumor and normal tissues were
statistically analyzed using SPSS 17.0 (SPSS, Inc., Chicago IL,
USA). Statistical comparisons were analyzed using the Student’s
t-test. Values of p<0.05 were considered to be statistically
significant.
Results
Reduction of hMOF gene expression level
is observed in CRC
To investigate the involvement of hMOF gene
expression in the pathogenesis of primary CRC, we first measured
hMOF mRNA using qPCR in 44 patients diagnosed with CRC. Compared to
matched normal tissues, the gene expression of hMOF was
significantly decreased in CRC tissues (p<0.05; n=44) (Fig. 1B). As shown in Fig. 1A, analysis of performed mRNA
expression of 44 samples revealed significant (>2-fold
decreased) downregulation of hMOF mRNA in 57% (25/44) of
patients, whereas 7% (3/44) of patients showed significant
(>2-fold increased) upregulation of hMOF. The
relationships between hMOF expression and clinical
parameters, including age and gender, tumor types, histological
grade, and lymph node metastasis were further analyzed. A
significant down-regulation of hMOF was only observed in
male (p<0.05), patients >60 years of age (<0.05) or
patients with lymph node metastasis (p<0.05) (Fig. 1B). In addition, obvious reduction
of hMOF gene expression was also detected in tissue with a
T4 tumor status (p<0.05) (Fig.
2A), but there was no association between low-expression of
hMOF and histological tumor types of CRC (Fig. 2B).
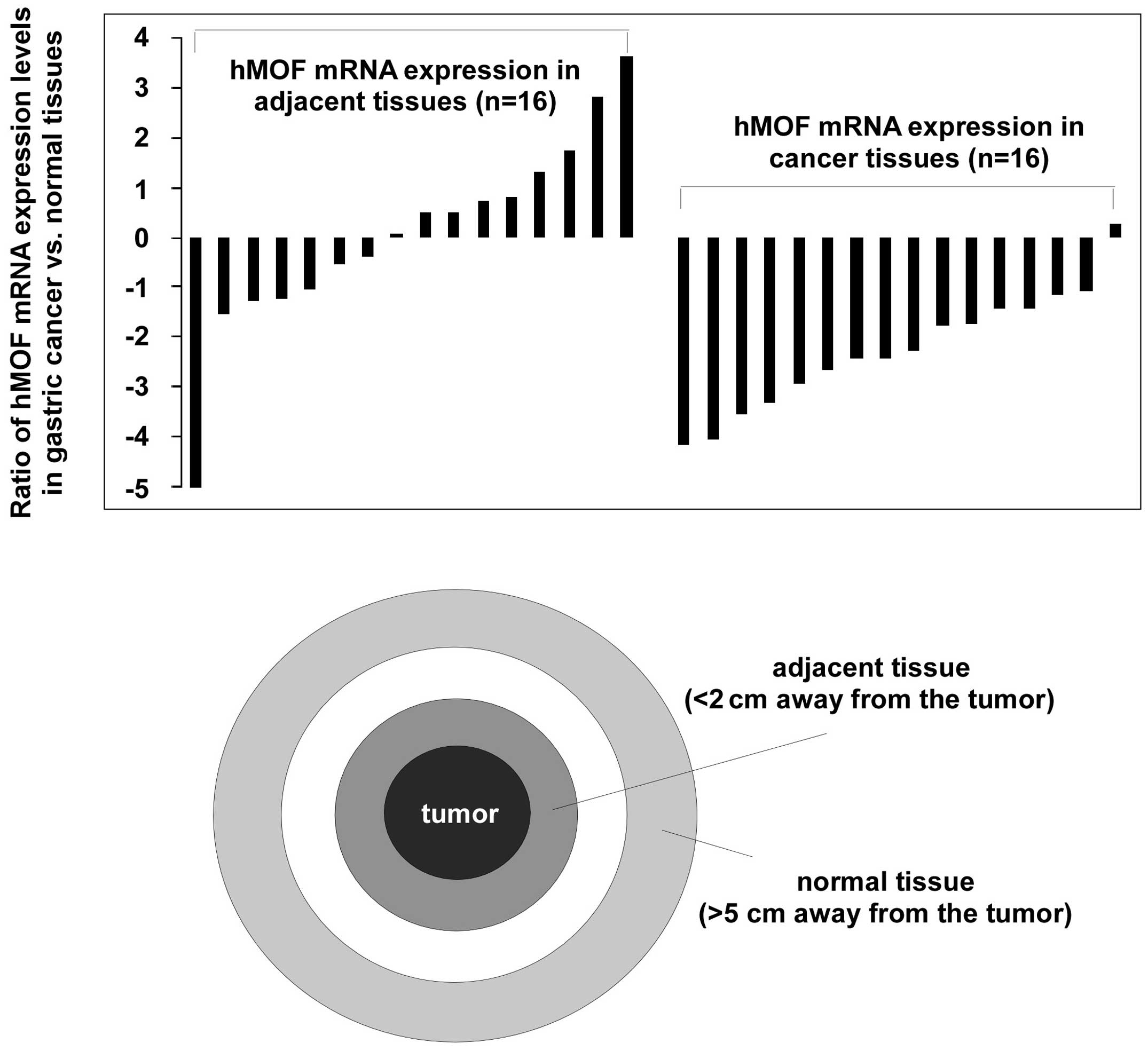
hMOF gene expression in gastric
cancer
Next, to understand the implication of hMOF gene
expression in the pathogenesis of primary gastric cancer, 16
clinical gastric cancer tissues and matched adjacent (<2 cm away
from the tumor) and/or normal (>5 cm away from the tumor)
tissues were used. As shown in Fig.
3, compared to matched normal tissues, hMOF mRNA
expression was significantly downregulated (>2-fold decrease) in
94% (15/16) of tumor tissue samples. Interestingly, hMOF
expression in adjacent tissues also showed great reduction
(>2-fold decrease) in 35% of samples. The summarization of the
clinical characteristics including age and gender, cell
differentiation and survival of patients is shown in Table I. Decreased hMOF expression
was observed in >70 years of age patients (p<0.01) and in the
female group (p<0.01), whereas, downregulation of hMOF
expression was strongly associated with poorly-differentiated tumor
tissue (p<0.01). In addition, significant connection was
observed between hMOF expression and <1-year survival of
patients (p<0.001). Similar results were obtained comparing
adjacent and cancer tissues. However, no statistically significant
difference was found between adjacent and normal tissues.
 | Table I.Relationship between hMOF gene
expression (qPCR) and clinicopathological characteristics of
gastric cancer. |
Table I.
Relationship between hMOF gene
expression (qPCR) and clinicopathological characteristics of
gastric cancer.
| Factor | Case (n) | Normal Mean ±
SD | Adjacent Mean ±
SD | Cancer Mean ±
SD | p-value Nor vs.
adj | p-value Nor vs.
can | p-value Adj vs.
can |
|---|
| All | 16 | 16.0±30.9 | 9.90±10.7 | 3.19±4.14 | 0.465 | 0.112 | 0.0257a |
| Age (years) | | | | | | | |
| ≤70 | 10 | 28.6±10.1 | 5.57±5.17 | 4.27±6.46 | 0.291 | 0.267 | 0.708 |
| >70 | 6 | 8.38±4.58 | 12.5±12.4 | 2.54±2.05 | 0.338 | 0.00172b | 0.0224a |
| Gender | | | | | | | |
| Female | 8 | 8.39±4.85 | 9.02±6.01 | 2.52±2.16 | 0.819 | 0.00737b | 0.0121a |
| Male | 8 | 23.5±9.59 | 10.8±8.39 | 3.86±5.58 | 0.445 | 0.226 | 0.224 |
|
Differentiation | | | | | | | |
| Moderate | 8 | 25.1±13.0 | 12.6±9.63 | 5.1±5.27 | 0.448 | 0.213 | 0.167 |
| Poorly | 8 | 6.84±3.93 | 7.18±6.42 | 1.27±0.754 | 0.902 | 0.00148b | 0.0215a |
| Survival of
patients | | | | | | | |
| >12
months | 8 | 25.0±12.3 | 9.72±6.25 | 4.89±5.35 | 0.336 | 0.211 | 0.119 |
| <12
months | 8 | 5.91±3.06 | 4.99±2.72 | 1.01±0.66 | 0.541 | 0.000581c | 0.00124b |
Correlation of hMOF expression with
clinicopathological characteristics of renal cell carcinoma
We previously found downregulation of hMOF
(91% of patients, 19/21) in RCC, resulting in decreased hMOF
protein, and this reduction is tightly correlated with histone
H4K16 acetylation (21). To
further confirm our results and to evaluate the relationship
between hMOF expression and clinicopathological parameters,
we measured hMOF mRNA using qPCR in 47 patients diagnosed
with RCC. Analysis of qPCR data revealed a significant (>2-fold
decreased) downregulation of hMOF mRNA in 74% (35/47) of
patients, whereas only 6% (3/47) of patients showed significant
(>2-fold increased) upregulation of hMOF (Fig. 4A). These results strongly support
our previous data. As shown in Fig.
4B, although low-expression of hMOF was found in both
>60 years of age (p<0.05) and <60 years of age (p<0.05)
groups, statistically significant reduction of hMOF was
recorded only in male patients. In addition, there was a
correlation between low-expression of hMOF and ccRCC
(p<0.05) or tissue with T1 tumor status (p<0.05). However,
declined hMOF expression showed no association with the RCC tissue
Fuhrman grading (data not shown).
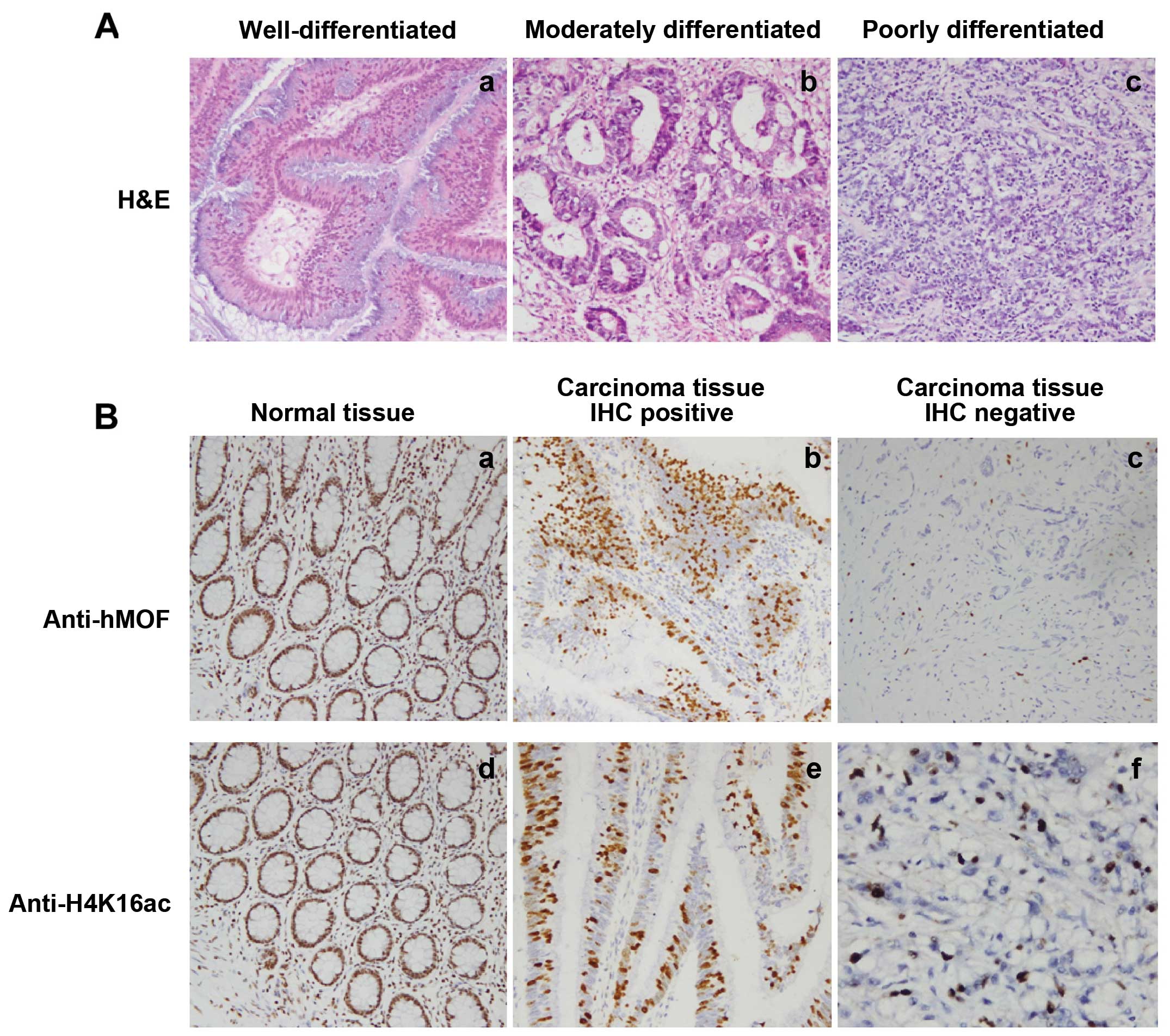
Reduction of hMOF protein levels in
primary human CRC
To determine whether the reduction of hMOF
gene expression resulted in reduced hMOF protein levels, western
blotting and IHC staining approaches were used. Aliquots of whole
cell extract from 22 paired, initially selected CRC and matched
normal tissues were analyzed by western blotting with hMOF
antibodies (GAPDH as internal reference protein). As expected,
frequent reduction of hMOF protein in CRC compared to matched
normal tissues was detected (Fig. 5,
bottom panel). The quantified protein levels (Quantity One
software) were analyzed by t-test. As shown in Fig. 5 (upper panel) hMOF protein
expression levels were significantly reduced in CRC tissues
(p<0.05). To determine whether the expression of hMOF was
tightly correlated with histone H4K16 acetylation, we performed
immunohistochemical staining (IHC) for hMOF and histone H4K16
acetylation in the formalin-fixed paraffin-embedded tissue sections
of selected CRC patients. The results revealed that hMOF protein
expression was correlated with acetylation of histone H4K16 in
parallel (Fig. 6).
Discussion
Histone acetylation status in cells is controlled by
HATs and HDACs. Any factor that creates an imbalance can lead to
abnormal cell function, even cancer. The HAT hMOF belongs to the
MYST (Moz-Ybf2/Sas3-Sas2-Tip60) family, and is believed to be
responsible for histone H4K16 acetylation in both Drosophila
and human cells (33,18,34).
It is known that hMOF participates in many biological processes,
including gene transcription, cell proliferation, differentiation,
DNA repair response (14,16,17,19,33).
Although little is known about the mechanism of hMOF in tumor
development and progress, the expression of hMOF in limited types
of clinical cancer tissues has been reported by several research
groups. Frequent downregulation of hMOF in primary breast
carcinomas, renal cell carcinoma and medulloblastomas was found,
and the reduction of hMOF protein expression tightly correlated
with acetylation of H4K16 in those tumors (20,21).
In contrast, the expression of hMOF in non-small cell lung
carcinoma tissues was frequently elevated (22,23).
In this study, we first investigated the expression
of HAT hMOF in primary CRC and gastric cancer tissues by qRT-PCR.
The results revealed that either hMOF mRNA expression or hMOF
protein expression was downregulated in human primary CRC (Figs. 1 and 5), and hMOF protein expression was
correlated with acetylation of histone H4K16 in parallel (Fig. 6). Further analysis of qPCR data
revealed that expression of hMOF was associated with the age of the
patients, gender, lymph node metastasis, and tissues with a T4
tumor status. It is noteworthy that the abnormal expression of hMOF
had already occurred in the adjacent tissues (<2 cm away from
the cancer tissue). Compared to matched normal tissues, although no
statistically significant difference was found between adjacent and
normal tissues, a significant reduction (>2-fold decrease) of
hMOF expression in adjacent tissues had already appeared in
35% of patients with gastric cancer. In addition, low-expression of
hMOF in gastric cancer was strongly associated with
poorly-differentiated tumor tissue and <1-year survival of
patients (Table I). Our results
demonstrate that abnormal expression of hMOF might be involved in
tumorigenesis of CRC and gastric cancer. In line with the results
obtained from a previous study (21), a significant (>2-fold decreased)
downregulation of hMOF mRNA in 74% (35/47) of patients with
RCC was observed. Downregulation of hMOF was strongly
associated with ccRCC and with T1 tumor status. This suggests that
abnormal expression of hMOF is not only associated with
tumor types, but might be also an early biomarker in RCC.
In conclusion, downregulation of hMOF was
observed in CRC, gastric cancer and RCC tissues. Although larger
series of clinical cases and analyses of overall survival are
needed, the molecular mechanism linking loss of hMOF expression may
be involved in the above described cancer progression.
Acknowledgements
This study was supported by the
Research Fund for the Doctoral Program of Higher Education of China
(20110061110020, J.J.) and the Key Scientific and Technological
Project of Jilin Province Science and Technology Development
Program (20130206005YY, J.J.).
References
|
1.
|
Jin J, Cai Y, Li B, Conaway RC, Workman
JL, Conaway JW and Kusch T: In and out: histone variant exchange in
chromatin. Trends Biochem Sci. 30:680–687. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Waldmann T and Schneider R: Targeting
histone modifications - epigenetics in cancer. Curr Opin Cell Biol.
25:184–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar
|
|
4.
|
Bannister AJ and Kouzarides T: Regulation
of chromatin by histone modifications. Cell Res. 21:381–395. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Hatzimichael E and Crook T: Cancer
epigenetics: new therapies and new challenges. J Drug Deliv.
2013:5293122013. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Haberland M, Montgomery RL and Olson EN:
The many roles of histone deacetylases in development and
physiology: implications for disease and therapy. Nat Rev Genet.
10:32–42. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Shi Y: Histone lysine demethylases:
emerging roles in development, physiology and disease. Nat Rev
Genet. 8:829–833. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Feinberg AP, Ohlsson R and Henikoff S: The
epigenetic progenitor origin of human cancer. Nat Rev Genet.
7:21–33. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Jones PA and Martienssen R: A blueprint
for a human epigenome project: the AACR human epigenome workshop.
Cancer Res. 65:11241–11246. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Fraga MF, Ballestar E, Villar-Garea A, et
al: Loss of acetylation at Lys 16 and trimethylation at Lys20 of
histone H4 is a common hallmark of human cancer. Nat Genet.
37:391–400. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Elsheikh SE, Green AR, Rakha EA, et al:
Global histone modifications in breast cancer correlate with tumor
phenotypes, prognostic factors, and patient outcome. Cancer Res.
69:3802–3809. 2009. View Article : Google Scholar
|
|
12.
|
Barlesi F, Giaccone G, Gallegos-Ruiz MI,
et al: Global histone modifications predict prognosis of resected
non small-cell lung cancer. J Clin Oncol. 25:4358–4364. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Mosashvilli D, Kahl P, Mertens C, et al:
Global histone acetylation levels: prognostic relevance in patients
with renal cell carcinoma. Cancer Sci. 101:2664–2669. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Sharma GG, So S, Gupta A, et al: MOF and
histone H4 acetylation at lysine 16 are critical for DNA damage
response and double-strand break repair. Mol Cell Biol.
30:3582–3595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Carrozza MJ, Utley RT, Workman JL and Côté
J: The divers functions of histone acetyltransferase complexes.
Trends Genet. 19:321–329. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Gupta A, Guerin-Peyrou TG, Sharma GG, et
al: The mammalian ortholog of Drosophla MOF that acetylates
histone H4 lysine 16 is essential for embryogenesis and
oncogenesis. Mol Cell Biol. 28:397–409. 2008.
|
|
17.
|
Smith ER, Cayrou C, Huang R, Lane WS, Côté
J and Lucchesi JC: A human protein complex homologous to the
Drosophila MSL complex is responsible for the majority of
histone H4 acetylation at lysine 16. Mol Cell Biol. 25:9175–9188.
2005.PubMed/NCBI
|
|
18.
|
Cai Y, Jin J, Swanson SK, et al: Subunit
composition and substrate specificity of a MOF-containing histone
acetyltransferase distinct from the male-specific lethal (MSL)
complex. J Biol Chem. 285:4268–4272. 2010. View Article : Google Scholar
|
|
19.
|
Mendjan S, Taipale M, Kind J, et al:
Nuclear pore components are involved in the transcriptional
regulation of dosage compensation in Drosophila. Mol Cell.
21:811–823. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Pfister S, Rea S, Taipale M, et al: The
histone acetyltransferase hMOF is frequently downregulated in
primary breast carcinoma and medulloblastoma and constitutes a
biomarker for clinical outcome in medulloblastoma. Int J Cancer.
122:1207–1213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Wang Y, Zhang R, Wu D, Lu Z, Sun W, Cai Y,
Wang C and Jin J: Epigenetic change in kidney tumor: downregulation
of histone acetyltransferase MYST1 in human renal cell carcinoma. J
Exp Clin Cancer Res. 32:82013. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Song JS, Chun SM, Lee JY, Kim DK, Kim YH
and Jang SJ: The histone acetyltransferase hMOF is overexpressed in
non-small cell lung carcinoma. Korean J Pathol. 45:386–396. 2011.
View Article : Google Scholar
|
|
23.
|
Zhao L, Wang DL, Liu Y, Chen S and Sun FL:
Histone acetyltransferase hMOF promotes S phase entry and
tumorigenesis in lung cancer. Cell Signal. 25:1689–1698. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Sameer AS: Colorectal cancer: molecular
mutations and polymorphisms. Front Oncol. 3:1142013. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Jemal A, Siegel E, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics. CA Cancer J Clin. 57:43–66.
2007.
|
|
26.
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Tamagawa H, Oshima T, Shiozawa M, et al:
The global histone modification pattern correlates with overall
survival in metachronous liver metastasis of colorectal cancer.
Oncol Rep. 27:637–642. 2012.PubMed/NCBI
|
|
28.
|
Tamagawa H, Oshima T, Numata M, et al:
Global histone modification of H3K27 correlates with the outcomes
in patients with metachronous liver metastasis of colorectal
cancer. EJSO. 39:655–661. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Liu N, Zhang R, Zhao X, et al: A potential
diagnostic marker for ovarian cancer: involvement of the histone
acetyltransferase, human males absent on the first. Oncol Lett.
6:393–400. 2013.
|
|
30.
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
Springer; Chicago, IL: 2010
|
|
31.
|
Choschzick M, Oosterwijl R, Muller V,
Woelber L, Simon R, Moch H and Tennstedt P: Overexpression of
carbonic anhydrase IX (CAIX) is an independent unfavorable
prognostic marker in endometrioid ovarian cancer. Virchows Arch.
459:193–200. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Liu X, Cao L, Ni J, et al: Differential
BCCIP gene expression in primary human ovarian cancer, renal cell
carcinoma and colorectal cancer tissues. Int J Oncol. 43:1925–1934.
2013.PubMed/NCBI
|
|
33.
|
Rea S, Xouri G and Akhtar A: Males absent
on the first (MOF): from flies to humans. Oncogene. 26:5385–5394.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Taiple M, Rea S, Richter K, Vilar A,
Lichter P, Imhof A and Akhtar A: hMOF histone acetyltransferase is
required for histone H4 lysine 16 acetylation in mammalian cells.
Mol Cell Biol. 25:6798–6810. 2005. View Article : Google Scholar : PubMed/NCBI
|