Introduction
Nasopharyngeal carcinoma (NPC) is a squamous cell
carcinoma derived from epithelial cells lining the nasopharynx with
highly malignant local invasion and early distant metastasis, which
is rare globally but common in southern China and Southeast Asia
(1). However, the cause of this
disease is still uncertain while genetic, ethnic and environmental
factors could all have a role. Although patients with early stage
NPC can achieve excellent local control by radiotherapy alone, the
5-year survival rate is ∼35% for patients with stage-IV(A–B)
disease (2–4). Once metastasis occurs, the prognosis
is even poorer. Therefore, better understanding of the pathogenesis
is essential for developing novel therapies for NPC.
MicroRNAs (miRNA) are short non-coding RNAs of 18–24
nucleotides that function as critical gene regulators in mammals
and other multicellular organisms. According to the results of
bioinformatic analysis, one single miRNA may target up to several
hundred mRNAs, indicating its potential influences on almost every
biological pathway (5,6). Recent studies have shown that >50%
of the miRNAs are frequently located at cancer associated regions
and fragile sites (7), playing a
role as tumor suppressor genes or oncogenes (8–10). A
number of miRNAs have been shown to have different biological
effects contributing to the development and progression of NPC
(11–16) and combination of some miRNAs is
useful in the diagnosis and prognosis of this disease (17,18),
suggesting the involvement of miRNAs in NPC tumorigenesis. Our
previous studies showed that miR-378 level in plasma was decreased
in NPC patients and miR-378 together with other 4 miRNAs may serve
as a set of biomarkers in NPC diagnosis (18). Nevertheless, the expression level
of miR-378 in NPC tissue and its functional role has not been
documented yet.
In this study, we investigated for the first time
the potential role of miR-378 in the NPC pathological process. The
miR-378 expression in NPC cell lines and tissues were examined and
its effects on cell growth, colony formation, cell migration and
invasion in vitro were tested. In addition, we investigated
the potential role of miR-378 on NPC tumor growth in a murine
model. Finally, the effect of miR-378 on its potential target gene
transducer of ERBB2 (TOB2) was tested. Our study may provide a new
strategy for the therapy of NPC.
Materials and methods
Clinical specimens
The NPC biopsy specimens (n=50) and normal
nasopharyngeal epithelium biopsy samples (n=31) were obtained from
patients in Nanfang Hospital (Southern Medical University,
Guangzhou, China) and were frozen in liquid nitrogen for further
study. None of the patients had received radiotherapy or
chemotherapy before biopsy sampling. Tumor samples were
pathologically confirmed as NPC. Informed written consent was
obtained from each patient. The research protocols were approved by
the Ethics Committee of Nanfang Hospital and registered in
Clinical.trials.gov (ID: NCT01171235).
Cell culture
The human nasopharyngeal carcinoma cell lines 5-8F,
6-10b, CNE1 and CNE2 were cultured in RPMI-1640 medium (HyClone,
Thermo scientific Inc, USA), supplemented with 10% fetal bovine
serum (FBS, HyClone) and 1% penicillin/streptomycin. The primary
nasopharyngeal epithelial cell NP69 (Cancer Research Center,
Southern Medical University, China) was cultured as the control
cell line in Keratinocyte-SFM (Gibco, Life Technologies Corp., USA)
according to the manufacturer’s instructions. HEK 293T cell line
(Cell Bank of Chinese Academy of Science in Shanghai, China) was
cultured in DMEM/High glucose medium (HyClone) with 10% FBS
(HyClone) and 1% penicillin/streptomycin. All cells were maintained
at 37°C with an atmosphere of 5% CO2.
RNA isolation, reverse transcription, and
quantitative real-time PCR
Total RNA was extracted from the samples using
RNAiso Plus (Takara, Shiga, Japan) and reversely transcribed to
cDNA using the All-in-One First-Strand cDNA Synthesis kit
(GeneCopoeia Inc., USA). Quantitative real-time PCR (qPCR) was
performed using All-in-One™ qPCR Mix (Applied GeneCopoeia Inc.) on
an ABI 7500HT System. U6 and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) snRNA were used as a miRNA and mRNA
endogenous control, respectively. All samples were normalized to
the internal control and the relative expression level of miR-378
and TOB2 was calculated using relative quantification assay.
Vector construction and lentivirus
transduction
To obtain stable cell lines overexpressing miR-378,
pre-miR-378 was cloned into the pLVTHM lentiviral vector
(Tronolab). The primers for cloning pre-miR-378 were: forward
sequence 5′-CGACGCGTCGGGCTGCGAGGAGTGAGCG-3′ and reverse sequence
5′-CCATCGATGGGAGTTCAAATGGCT TGCTCC-3′. The lentivial vectors and
packing system (psPAX2 and Pmd2G) were co-transfected by calium
phosphate precipitation. The 5-8F and 6–10B transduced by the
packaged lentivirus were named lv-miR-378/5-8F and
lv-miR-378/6–10B. The control cells transduced by lentivirus using
the original pLVTHM lentiviral vector were named lv-control/5-8F
and lv-control/6–10B. The stably transduced cells were then
selected using flow cytometry. The positive clones with miR-378
overexpression were confirmed by real-time PCR.
Cell proliferation and colony-formation
assay
To determine the effect of mir-378 on cell
proliferation, the cells with stable overexpression of miR-378 were
seeded into 96-wells plates at a density of 1×103
cells/well with 5 replicate wells of each condition. Cell Counting
Kit-8 (CCK-8, Kaigene Inc., China) was added to the maintenance
cell medium, and all cells were incubated at 37°C for an additional
2 h before being measured. Absorbance values were measured daily
for four consecutive days at a wavelength of 450 nm.
In measuring colony-forming activity, cells were
counted and seeded into 12-well plates at 100 cells per well.
Twelve days after seeding, the numbers of colonies containing
>50 cells were counted after dyed crystal violet.
Migration and invasion assays
Transwell migration assays and invasion assays were
conducted to determine the functional effects of miR-378 on cell
migration and invasion. Briefly, for transwell migration assays,
5×104 cells were plated in the top chamber (24-well
insert; pore size, 8 μm; Corning). For invasion assays,
1×105 cells were plated in the top chamber with
Matrigel-coated membrane (24-well insert; pore size, 8 μm;
BD Biosciences). Cells were then cultured at 37°C for 24 h. After
that, the cells on the surface of the up chamber were swapped with
cotton swap and the cells under the surface of the low chamber were
stained with crystal violet (0.1%) and counted under a
microscope.
Tumor growth assay
To further investigate the proliferation efficacy of
miR-378 in vivo, ten female BALB/C nude mice about four to
five-week-old were purchased from Laboratory Animal Center of
Southern Medical University. The animal protocol was approved by
the Animal Investigation Committee of Southern Medical University.
Ten nude mice were randomly divided into two groups.
lv-miR-378/5-8F or lv-control/5-8F (1×105) cells were
injected into the dorsal flank of each mouse. Tumor size was
measured every other day. Mice were sacrificed and tumors were
dissected and weighed. Tumor volumes were calculated as follows:
volume = (D×d2)/2, where D = the longest diameter and d
= the shortest diameter.
miRNA target prediction and
validation
Three microRNA target databases (miRBase, PicTar and
TargetScan) were utilized to predict mir-378 target genes, and a
total of 16 genes were selected (all the primers are listed in
Table I). Then, the predicted
targets were validated in NPC cell line 5-8F using RT-qPCR.
 | Table I.Targets of miR-378 predicted by
bioinformatic prediction. |
Table I.
Targets of miR-378 predicted by
bioinformatic prediction.
| Gene | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) |
|---|
| TNS3 |
AAGAGAACAAGCCATCGCCA |
AAGTGCCGGACGAGTTCATT |
| SUFU |
TCCCAAAGAGTACAGCTGGC |
TGTAGCGGACTGTCGAACAC |
| VASH2 |
GAGGAGGAGGACAAAGACGG |
GAAGGCTTTGCCAAGAAGGC |
| FOXN3 |
TCGTTGTGGTGCATAGACCC |
GTGGACCTGATGTGCTTTGATA |
| TOB2 |
ATGCAGCTAGAGATCAAAGTGGC |
CCAATGTGAACACAGCGGAAG |
| NF1 |
AAAACCAGCGGAACCTCCTT |
GCTGGCTAACCACCTGGTATAAA |
| VAT1 |
AGTCCCACAACACTGCCAAA |
ACCACCATAGAGACGGGACA |
| TRIB2 |
TGGGAGATCGCGGAACAAAA |
AGGTTCGGGCTGAAACTCTG |
| SRGAP2 |
AGCCCGGAATGAGTACTTGC |
GGTAGCCTAAGTCACAACACTGA |
| MARVELD1 |
AGGATGAGCGACGAGTTTGG |
CCAAGACAACCGAGCACAGA |
| KLF9 |
GGGGTTTGGTTTGTGACGTG |
TTTTCCCGAGTCCACTGACG |
| PRDM7 |
GAGTGCCACGCTCTTTCTGA |
TGAGCAGCACAGCTGTCAAT |
| SMARCB1 |
GCGAGTTCTACATGATCGGCT |
CACAGTGGCTAGTCGCCTC |
| PA2G4 |
CAGGAGCAAACTATCGCTGAG |
GGACCGAAGTACCCTGTTGG |
| RBL1 |
CTGGACGACTTTACTGCCATC |
TCCAACCGTGGGAATAATGCT |
| DLC1 |
GCACGTTCCTGATCTGGAATC |
CCAAAATGTCAACTTACCAGCCT |
miRNA transfection and western
blotting
MiRNAs were transfected at a working concentration
of 80 nmol/l using Lipofectamine 2000 reagent (Invitrogen). The
miR-378 mimic, a nonspecific miR control, anti-miR378 and a
nonspecific anti-miR control were all purchased from GenePharma
(Shanghai, China). Sixty hours after transfection, the cells were
harvested for protein extraction. Protein lysates extracted from
stable cell lines and miRNA transfected cell lines were separated
by 10% SDS-PAGE, and electrophoretically transferred to PVDF
(polyvinylidene difluoride) membrane (Millipore). Then, the
membrane was incubated with goat polyclonal antibody against human
TOB2 (Santa Cruz Biotechnology) followed by HRP (horseradish
peroxidase)-labeled goat anti-mouse IgG (Santa Cruz Biotechnology)
and detected by chemiluminescence. GAPDH was used as a protein
loading control. The intensity of protein fragments was quantified
with the Quantity One software (4.5.0 basic, Bio-Rad).
Statistical analysis
Statistical analyses were performed using SPSS 13.0
statistical software. All experiments were performed three times.
Two-tailed Student’s t-test was used for comparison of two
independent groups. MiR-378 expression between tumor and normal
samples was analyzed by the Mann-Whitney U test. P-values <0.05
were considered statistically significant.
Results
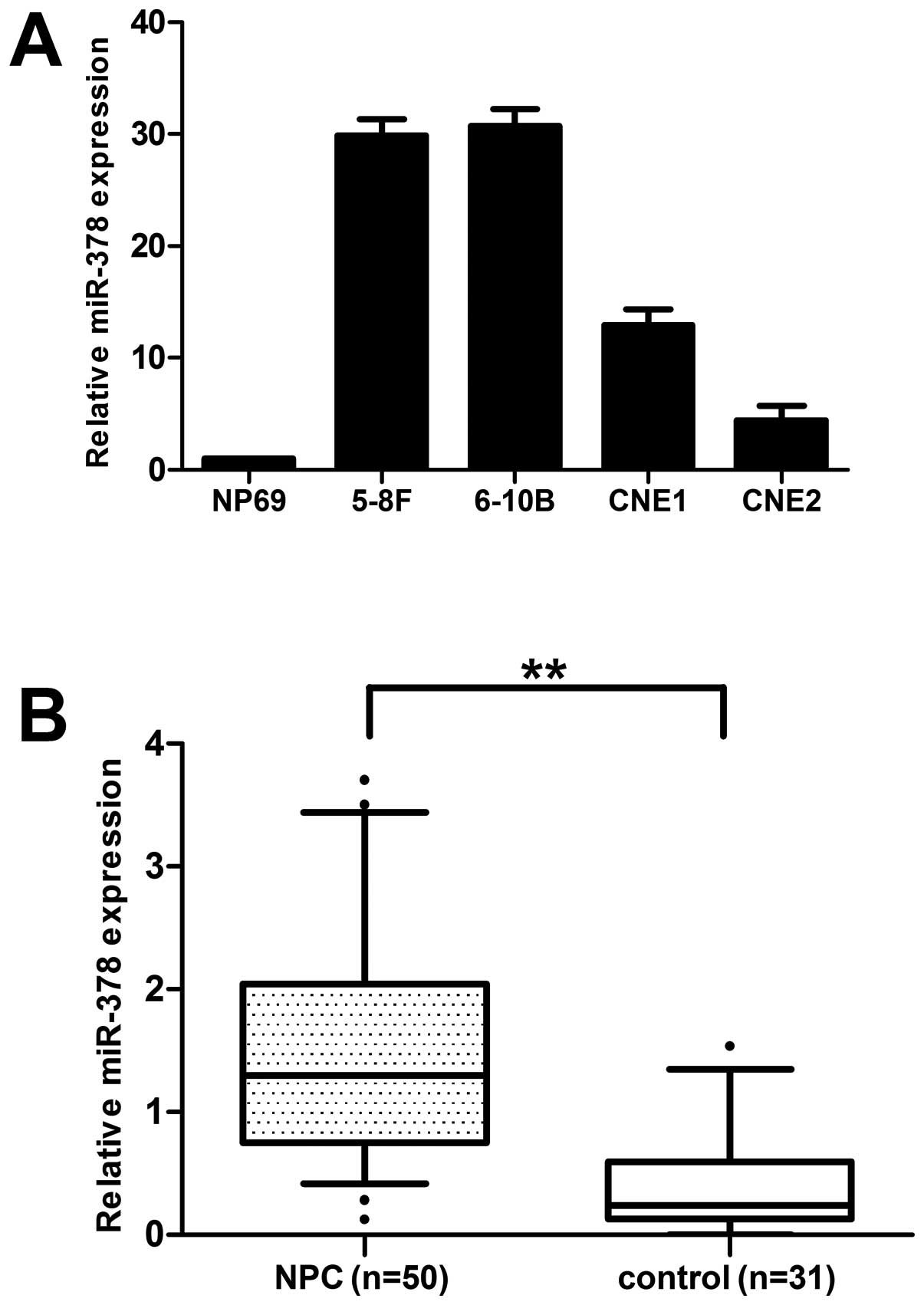
miR-378 is upregulated in NPC cell lines
and clinical specimens
5-8F, 6–10B, CNE1 and CNE2 cell lines were selected
to measure the miR-378 expression level using qRT-PCR. The results
showed that the basal expression level of miR-378 was generally
upregulated in 4 NPC cell lines compared with the immortalized, but
not tumorigenic nasopharyngeal epithelial cell line NP69 (Fig. 1A).
In order to further test and verify the expression
level of miR-378, we tested its transcript levels in 50 tumor
specimens relative to 31 normal nasopharyngeal tissues. A
subsequent analysis confirmed that the expression of miR-378 had a
3.63-fold increase compared with normal tissues (Fig. 1B). On the basis of these results,
we focused on miR-378 for further functional studies to evaluate
its roles in NPC pathogenesis.
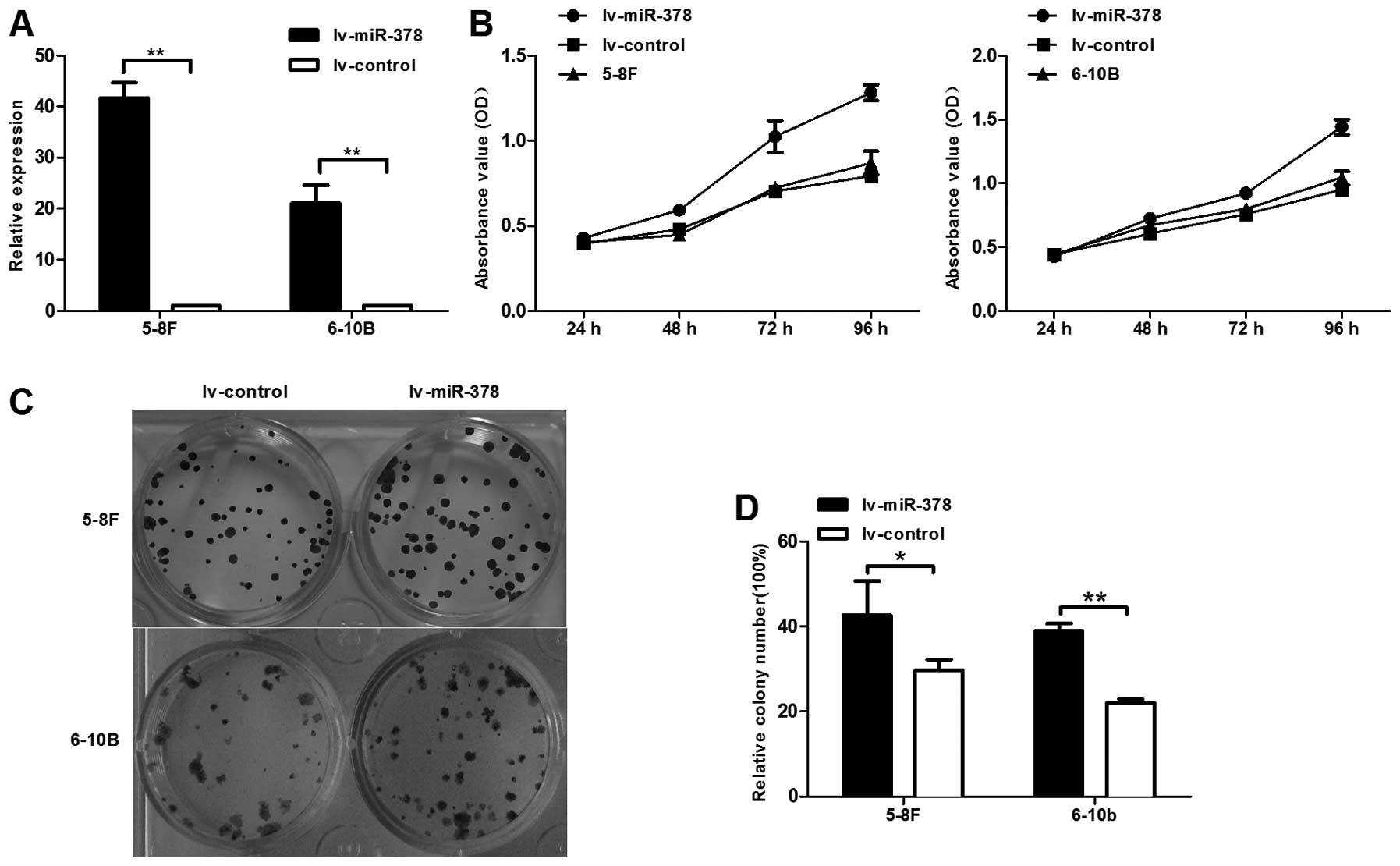
miR-378 enhances the proliferation
capacity of NPC cells in vitro
To determine whether miR-378 affected the
proliferation of NPC cells, we used lentiviral vectors to stably
upregulate the expression level of miR-378 in 5-8F and 6–10B cell
lines. The results demonstrated that the expression level of
miR-378 was upregulated ∼45-fold and 20-fold in 5-8F and 6–10B
cells, respectively, after transducted with lentiviral vectors
containing miR-378 (Fig. 2A).
CCK-8 was utilized to measure cell proliferation in
lv-miR-378/5-8F and lv-miR-378/6–10B cells. The results showed that
miR-378 increased the capability of cell proliferation by 61.6 and
51.9% in lv-miR-378/5-8F and lv-miR-378/6–10B than lv-control cell,
respectively (Fig. 2B). Colony
formation experiment was also performed to evaluate NPC cell
proliferation. Similar results indicated that miR-378 increased the
colony formation ability by 43.8 and 77% in lv-miR-378/5-8F and
lv-miR-378/6–10B than lv-control cell, respectively (Fig. 2C and D). Thus, upregulation of
miR-378 plays a crucial role in NPC cell growth, survival and
colony formation.
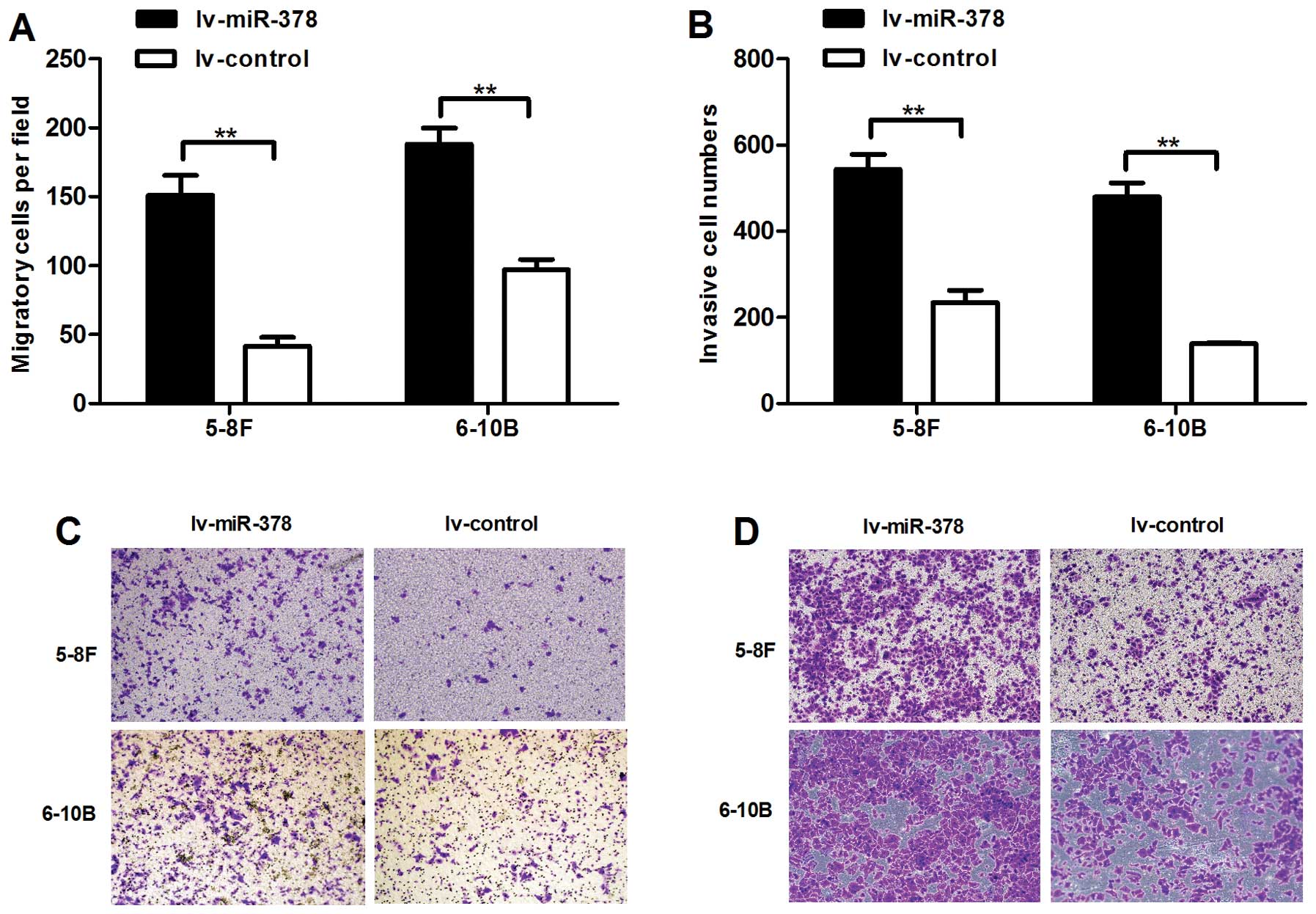
miR-378 promoted the migratory and
invasive capacities of NPC cells in vitro
To investigate the effects of upregulated miR-378 in
cell invasion and migration, we conducted cell migration and cell
invasion assays. We found that upregulated expression of miR-378
significantly promoted the migratory and invasive abilities of
lv-miR-378/5-8F cells and lv-miR-378/6–10B cells. The numbers of
migrated cells increased by 2.61- and 0.94-fold, respectively
(Fig. 3A and C). The numbers of
invasive cells increased by 1.32- and 2.47-fold in lv-miR-378/5-8F
cells and lv-miR-378/6–10B cells than lv-control cells,
respectively (Fig. 3B and D).
These results highlighted the vital role of miR-378 in NPC
metastasis.
miR-378 accelerates tumor growth in
vivo
Given the role of miR-378 in vitro in
proliferation, we next determined whether miR-378 could affect
tumor growth in vivo. Lv-miR-378/5-8F cells and lv-control
cells were used for evaluating the influence of miR-378
overexpression on the growth of tumor xenografts. We implanted
lv-miR-378/5-8F or lv-control/5-8F cells subcutaneously in nude
mice, respectively (n=5 per group). As shown in Fig. 4, lv-miR-378/5-8F cells resulted in
an approximately 2.8-fold increase in tumor size relative to
lv-control/5-8F cells (989±327 mm3 vs. 353±184
mm3 after 15 days, P<0.05). After the tumors were
denuded and weighed, the results of the weight were similar to
those of tumor volume. Thus, these results showed that miR-378
accelerated tumor growth in vivo.
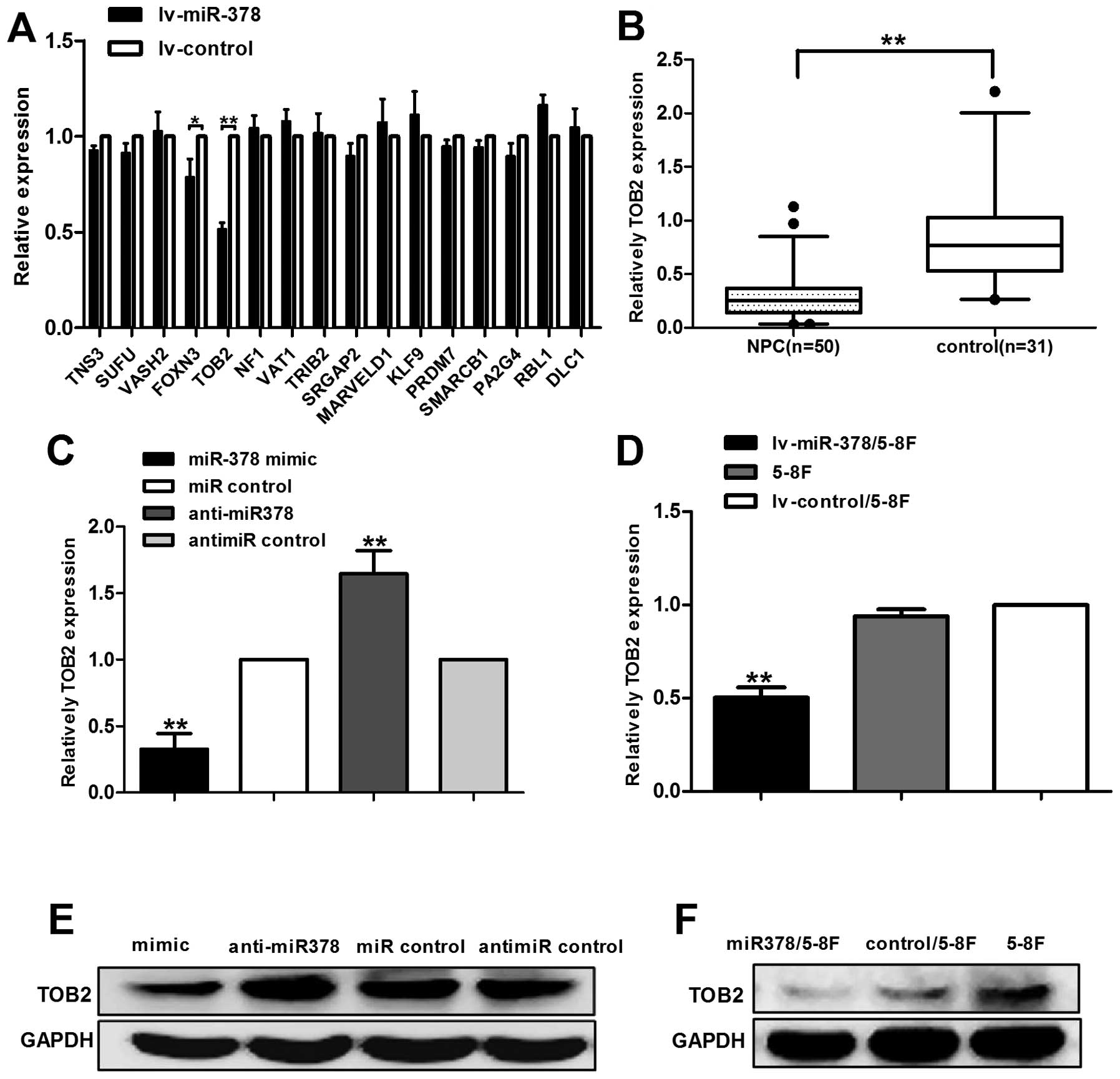
TOB2 is decreased in NPC cell lines and
tissues and is down-regulated by miR-378
By bioinformatic analysis, we focused on 16 possible
target genes of mir-378. Primers were designed using Primer-BLAST
and the selected targets were validated by RT-qPCR in
lv-miR-378/5-8F cells. The result of RT-qPCR showed that the
expression level of TOB2 decreased more significantly, compared
with other 15 possible target genes (Fig. 5A). To further determine whether
miR-378 expression was associated with TOB2 in human NPC tissues,
we examined the expression of TOB2 in 50 tumor specimens and 31
normal nasopharyngeal tissues. Notably, the results showed that
TOB2 was significantly downregulated in NPC tissues as compared
with non-cancer nasopharyngeal tissues (Fig. 5B).
To further investigate the relation between miR-378
and TOB2, we conducted transient transfection in 5-8F cells to
enhance or inhibit the expression of miR-378. As a result, up- or
downregulation of miR-378 led to a dramatic change in TOB2 mRNA and
protein expression (Fig. 5C and
E). Moreover, similar results were obtained in the stably
transfected cell lv-miR-378/5-8F (Fig.
5D and F).
Discussion
MiR-378 was first reported expressed in a number of
cancer cell lines and took part in the expression of vascular
endothelial growth factor (19,20).
No biological function of miR-378 was documented until 2007, Lee
and his colleges reported that miR-378 overexpression enhanced cell
survival, tumor growth and angiogenesis through repression of Sufu
and Fus-1, two tumor suppressors (21). Since then, an increasing number of
studies were conducted to shed light on the function of miR-378 in
cancers. However, despite the progress that has been made in recent
years regarding the role of miR-378 in the pathological process of
cancers, our knowledge of miR-378 functions in NPC is limited.
Emerging evidence clearly indicates that miR-378 is
abnormally expressed and has been implicated in various tumors such
as breast cancer (22), colorectal
cancer (23) and diffuse large
B-cell lymphoma (24). It was
significantly upregulated in the serum of patient with GC and renal
carcinoma, indicating its potential role as a serum biomarker in
early cancer diagnosis (25,26).
Mir-378 was also remarkably elevated in adjacent non-tumorous
tissues and decreased in cancer tissues and cell lines in gastric
cancer (27–29). Previous functional studies reported
that ectopic overexpression of mir-378 can promote cell
proliferation, angiogenesis, invasion and induced chemoresistance
in a number of solid tumors including glioblastoma, non-small cell
lung cancer (NSCLC), and breast cancer (21,22,30,31),
suggesting its role as onco-miRNA. On the contrary, however, the
expression level of miR-378 in colorectal cancer tissue was
significantly decreased and restoration of miR-378 inhibited G1/S
transition (23). In the study of
miRNA profile in basal cell carcinoma and cutaneous squamous cell
carcinoma, mir-378 was also among the downregulated miRNAs
(32,33). In addition, miR-378 was abnormally
expressed and epigenetically regulated in gastric cancer cell lines
and tissues via the suppression of vascular endothelial growth
factor (VEGF) signaling (29).
These findings indicated that miR-378 may play an important role in
certain tumors as tumour suppressors.
Our results showed that miR-378 was overexpressed in
NPC cell lines and cancer tissues. Functional studies revealed that
overexpression of miR-378 in NPC cell lines was able to enhance
cell growth and colony formation in vitro and promote
tumorigenesis in a murine model of NPC xenografts. Results of the
cell migration and invasion assays showed an enhancement in cell
migration and invasion ability in NPC cells overexpressing miR-378.
In our murine model of NPC xenografts, tumors derived from
lv-miR-378/5-8F cells had a significantly larger volume and weight
compared to the control group. These findings revealed the role of
miR-378 as onco-miRNA in NPC. These controversial results suggested
that the role of mir-378 was possibly tumor specific and highly
dependent on its targets in different cancer cells. The
tissue-dependent expression of miRNAs may influence protein
translation during diverse cellular processes. Moreover, the
aberrant expression of their target genes affected different
biological pathways with diverse functions (34).
It is worth mentioning that in our previous studies
(18), miR-378 was found to
decrease in the plasma of NPC patients. It increased to a certain
extent in patients without relapse but further decreased when
cancer relapsed and with metastasis. Compared with the results in
the present study, the expression level of miR-378 between NPC
patient plasma and tissue changed in the opposite direction. The
situation was similar to other studies showing a contrary change of
miRNAs between tissues and plasma as well (35,36),
suggesting that there may exist a cellular selection mechanism of
miRNA intake and release. Thus, we speculate that NPC cells may
selectively capture certain miRNAs such as mir-378, resulting an
elevated level in tissue. The mir-378 downregulated in tumor
tissues showed higher levels in the plasma or serum, thus cancer
may selectively secret mir-378 from the cells. Yet the reason why
there is such a remarkable difference between extracellular and
cellular miRNA expression remains unknown. Clarification of this
requires further efforts.
It is well known that a single miRNA can have
hundreds of target sites and regulates numerous protein-coding
genes, which form a regulatory network (6). To explore the potential target genes
of miR-378, three major miRNA databases (miRBase, PicTar and
TargetScan) were used to perform bioinformatic prediction. The
target genes with the highest context scores were selected and
validated in lv-miR-378/5-8F and 5-8F using qRT-PCR. Among the 16
selected target genes, transducer of ERBB2 (TOB2) was top ranked
and its mRNA level was the most downregulated one in
lv-miR-378/5-8F compared to the control group, whereas, the
expression level of TOB2 in tumor specimens was downregulated
significantly compared with normal tissues. Additionally, knockdown
of miR-378 by transient transfection of miR-378 inhibitor
significantly upregulated TOB2 mRNA and protein expression.
Therefore, all these results suggested that TOB2 may be a target
regulated by miR-378 in NPC.
TOB2 belongs to the anti-proliferative Tob/BTG
family proteins (37) which
regulate cell cycle progression in a variety of cell types and may
have important roles in preventing tumourigenesis and cancer
(38,39). It can inhibit cell proliferation
through suppressing cell cycle progression from the G0/G1 to S
phases (40). Accumulating
evidence indicates that TOB2 together with its family members are
expressed in various adult tissues and act primarily as
transcriptional repressors in several signaling pathways. However,
few mechanistic investigations have been conducted on TOB2 function
in cancers. Feng et al investigated the interactions between
miR-378 and TOB2 and their potential downstream signal pathway in
breast cancer. They confirmed that TOB2 was a functional target of
miR-378 and its expression was directly inhibited by miR-378. In
addition, the candidate tumor suppressor TOB2 achieved its function
by transcriptionally repressing the proto-oncogene cyclin D1
(41). As in the present study,
overexpression of miR-378 promotes NPC cell proliferation and tumor
growth and these may be mediated by downregulation of TOB2.
Therefore, we assumed that the function of miR-378 in NPC may be
similar to that in breast cancer and TOB2 is probably a direct
target gene of miR-378 in NPC. Besides, as our results indicated
that miR-378 overexpression could also enhance the migration and
invasion ability of NPC cells, it is very possible that miR-378 has
multiple target genes and their interactions result in the
oncogenic effects in NPC.
In conclusion, this study identified for the first
time that miR-378 is upregulated in NPC tissues and cell lines. It
can promote cell proliferation, migration, invasion and tumor
growth in NPC in vitro and in vivo. The oncogenic
effects of miR-378 were probably through downregulation of TOB2.
Since miR-378 is overexpressed in NPC, knockdown of this miRNA in
tumor tissue may provide us a new therapeutic strategy. Although
miRNA-based therapies remain in their infancy, our findings on
miR-378 are encouraging and suggest that this specific miRNA may be
a potential target for the treatment of NPC.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (U1132003, 81172053 to
X.P.L.), the National Natural Science Foundation of China (Grant
no. 81172053, to X.P.L.) and the Foundation for Distinguished Young
Talents in Higher Education of Guangdong, China (2012LYM_0039).
References
|
1.
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Heng DM, Wee J, Fong KW, et al: Prognostic
factors in 677 patients in Singapore with nondisseminated
nasopharyngeal carcinoma. Cancer. 86:1912–1920. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Hong MH, Mai HQ, Min HQ, Ma J, Zhang EP
and Cui NJ: A comparison of the Chinese 1992 and fifth-edition
International Union Against Cancer staging systems for staging
nasopharyngeal carcinoma. Cancer. 89:242–247. 2000. View Article : Google Scholar
|
|
4.
|
Ma J, Mai HQ, Hong MH, et al: Is the 1997
AJCC staging system for nasopharyngeal carcinoma prognostically
useful for Chinese patient populations? Int J Radiat Oncol Biol
Phys. 50:1181–1189. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar
|
|
9.
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
10.
|
Wiemer EAC: The role of microRNAs in
cancer: no small matter. Eur J Cancer. 43:1529–1544. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Sengupta S, den Boon JA, Chen IH, et al:
MicroRNA 29c is down-regulated in nasopharyngeal carcinomas,
up-regulating mRNAs encoding extracellular matrix proteins. Proc
Natl Acad Sci USA. 105:5874–5878. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Yu L, Lu J, Zhang B, et al: miR-26a
inhibits invasion and metastasis of nasopharyngeal cancer by
targeting EZH2. Oncol Lett. 5:1223–1228. 2013.PubMed/NCBI
|
|
13.
|
Luo Z, Dai Y, Zhang L, et al: miR-18a
promotes malignant progression by impairing microRNA biogenesis in
nasopharyngeal carcinoma. Carcinogenesis. 34:415–425. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Liu Y, Cai H, Liu J, et al: A miR-151
binding site polymorphism in the 3′-untranslated region of the
cyclin E1 gene associated with nasopharyngeal carcinoma. Biochem
Biophys Res Commun. 432:660–665. 2013.PubMed/NCBI
|
|
15.
|
Li G, Liu Y, Su Z, et al: MicroRNA-324-3p
regulates nasopharyngeal carcinoma radioresistance by directly
targeting WNT2B. Eur J Cancer. Apr 10–2013.Epub ahead of print.
|
|
16.
|
Deng M, Ye Q, Qin Z, et al: miR-214
promotes tumorigenesis by targeting lactotransferrin in
nasopharyngeal carcinoma. Tumour Biol. 34:1793–1800. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Liu N, Chen N-Y, Cui R-X, et al:
Prognostic value of a microRNA signature in nasopharyngeal
carcinoma: a microRNA expression analysis. Lancet Oncol.
13:633–641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Liu X, Luo HN, Tian WD, et al: Diagnostic
and prognostic value of plasma microRNA deregulation in
nasopharyngeal carcinoma. Cancer Biol Ther. 14:Aug 23–2013.Epub
ahead of print.
|
|
19.
|
Jiang J, Lee EJ, Gusev Y and Schmittgen
TD: Real-time expression profiling of microRNA precursors in human
cancer cell lines. Nucleic Acids Res. 33:5394–5403. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Hua Z, Lv Q, Ye W, et al: MiRNA-directed
regulation of VEGF and other angiogenic factors under hypoxia. PLoS
One. 1:e1162006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA.
104:20350–20355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Eichner LJ, Perry MC, Dufour CR, et al:
miR-378(*) mediates metabolic shift in breast cancer cells via the
PGC-1β/ERRγ transcriptional pathway. Cell Metab. 12:352–361.
2010.
|
|
23.
|
Faltejskova P, Svoboda M, Srutova K, et
al: Identification and functional screening of microRNAs highly
deregulated in colorectal cancer. J Cell Mol Med. 16:2655–2666.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Qian J, Lin J, Qian W, et al:
Overexpression of miR-378 is frequent and may affect treatment
outcomes in patients with acute myeloid leukemia. Leuk Res.
37:765–768. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Redova M, Poprach A, Nekvindova J, et al:
Circulating miR-378 and miR-451 in serum are potential biomarkers
for renal cell carcinoma. J Transl Med. 10:552012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Liu H, Zhu L, Liu B, et al: Genome-wide
microRNA profiles identify miR-378 as a serum biomarker for early
detection of gastric cancer. Cancer Lett. 316:196–203. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Yao Y, Suo AL, Li ZF, et al: MicroRNA
profiling of human gastric cancer. Mol Med Rep. 2:963–970.
2009.PubMed/NCBI
|
|
28.
|
Guo J, Miao Y, Xiao B, et al: Differential
expression of microRNA species in human gastric cancer versus
nontumorous tissues. J Gastroenterol Hepatol. 24:652–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Deng H, Guo Y, Song H, et al: MicroRNA-195
and microRNA-378 mediate tumor growth suppression by epigenetical
regulation in gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF
and Wang SF: MicroRNA-378 is associated with non-small cell lung
cancer brain metastasis by promoting cell migration, invasion and
tumor angiogenesis. Med Oncol. 29:1673–1680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Wu QP, Xie YZ, Deng Z, et al: Ergosterol
peroxide isolated from Ganoderma lucidum abolishes microRNA
miR-378-mediated tumor cells on chemoresistance. PLoS One.
7:e445792012.PubMed/NCBI
|
|
32.
|
Sand M, Skrygan M, Sand D, et al:
Expression of microRNAs in basal cell carcinoma. Br J Dermatol.
167:847–855. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Sand M, Skrygan M, Georgas D, et al:
Microarray analysis of microRNA expression in cutaneous squamous
cell carcinoma. J Dermatol Sci. 68:119–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Wang K, Zhang S, Marzolf B, et al:
Circulating microRNAs, potential biomarkers for drug-induced liver
injury. Proc Natl Acad Sci USA. 106:4402–4407. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Tanaka M, Oikawa K, Takanashi M, et al:
Down-regulation of miR-92 in human plasma is a novel marker for
acute leukemia patients. PLoS One. 4:e55322009. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J,
et al: Tob2, a novel anti-proliferative Tob/BTG1 family member,
associates with a component of the CCR4 transcriptional regulatory
complex capable of binding cyclin-dependent kinases. Oncogene.
18:7432–7441. 1999. View Article : Google Scholar
|
|
38.
|
Suzuki T, K-Tsuzuku J, Ajima R, Nakamura
T, Yoshida Y and Yamamoto T: Phosphorylation of three regulatory
serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell
proliferation and transformation. Genes Dev. 16:1356–1370. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Boiko AD, Porteous S, Razorenova OV,
Krivokrysenko VI, Williams BR and Gudkov AV: A systematic search
for downstream mediators of tumor suppressor function of p53
reveals a major role of BTG2 in suppression of Ras-induced
transformation. Genes Dev. 20:236–252. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Winkler GS: The mammalian
anti-proliferative BTG/Tob protein family. J Cell Physiol.
222:66–72. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Feng M, Li Z, Aau M, Wong CH, Yang X and
Yu Q: Myc/miR-378/TOB2/cyclin D1 functional module regulates
oncogenic transformation. Oncogene. 30:2242–2251. 2011. View Article : Google Scholar : PubMed/NCBI
|