Introduction
Breast malignancy is the most prevalent type of
cancer and the leading cause of cancer death in females around the
world (1). According to the
presence of ER, PR and HER2, breast cancer can be classified into
two molecular subtypes: the triple-negative subtype (all three of
ER, PR, HER2 are absent, aka TNBC) and non-triple-negative subtype
(at least one receptor remains, aka NTNBC) (2,3).
Traditional anticancer methods, surgery,
radiotherapy and chemotherapy are generally used for breast cancer,
but they are not so efficient within specific subtypes. For hormone
receptor-positive breast cancer, endocrine therapy is a better
choice in patients with chronic disease progression. In recent
years, with the development of targeted therapies, antibody-based
drugs aiming at cancer-specific molecules has been widely applied
and turned out to be effective, such as Trastuzumab (Herceptin)
against ERBB2 in HER2+ subtypes (4). However, no clear consensus has been
reached for the treatment of TNBC (5,6).
Plenty of clinical reports showed that TN is the subtype of higher
malignancy and has a poorer expectation of prognosis and overall
survival in breast cancer (2,7,8). Due
to the absence of ER, PR and HER2, endocrine treatment and
Trastuzumab were invalid for TNBC patients. Current options such as
chemotherapy (by platinum compounds or doxorubicin), EGFR
antagonists, PARP inhibitors and anti-angiogenesis therapy do not
act so well against TNBC, and combinations of these agents require
further investigation (5,9). As a result, searching for new
therapeutic targets for TNBC is of great clinical value.
To investigate genes correlated with the poor
prognosis of TNBC and searching for potential TNBC-specific
therapeutic targets, we analyzed 5 breast cancer expressing
profiling datasets from TCGA and GEO databases (10) and generated a primary set whose
expression level was much higher in TNBC patients than in normal
controls. To ensure the specificity of these genes in TNBC,
corresponding NTNBC expression data were then compared by
independent sample T-test. The expression level of 45 genes were
found to be statistically higher in TNBC than in NTNBC.
Kaplan-Meier analysis for the 45-gene TNBC specific cohort was
performed with further two clinical datasets, and within these
cohort genes, BIRC5, CENPA and FAM64A were found significantly
associated with poor breast cancer survival. This correlation was
also seen in patients of early cancer stages and grades, and
synchronous upregulation of three genes is correlated with the
worst prognosis. Analysis of pathway revealed that 3 hub genes were
largely co-expressed with many other molecules related to survival,
and potential interactions and the related pathways are also
discussed. These findings suggested that the high expressions of
BIRC5, CENPA and FAM64A are involved with the poor clinical outcome
of TNBC, and they are potential prognostic markers and therapeutic
targets of triple-negative breast cancer.
Materials and methods
Ethics statement
The databases searched in our study are available
online. We were free to use TCGA breast cancer databases from The
Cancer Genome Atlas (https://tcga-data.nci.nih.gov) by meeting its
freedom-to-publish criteria. All the Gene Expression Omnibus
datasets are also accessible to public from the GEO website
(www.ncbi.nlm.nih.gov/geo) and the ROCK breast
cancer functional genomics (http://www.rock.icr.ac.uk), an integrated online
breast cancer knowledgebase which provides and updates detailed
microarray databases and information from reports on breast cancer
(11).
Selection of databases
Five breast cancer microarray databases from
published studies were used to identify TNBC specific genes.
Normalized gene expression profiling data were downloaded from TCGA
and GEO websites, while the clinical information was extracted from
corresponding reports, TCGA and ROCK websites. Samples were
deposited into TNBC, NTNBC and normal groups according to molecular
subtyping and immunohistochemical staining results, as presented in
Table I.
 | Table I.Statistics of the five microarray
databases that generated the 48-gene primary set. |
Table I.
Statistics of the five microarray
databases that generated the 48-gene primary set.
| Dataset ID | TNBC | NTNBC | N | Total number |
|---|
| GSE1992 | 25 | 125 | 8 | 158 |
| GSE2607 | 8 | 36 | 6 | 50 |
| GSE3744 | 18 | 22 | 7 | 47 |
| GSE47561 | 73 | 1457 | 40 | 1570 |
| TCGA | 71 | 398 | 61 | 530 |
| Total number | 195 | 2038 | 122 | 2355 |
The two further online clinical datasets were chosen
to analyze the association of TNBC specific genes with breast
cancer prognosis. NKI data were extracted from a previous study of
van de Vijver et al (12),
while 2006CC data from the study of Chin et al (13). The median age of patients in NKI
and 2006CC data are 44 and 51, respectively, and each dataset
contains patients of different lymph status and molecular subtypes.
Except for microarray data, NKI dataset contains overall survival
(OS), distant metastasis-free survival (DMFS), disease-free
survival (DFS) and Nottingham Grading Score (NGS) information,
while 2006CC dataset contains OS, DMFS, relapse-free survival (RFS)
and TNM staging information.
Identification of TNBC specific
genes
Triple-negative subtype constitutes only a small
fraction of all breast cancer cases, hence to investigate as many
breast cancer patients as possible while avoiding cross-study batch
effects, we separately carried out statistical analysis in each of
these 5 databases (14).
Significant analysis of microarray (SAM) method (p<0.05,
q<0.25) and independent sample t-test (p<0.05) were used to
compare the expression level of each gene between patients and
healthy persons. Since the platform and corresponding gene sums of
the five datasets were not in concert, to avoid batch effects
between experiments while excluding as many heterogenetic genes as
possible, statistical analysis was performed for each dataset, and
the overlapping part was output. Overlapping genes upregulated
>2-fold in patients while with statistical significance
(p<0.05) were deposited into the primary set.
To ensure that members of this gene set were
specifically overexpressed in TNBC, we compared corresponding TNBC
expression data of genes in the primary set with NTNBC in each
database via SAM method or independent sample t-test with the
judgment criteria of p<0.05. A cohort of genes statistically
upregulated in triple-negative breast cancer was generated and then
annotated by DAVID (http://david.abcc.ncifcrf.gov/). The expression
intensity of microarray data was demonstrated as heatmap by Mev 4.0
software.
Association of TNBC-upregulated genes
with patient prognosis
In the five datasets applied to generate a 45-gene
cohort, four GEO datasets contain no clinical follow-ups, while a
large fraction of TCGA survival information were censored or still
upgrading. For further analysis, we chose NKI and 2006CC clinical
sets to investigate the association between TNBC cohort genes and
breast cancer prognosis. The median of TNBC-upregulated cohort gene
levels across samples were calculated to define the attribution of
each sample. For each gene, patients with an expression level
higher than median was classified as ‘high’, while those with a
level lower than median was classified as ‘low’. The relationship
between the level of each gene and patient survival [including
disease-free survival (DFS), distant metastasis-free survival
(DMFS), relapse-free survival (RFS) and overall survival (OS)] was
described via Kaplan-Meier analysis in both clinical sets. The
survival curves were drawn via GraphPad Prism 5, patients of ‘high’
or ‘low’ categories were displayed by curves of different colors,
while censored value were displayed as small bars. Genes whose
expression level statistically correlated with breast cancer
survival in both clinical sets were defined as hub genes.
Tumor grade/stage differentiated
relationship of hub genes with survival
To investigate the role of hub genes in survival
within different tumor periods, we divided the patients into
subgroups according to tumor grade and stage. In two clinical sets
applied in this study, NKI contains Nottingham grading score (NGS)
information, patients in this set were classified as grade 1–3,
while 2006CC contains TNM staging information, patients in this set
were classified as stage I–II and stage III–IV. Expression level of
each hub gene in each sample were defined as ‘high’ or ‘low’ by
using median as the threshold as mentioned above. Kaplan-Meier
analysis of hub gene-related survival was then performed.
Grouped survival analysis of three hub
genes
To find out whether identified hub genes were
indispensable factors influencing patient survival, we classified
samples of both clinical sets into groups according to the level of
hub genes. Samples with all hub genes highly expressed were defined
as ‘triple-positive’, samples with all hub genes downregulated were
defined as ‘triple-negative’, while those in the middle were
further grouped. Kaplan-Meier analysis was made for each group.
Cox proportional hazards regression
analysis of the genes of interest
To weigh the relative influence of the genes on
prognosis, Cox proportional hazards regression analysis was
performed in two clinical sets using OS and DMFS. Analysis was
performed by SPSS 19.0. All the genes of interest were used as
covariates, and the ‘Forward: LR’ method was used.
Pathway analysis of survival associated
TNBC genes
In order to further investigate the role of these
survival-related genes in breast cancer, we constructed an
interaction network using GNCpro online tools (15). Genes and pathways shown in this
network were analyzed in NCBI and KEGG, and related reports were
downloaded from PubMed.
Results
Generation of triple-negative breast
cancer-specific gene cohort
Definitely molecular subtyped TNBC class covers only
∼12–17% of all breast cancer patients, exclusive of basal-like and
BRCA1-related subtypes (5). To
expand the sample pool as much as possible, we browsed all
available public databases, and those with TNBC samples fewer than
5 were discarded. Only 5 databases were chosen, and the total
number of TNBC, NTNBC and normal were 211, 2038 and 122,
respectively (16) (Table I). The expression level of
triple-negative samples versus normal in GSE3744 (17) and GSE47561 (11) data were compared based on SAM
methods (18) with p<0.05 and
q<0.25, but for TCGA, GSE2607 (19) and GSE1992 (20) data, independent sample t-test
(p<0.05) was applied instead of SAM because of the existence of
numerous databases. A primary set of 48 genes were found to be
expressed ≥2-fold higher in TNBC than in healthy controls across
the five databases. A heatmap for the 48-gene primary set from TCGA
database is shown as Fig. 1.
With a judgment criteria of p<0.05, in all 48
members, a cohort of 45 genes was identified to be statistically
upregulated in TNBC compared to NTNBC, 2 genes were of similar
levels in TNBC and NTNBC, and 1 gene was downregulated in NTNBC,
not in TNBC. These 45 TNBC specific genes were annotated to be
related to important cellular mechanisms such as cell cycle
regulation, centrosome function, enzymatic canalization and
maintenance of chromosome integrity, all of which were potential
breach points of carcinogenesis (Table
II).
 | Table II.Fold changes and annotations of 45
TNBC-upregulated genes from TCGA breast cancer data. |
Table II.
Fold changes and annotations of 45
TNBC-upregulated genes from TCGA breast cancer data.
| Fold change
| |
|---|
| Gene symbol | TNBC vs. N | TNBC vs. NTNBC | The gene name |
|---|
| ANP32E | 3.12 | 2.78 | Acidic (leucine-rich)
nuclear phosphoprotein 32 family, member E |
| ATAD2 | 4.92 | 1.62 | ATPase family, AAA
domain containing 2 |
| BIRC5 | 9.55 | 2.01 | Baculoviral IAP
repeat-containing 5 (survivin) |
| BUB1 | 22.53 | 2.94 | BUB1 budding
uninhibited by benzimidazoles 1 homolog (yeast) |
| CCNB1 | 5.09 | 1.45 | Cyclin B1 |
| CCNB2 | 8.32 | 2.23 | Cyclin B2 |
| CDC2 | 6.29 | 1.58 | Cell division cycle
2, G1→S and G2→M |
| CDC20 | 7.83 | 2.69 | CDC20 cell division
cycle 20 homolog (S. cerevisiae) |
| CDKN3 | 7.35 | 1.56 | Cyclin-dependent
kinase inhibitor 3 |
| CENPA | 12.34 | 2.93 | Centromere protein
A |
| CENPF | 15.12 | 2.35 | Centromere protein
F, 350/400 ka (mitosin) |
| CEP55 | 24.52 | 2.74 | Centrosomal protein
55 kDa |
| CHEK1 | 9.64 | 2.88 | CHK1 checkpoint
homolog (S. pombe) |
| CKS2 | 7.3 | 1.59 | CDC28 protein
kinase regulatory subunit 2 |
| CXCL10 | 3.19 | 1.28 | Chemokine (C-X-C
motif) ligand 10 |
| E2F3 | 2.82 | 1.96 | E2F transcription
factor 3 |
| EXO1 | 8.54 | 2.38 | Exonuclease 1 |
| FAM64A | 9.97 | 2.59 | Family with
sequence similarity 64, member A |
| GPSM2 | 5.91 | 2.38 | G-protein
signalling modulator 2 (AGS3-like, C. elegans) |
| H2AFZ | 2.36 | 1.41 | H2A histone family,
member Z |
| HMMR | 5.23 | 1.49 | Hyaluronan-mediated
motility receptor (RHAMM) |
| HN1 | 5.46 | 1.51 | Hematological and
neurological expressed 1 |
| KIAA0101 | 4.26 | 1.17 | KIAA0101 |
| KIF20A | 9.7 | 2 | Kinesin family
member 20A |
| LAMP3 | 8.54 | 4.62 |
Lysosomal-associated membrane protein
3 |
| LMNB1 | 6.98 | 1.69 | Lamin B1 |
| LRP8 | 6.87 | 3.43 | Low density
lipoprotein receptor-related protein 8, apolipoprotein E
receptor |
| MAD2L1 | 5.15 | 1.76 | MAD2 mitotic arrest
deficient-like 1 (yeast) |
| MCM4 | 5.76 | 1.94 | MCM4 minichromosome
maintenance deficient 4 (S. cerevisiae) |
| MLF1IP | 5.59 | 1.44 | MLF1 interacting
protein |
| MMP1 | 28.61 | 3.46 | Matrix
metallopeptidase 1 (interstitial collagenase) |
| NEK2 | 17 | 1.74 | NIMA (never in
mitosis gene a)-related kinase 2 |
| NFE2L3 | 5.26 | 2.99 | Nuclear factor
(erythroid-derived 2)-like 3 |
| OASL | 4.15 | 1.29 |
2′-5′-oligoadenylate synthetase-like |
| PARP1 | 2.98 | 1.28 | Poly(ADP-ribose)
polymerase family, member 1 |
| PBK | 11.92 | 1.88 | PDZ binding
kinase |
| RACGAP1 | 3.88 | 1.39 | Rac GTPase
activating protein 1 |
| RECQL4 | 4.37 | 1.48 | RecQ protein-like
4 |
| RRM2 | 9.81 | 1.81 | Ribonucleotide
reductase M2 polypeptide |
| TAP1 | 2.87 | 1.44 | Transporter 1,
ATP-binding cassette, sub-family B (MDR/TAP) |
| TMEM48 | 2.83 | 1.46 | Transmembrane
protein 48 |
| TOP2A | 8.8 | 1.58 | Topoisomerase (DNA)
IIα 170 kDa |
| TRIP13 | 7.06 | 2.08 | Thyroid hormone
receptor interactor 13 |
| TTK | 13.81 | 3.27 | TTK protein
kinase |
| TYMS | 7.71 | 2.29 | Thymidylate
synthetase |
Cohort genes correlated with poor
prognosis
The poor clinical outcome and short survival of TNBC
might be related to the overexpression of some TNBC specific genes
(21). The association between
cohort genes and patient survival were analyzed using NKI and
2006CC clinical sets according to procedures mentioned in Materials
and methods. We found that among the 45 TNBC-upregulated genes, the
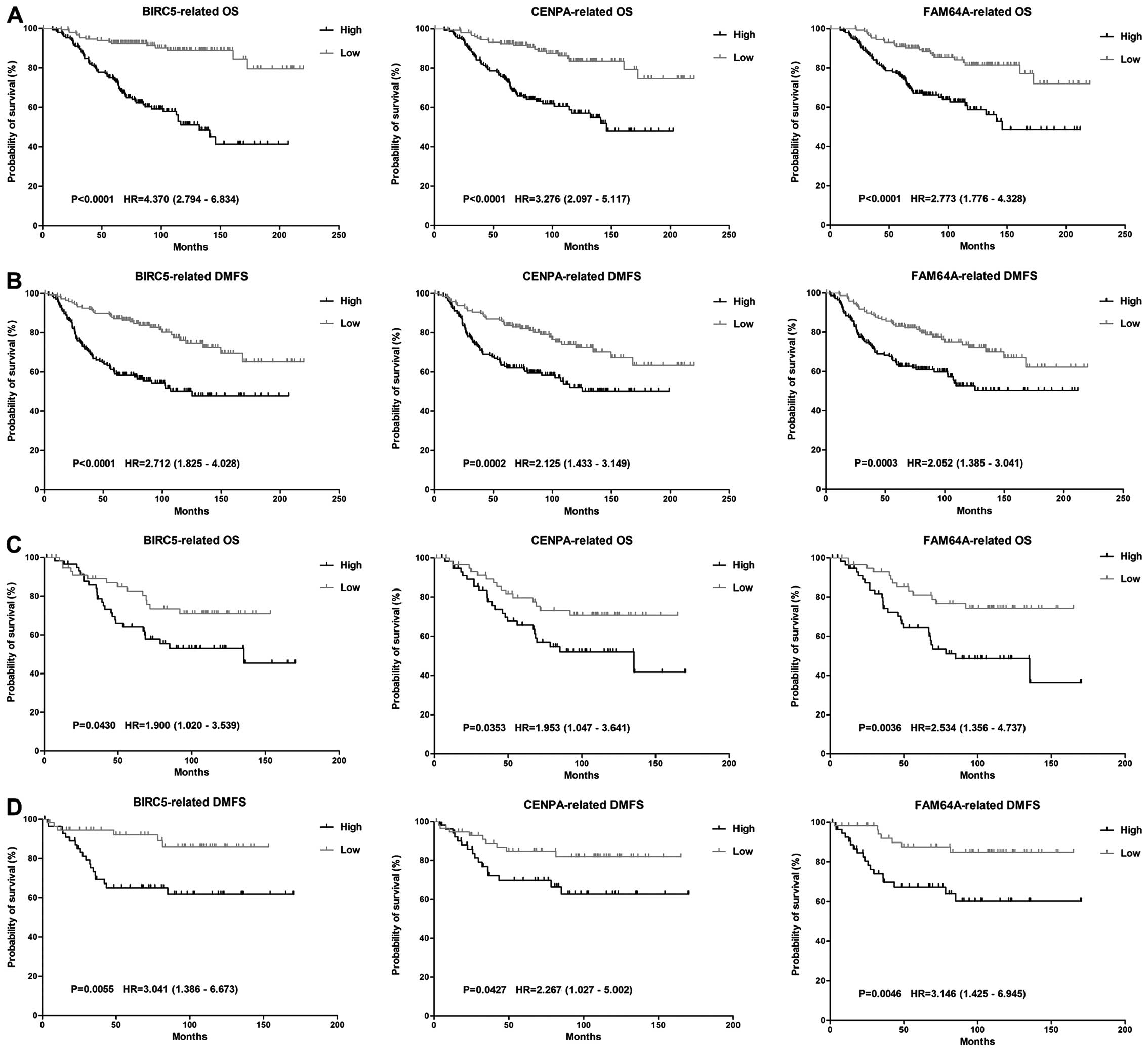
overexpression of BIRC5, CENPA and FAM64A showed a significant
correlation with poor OS and DMFS (p<0.05) despite the molecular
subtypes of breast cancer (Fig.
2). High expression of these genes were also related with poor
DFS and RFS. Therefore, these three genes were classified as hub
genes.
Apart from BIRC5, CENPA and FAM64A, some other genes
in the cohort were found to be co-expressed with these three hub
members and were potentially involved in breast cancer survival as
well. In these candidates, the level of BUB1 and CCNB2 genes were
also negatively associated with clinical survival, though their
relationship with OS in 2006CC data were not statistically
supported by log-rank test as with DFS, DMFS or RFS.
Three hub genes are related to survival
in early tumor stage/grade
According to available clinical information,
patients in 2006CC were divided as stage I–II and stage III–IV,
while patients in NKI clinical set were divided as grade 1–3.
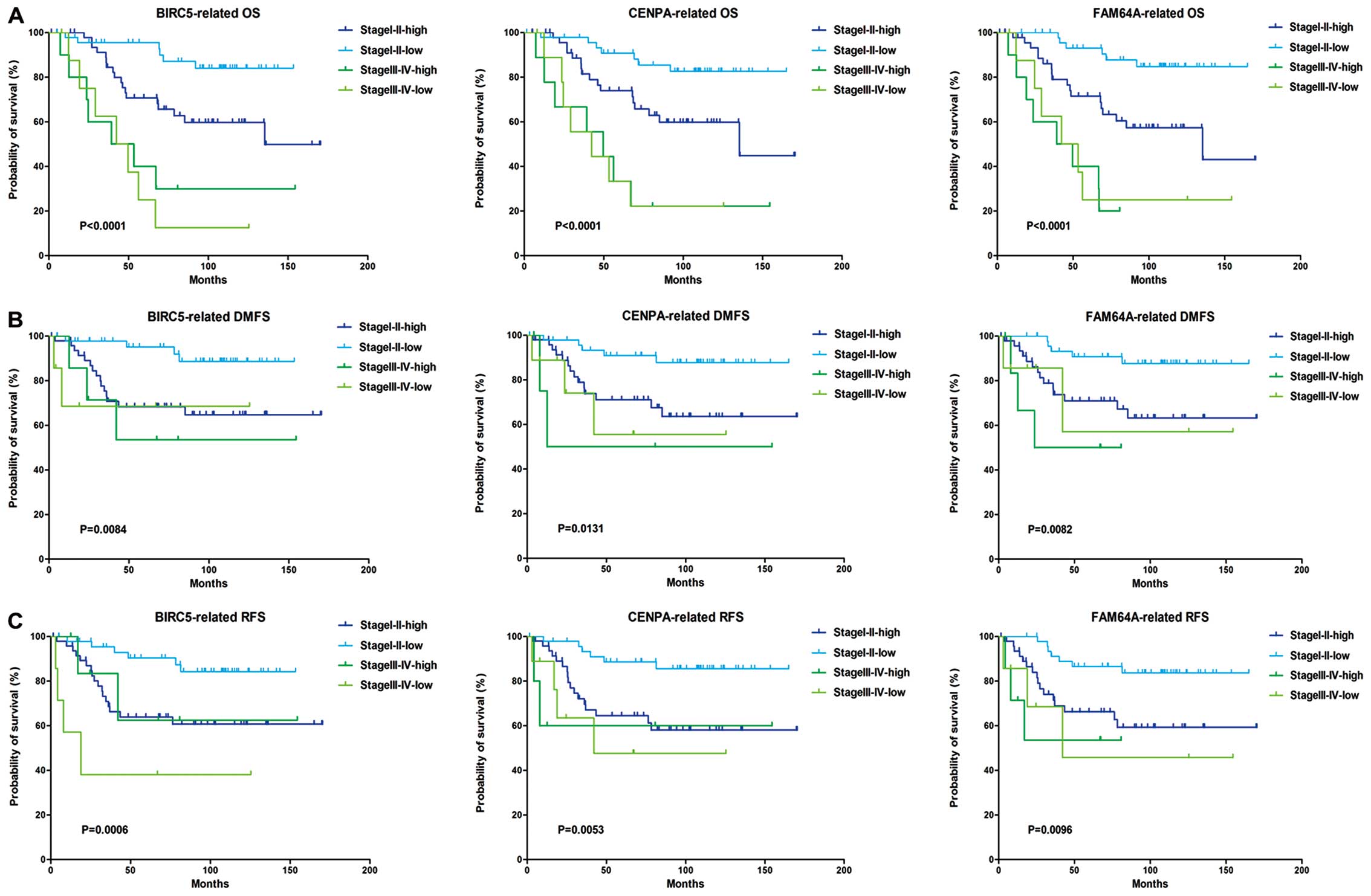
Kaplan-Meier analysis showed that patients of stage III–IV have
poorer survival than stage I–II, while the rank of survival with
NGS was grade 3>2>1, these results agreed with clinical
observations. Furthermore, high level of BIRC5, CENPA and FAM64A
were correlated with poor survival in stage I–II patients (Fig. 3). Since the number of advanced
tumor samples was too few to perform convincible statistic
analysis, we are not sure whether this relationship exists in stage
III–IV patients. Similar correlation was also found in patients
classified as grade 1–3 by Nottingham grading system.
Synchronous hub gene upregulation is
correlated with the worst prognosis
To find out whether these three hub genes were
indispensable factors influencing patient survival, we defined
patients with all hub genes highly expressed as ‘triple-positive’
(TP), two genes highly expressed as ‘double-positive’ (DP), one
gene highly expressed as ‘double-negative’ (DN), and all 3 genes of
low level as ‘triple-negative’ (TN). Relationships of these 4
categories with survival were described by Kaplan-Meier analysis.
We found that in NKI data, the TN group had the best OS, DMFS and
DFS, while TP was of the worst OS but with similar level of
DMFS/DFS as DP and DN groups. In 2006CC data, due to the sample
number in each group being limited, survival of TP was similar with
DP and NP, but TN still had the best OS, DMFS and RFS (Fig. 4). These results suggested that
BIRC5, CENPA and FAM64A are all important molecules influencing
survival in breast cancer. In other words, synchronous lower level
of three hub genes indicates the best prognosis in breast cancer
patients, while synchronous upregulation of three hub genes
indicates the worst. Since three hub genes has been previously
validated to be upregulated in TNBC subtypes, these findings
suggested that the overexpression of BIRC5, CENPA and FAM64A may be
the reason for the poor clinical outcome of TNBC, and they are
potential therapeutic targets for triple-negative breast
cancer.
Cox analysis indicates BIRC5 is an
independent prognostic factor in breast cancer
To evaluate the independence of five TNBC-specific
genes in inducing poor prognosis, Cox proportional hazards
regression analysis was performed in NKI and 2006CC clinical sets
according to OS and DMFS. For NKI data, BIRC5 and CCNB2 were
considered to be independent prognostic markers for OS and DMFS,
while for 2006CC data, BIRC5 and CENPA were considered as
independent prognostic markers for OS and DMFS (data not shown),
giving a hint that BIRC5 is the more important of these genes in
the generation of TNBC high malignancy.
An interaction network of survival
related cohort genes
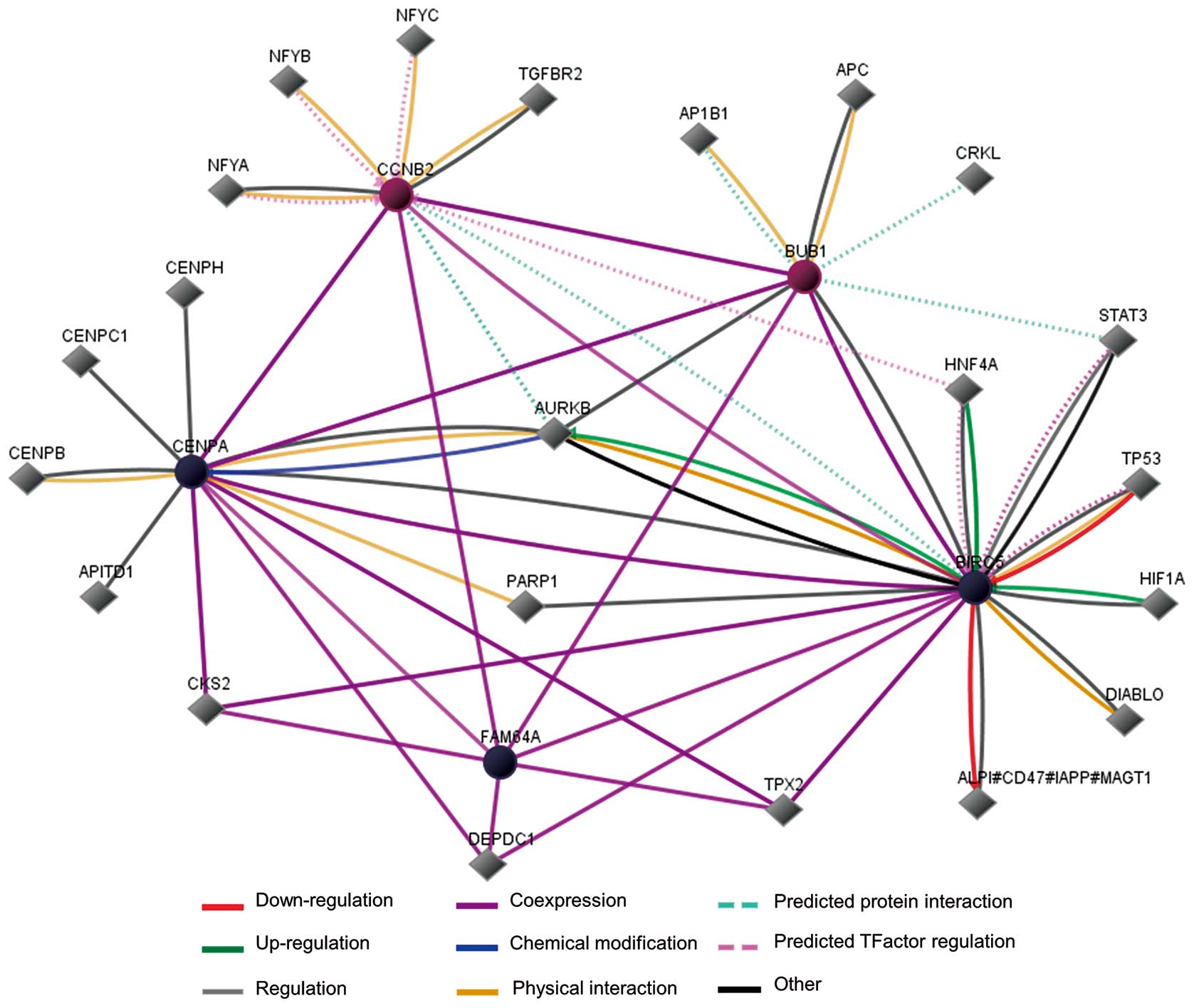
Pathway analysis indicated that BIRC5, CENPA and
FAM64A were co-expressed with BUB1 and CCNB2 (Fig. 5). Among the 5 genes, the function
of BIRC5, CENPA, BUB1 and CCNB2 have been well studied and proved
to be involved with cancer development, while FAM64A is a
relatively new molecule with fewer details from NCBI and a limited
number of reports. FAM64A was reported to be highly expressed in
cancer cell lines and associated with cancer cell cycle regulation
and proliferation. In addition, the aurora kinase B gene (aka
AURKB, whose protein product regulates chromosome segregation as a
serine/threonine kinase) on the map has been confirmed to link
BIRC5 and CENPA mechanistically, indicating that AURKB plays an
important role among our TNBC-specific genes (Fig. 5).
Discussion
Although endocrine treatment and targeted drugs
against breast cancer have achieved remarkable therapeutic
outcomes, the non-triple-negative subtype remains more aggressive
and difficult to treat (22,23).
The most distinctive difference between TNBC and NTNBC is the
status of ER, HER2 and PR (2),
which gives the impression that TNBC patient have a more similar
gene expression signature with healthy people. However, TNBC is
indeed one of the most malignant breast subtypes, there might be
some unknown molecules and pathways not yet identified. Current
therapeutic options of TNBC treatment are chemotherapy and some
auxiliary methods (anti-angiogenesis or anti-EGFR) (9), only a small number of previous
studies have provided analytical routes and possible gene
candidates for targeted treatment of TNBC. In this study, we aimed
to find some genes related to prognosis of TNBC by bioinformatic
analysis, and provide instructive information for the targeted
therapy of TNBC.
After analyzing five online breast cancer microarray
datasets, we found a cohort of 45 genes overexpressed in TNBC
samples comparing with NTNBC, and in the controls. We checked the
relationship between gene level and patient survival. Kaplan-Meier
analysis was performed for each gene in the TNBC cohort, and the
overexpression of BIRC5, CENPA and FAM64A were found to be
associated with poorer OS, DFS, DMFS and RFS. The relationship of
the hub genes with survival was also explored according to TNM
stage and NGS grade. Since the majority of samples in two clinical
sets were located in early or middle stage/grade, as a result, with
limited stage III–IV or grade 3 data, it is hard to judge whether
the hub genes were related to prognosis in advanced breast cancer.
However, this association was clearly seen in patients of stage
I–II or grade 1–2, proving that the three hub genes were related to
poor prognosis at least in early cancer period. Furthermore,
synchronous high expression of these three hub genes indicates the
worst clinical outcome while synchronous low level indicates the
best, emphasizing their contributions to poor prognosis. On the
other hand, two other genes, BUB1 and CCNB2, were also found to be
related to DFS, DMFS and RFS. A tendency of correlation with OS was
also found, though the log-rank p-value was >0.05. Cox model
analysis revealed that BIRC5 was more likely to be an independent
prognostic factor.
Since the expression of BIRC5, CENPA and FAM64A are
significantly upregulated in triple-negative breast cancer, we
consider that the overexpression of these genes may be a reason to
the high malignancy of TNBC, and they are potential prognostic
markers and therapeutic targets in anti-TNBC treatment. BUB1 and
CCNB2 may also play some roles influencing TNBC survival.
As the pathway network shows, our interested genes
are mainly involved with cell-cycle and mitosis. Among the hub
genes, BIRC5 (survivin) has been reported to be related with
unfavorable prognostic state (24,25)
in cancers of breast (25) and
neuroblastoma (26), but its role
with survival has not been linked to TNBC. CENPA is known to be a
key factor in centrosome functions, but the relationship with
breast cancer and prognosis is not well characterized yet.
According to its primary functions in mitosis (27), we speculate that CENPA influences
survival in cancer by promoting chromosomal segregation,
kinetochore association and genomic instability (28). FAM64A is a relatively new molecule,
only a few reports exist on its involvement with ovarian cancer
cell cycle and proliferation (29), therefore specific mechanisms and
pathways on this molecule merit investigation. As for BUB1 and
CCNB2, though the relationships with breast cancer has been
reported (30,31), their role in TNBC still needs to be
further addressed.
According to the interaction map drawn by us, AURKB
gene seems to be located at the center of the prognosis-related
gene network. This gene encodes a member of the aurora kinase
subfamily of serine/threonine kinases, which participate in the
regulation of segregation of chromosomes during mitosis and meiosis
through association with microtubules, and cancer as well (32). Since AURKB has been reported to be
a risk factor in lung and breast malignancy (33,34),
it is quite possible that AURKB participates in the regulation of
TNBC prognosis. We performed Pearson bivariate correlation analysis
for AURKB and the five genes we identified (BIRC5, CENPA, FAM64A,
BUB1 and CCNB2). SPSS calculation revealed that the Pearson
correlation coefficients between AURKB and each of the five genes
were all of statistical significance (data not shown). Exploration
of the association between AURKB and patient survival revealed that
the level of this gene was negatively correlated with survival
similarly to BIRC5, CENPA or FAM64A (data not shown). However,
since AURKB remains at a relatively even level among cancer
patients, it may play the role of a ‘passenger’ instead of a
‘driver’ in TNBC cases.
FAM64A is the substrate of the kinase-interacting
stathmin (KIS or UHMK1), a serine/threonine protein kinase that
promotes cell cycle progression through G1 by phosphorylation of
the cyclin-dependent kinase inhibitor 1B (p27Kip1)
(29). Since AURKB and BUB1 are
also serine/threonine kinases, we doubt whether FAM64A is a
substrate of AURKB or BUB1. It has been proven that BIRC5 can
upregulate AURKB, while the latter chemically modifies CENPA, so it
is possible that BIRC5 exerts the same influence on FAM64A via
AURKB or BUB1. We hypothesize that in TNBC patients, except causing
poor prognosis, BIRC5 can also promote FAM64A function via AURKB or
BUB1 mediated phosphorylation, thus bringing further impairment to
clinical outcome in TNBC.
Acknowledgements
We thank Tiantian She, Hongying Duan
and Ting Ma for helpful suggestions. This study was supported by
the National Natural Science Foundation of China (81071732).
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Dent R, Trudeau M, Pritchard KI, et al:
Triple-negative breast cancer: clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Rakha EA, El-Sayed ME, Green AR, Lee AH,
Robertson JF and Ellis IO: Prognostic markers in triple-negative
breast cancer. Cancer. 109:25–32. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Romond EH, Perez EA, Bryant J, et al:
Trastuzumab plus adjuvant chemotherapy for operable HER2-positive
breast cancer. N Engl J Med. 353:1673–1684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Mahamodhossen YA, Liu W and Rong-Rong Z:
Triple-negative breast cancer: new perspectives for novel
therapies. Med Oncol. 30:6532013. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Cleator S, Heller W and Coombes RC:
Triple-negative breast cancer: therapeutic options. Lancet Oncol.
8:235–244. 2007. View Article : Google Scholar
|
|
8.
|
Schneider BP, Winer EP, Foulkes WD, et al:
Triple-negative breast cancer: risk factors to potential targets.
Clin Cancer Res. 14:8010–8018. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Bosch A, Eroles P, Zaragoza R, Viña JR and
Lluch A: Triple-negative breast cancer: molecular features,
pathogenesis, treatment and current lines of research. Cancer Treat
Rev. 36:206–215. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Balko JM, Cook RS, Vaught DB, et al:
Profiling of residual breast cancers after neoadjuvant chemotherapy
identifies DUSP4 deficiency as a mechanism of drug resistance. Nat
Med. 18:1052–1059. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ur-Rehman S, Gao Q, Mitsopoulos C and
Zvelebil M: ROCK: a resource for integrative breast cancer data
analysis. Breast Cancer Res Treat. 139:907–921. 2013.PubMed/NCBI
|
|
12.
|
van de Vijver MJ, He YD, van’t Veer LJ, et
al: A gene-expression signature as a predictor of survival in
breast cancer. N Engl J Med. 347:1999–2009. 2002.
|
|
13.
|
Chin K, DeVries S, Fridlyand J, et al:
Genomic and transcriptional aberrations linked to breast cancer
pathophysiologies. Cancer Cell. 10:529–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Butte A: The use and analysis of
microarray data. Nat Rev Drug Discov. 1:951–960. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Han Y, Huang H, Xiao Z, et al: Integrated
analysis of gene expression profiles associated with response of
platinum/paclitaxel-based treatment in epithelial ovarian cancer.
PLoS One. 7:e527452012. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Konstantinopoulos PA, Cannistra SA,
Fountzilas H, et al: Integrated analysis of multiple microarray
datasets identifies a reproducible survival predictor in ovarian
cancer. PLoS One. 6:e182022011. View Article : Google Scholar
|
|
17.
|
Richardson AL, Wang ZC, De Nicolo A, et
al: X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Brown MP, Grundy WN, Lin D, et al:
Knowledge-based analysis of microarray gene expression data by
using support vector machines. Proc Natl Acad Sci USA. 97:262–267.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Perreard L, Fan C, Quackenbush JF, et al:
Classification and risk stratification of invasive breast
carcinomas using a real-time quantitative RT-PCR assay. Breast
Cancer Res. 8:R232006. View
Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Hu Z, Fan C, Oh DS, et al: The molecular
portraits of breast tumors are conserved across microarray
platforms. BMC Genomics. 7:962006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Gluz O, Liedtke C, Gottschalk N, Pusztai
L, Nitz U and Harbeck N: Triple-negative breast cancer - current
status and future directions. Ann Oncol. 20:1913–1927. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Rouzier R, Perou CM, Symmans WF, et al:
Breast cancer molecular subtypes respond differently to
preoperative chemotherapy. Clin Cancer Res. 11:5678–5685. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Vogel CL, Cobleigh MA, Tripathy D, et al:
Efficacy and safety of trastuzumab as a single agent in first-line
treatment of HER2-overexpressing metastatic breast cancer. J Clin
Oncol. 20:719–726. 2002. View Article : Google Scholar
|
|
24.
|
Hinnis AR, Luckett JC and Walker RA:
Survivin is an independent predictor of short-term survival in poor
prognostic breast cancer patients. Br J Cancer. 96:639–645. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Span PN, Tjan-Heijnen VC, Heuvel JJ, de
Kok JB, Foekens JA and Sweep FC: Do the survivin (BIRC5) splice
variants modulate or add to the prognostic value of total survivin
in breast cancer? Clin Chem. 52:1693–1700. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Islam A, Kageyama H, Takada N, et al: High
expression of Survivin, mapped to 17q25, is significantly
associated with poor prognostic factors and promotes cell survival
in human neuroblastoma. Oncogene. 19:617–623. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Regnier V, Vagnarelli P, Fukagawa T, et
al: CENP-A is required for accurate chromosome segregation and
sustained kinetochore association of BubR1. Mol Cell Biol.
25:3967–3981. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Amato A, Schillaci T, Lentini L and Di
Leonardo A: CENPA overexpression promotes genome instability in
pRb-depleted human cells. Mol Cancer. 8:1192009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Archangelo LF, Greif PA, Maucuer A, et al:
The CATS (FAM64A) protein is a substrate of the Kinase Interacting
Stathmin (KIS). Biochim Biophys Acta. 1833:1269–1279. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Takagi K, Miki Y, Shibahara Y, et al: BUB1
immunolocalization in breast carcinoma: its nuclear localization as
a potent prognostic factor of the patients. Horm Cancer. 4:92–102.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Li L, Xu DB, Zhao XL and Hao TY:
Combination analysis of Bub1 and Mad2 expression in endometrial
cancer: act as a prognostic factor in endometrial cancer. Arch
Gynecol Obstet. 288:155–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Hegyi K and Mehes G: Mitotic failures in
cancer: Aurora B kinase and its potential role in the development
of aneuploidy. Pathol Oncol Res. 18:761–769. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Takeshita M, Koga T, Takayama K, et al:
Aurora-B overexpression is correlated with aneuploidy and poor
prognosis in non-small cell lung cancer. Lung Cancer. 80:85–90.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Tchatchou S, Wirtenberger M, Hemminki K,
et al: Aurora kinases A and B and familial breast cancer risk.
Cancer Lett. 247:266–272. 2007. View Article : Google Scholar : PubMed/NCBI
|