Introduction
Prostate cancer (PCa) is the second leading cause of
male cancer-related death in the United States, with 238,590
estimated new cases that occurred in 2013 and almost 29,720 deaths
(1). Localized PCa patients have a
long-term survival due to the combination of radiation and androgen
deprivation therapy (2). However,
clinical observation shows that after an initial responsiveness to
androgen withdrawal treatment, almost all PCa will inevitably
progress to recurrent castration-resistant prostate cancer (CRPC),
and acquires the potential of metastasis, for which few therapeutic
options with limited durability are available (3). The underlying mechanisms during the
transition from androgen-dependent (AD) to androgen-independent
(AI), from localized status to metastasis, remain to be elucidated.
Identification of genes involved in this transition may provide
insight into finding novel therapeutic strategies for CRPC.
Epidermal growth factor receptor (EGFR) is a member
of the human epidermal growth factor receptor or ErbB family of
receptor tyrosine kinases (4).
EGFR is overexpressed in 40–80% of prostate cancer cells, and
overexpression more common in African American men with prostate
cancer (5). Furthermore, previous
clinical studies suggested a correlation of EGFR expression with
androgen-independence (6). More
importantly, EGFR itself may be under the regulation of androgen
signaling pathway, being negative in normal prostate cells but
positive in prostate cancer cells, especially in
androgen-independent cancer cells (7). In an effort to overcome
castration-resistance, trials combining EGFR or dual kinase
inhibitors with other novel agents are in development (8).
Emerging evidence indicates that LncRNAs which are
RNA specis >200 bp in length (9) and frequently polyadenylated (10), are involved in physiological
aspects of cell-type determination and tissue homeostasis (11), and in cancer lncRNAs are known to
play important roles in carcinogenesis and tumor progression
(12–16). One of the overexpressed lncRNAs in
prostate cancer, PCGEM1, is tissue specific and PCa-associated
lncRNA gene (15), whereas another
highly expressed lncRNA, PRNCR1 (PCAT8), is pervasively transcribed
from the critical region of 8q24 Region 2, which is significantly
associated with PCa susceptibility (17). Previous studies also demonstrated
that a novel long non-coding RNA (named PlncRNA-1) was frequently
overexpressed in PCa cell lines and tissues and associated with
cell viability and cell apoptosis (18). Recent studies have revealed the
contribution of other lncRNAs as proto-oncogenes, drivers of
metastatic transformation and tumor suppressor genes in prostate
cancer, such as prostate cancer gene 3 (PCA3), metastasis
associated in lung adenocarcinoma transcript 1 (MALAT-1), and
PCAT-1 (13,19–21).
However, the global expression profile of lncRNAs in
androgen-dependent (AD) to androgen-independent (AI) prostate
cancer is not fully uncovered.
Prostate cancer cell line LNCaP and C4-2 have the
same genetic background and the unique advantage of remarkably
mimicking the phenotypic and genotypic changes observed in clinical
human PCa (22). In the current
study, we used micro-array technology to compare the lncRNA and
mRNA expression between LNCaP cell line which was an
androgen-dependent, non-metastasis (23) and the lineage-related C4-2 cell
line which acquired characteristic of androgen-independence and
possessed the capacity of metastasizing to lymph nodes and bone
(24). As a result, 257
PCa-associated lncRNA transcripts were identified to be differently
expressed in LNCaP and C4-2, including one well-known lncRNA,
PlncRNA-1 (18). Among them, we
further identified long intergenic non-protein coding RNA 963
(Linc00963) as the lncRNA with the most significantly different
expression in LNCap and C4-2 cell lines using real-time PCR.
Additionally, we identified EGFR as the putative target molecule by
bioinformatics prediction. We further showed that suppression of
Linc00963 by siRNA could reduce the capacity of cell viability and
invasion and the expression of EGFR in C4-2 cells in
vitro.
In the present study we demonstrated for the first
time that Linc00963 was significantly associated with the capacity
of cell metastasis in prostate cancer cells, and EGFR was the
putative target molecule of Linc00963. Taken together, our data
suggested that Linc00963 plays an important role in the transition
from AD to AI, and it could be a useful therapeutic target to
prevent PCa metastasis.
Materials and methods
Cell culture
LNCaP and C4-2 cell lines were cultured in RPMI-1640
(Invitrogen, Carlsbad, CA, USA) supplemented with 8% fetal bovine
serum (FBS; Hyclone, Logan, UT, USA), 10 mM HEPES, 1.0 mM sodium
bicarbonate and 1% antibiotic/antimycotic solutions. All the cells
were cultured at 37°C in a humidified atmosphere of 5%
CO2.
RNA extraction and microarray target
preparation
Total RNA was extracted from C4-2 and LNCaP cell
lines using TRIzol reagent (Invitrogen) according to the
manufacturers’ protocols. RNA cleanup including a DNase I digestion
step was performed using RNeasy spin columns (Qiagen). RNA
integrity was measured by the relative abundance of 28S/18S
ribosomal subunits, verified through micro-fluid capillary
electrophoresis (Agilent Bioanalyzer 2100).
Microarray analysis
For microarray analysis, qualified total RNA was
further purified by RNeasy mini kit (Qiagen) and RNase-free DNase
set (Qiagen). Total RNA was then amplified and labeled by Low Input
Quick Amp Labeling kit (Agilent), following the manufacturer’s
instructions. Labeled cRNA were purified by Qiagen
RNeasy® mini kit. Each Slide was hybridized with 600 ng
Cy3/Cy5-labeled cRNA using Gene Expression Hybridization kit
(Agilent) in Hybridization Oven (Agilent), according to the
manufacturer’s instructions. After 17 h of hybridization, slides
were washed in staining dishes (Thermo Shandon) with Gene
Expression Wash Buffer kit (Agilent). Slides were scanned with
Agilent C Scanner Settings, Dye channel: Green, scan resolution = 3
μm, 20 bit; Red, scan resolution = 5 μm, 20 bit. Data
were extracted with Feature Extraction software 10.7 (Agilent). Raw
data were normalized by Quantile algorithm, Gene Spring Software
11.0 (Agilent). Hierarchical clustering was performed based on
differentially expressed mRNAs and lncRNAs using Cluster_Treeview
software from Stanford University.
Transfection and gene silencing
For small interfering RNA (siRNA) transfection, the
following siRNA duplexes were synthesized (Genepharma, Shanghai,
China): si-Linc00963-1 (5′-GGCAAGUGCUUUCAACUCUTT-3′), and
si-Linc00963-2 (5′-GCUCACUGAACUUUCUGAATT-3′), targeting the
Linc00963 gene, and the negative control duplex, (5′-UUC
UCCGAACGUGUCACGUTT-3′). These siRNA duplexes (100 nmol/l) were
transfected into C4-2 cells using Lipofectamine 2000 (Invitrogen)
according to the manufacturer’s instructions. C4-2 cells were
harvested 48 h post-transfection for gene analysis.
Real-time quantitative
reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cell lines using TRIzol
reagent (Invitrogen), and subsequent synthesis of cyclic DNA (cDNA;
Takara, Japan), were carried out according to the manufacturers’
protocols. qRT-PCR was performed using the CFX96TM
Real-time PCR system (Bio-Rad, Hercules, CA, USA) with the SYBR
Green II kit (#DRR041A; Takara) according to the manu facturer’s
instructions. qRT-PCR analysis was carried out in a total volume of
20 μl with the following amplification steps: an initial
denaturation step at 95°C for 10 min; followed by 40 cycles of
denaturation at 95°C for 30 sec; and then elongation at 55°C for 30
sec. The expressions were normalized to the human β-actin gene. The
following primer sequences were used: 5′-GGTAAATCGAGGCCCAGAGAT-3′
(sense) and 5′-ACGTGGATGACAGCGTGTGA-3′ (antisense) for Linc00963;
5′-CGTCTTCCCCTCCATCGT-3′ (sense) and 5′-GAAGGTGTGGTGCCAGATTT-3′
(antisense) for β-actin.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell proliferation in vitro was analyzed with
MTT proliferation assay. The yellow dye MTT is reduced to a blue
formazan product by respiratory enzymes that are active only in
viable cells, making the amount of color change indicative of cell
proliferation. C4-2 cells were transfected with no siRNA
(parental), specific siRNA (si-Linc00963-1, or si-Linc00963-2) and
control siRNA (si-scramble) for 24 h and suspended in RPMI-1640
with 10% FBS. Briefly, 2000–3000 cells of each clone (parental,
si-scramble, si-Linc00963-1 or si-Linc00963-2) were plated per well
in five 96-well plates in 200 μl of medium. For analysis, 20
μl of MTT substrate of a 2.5 mg/ml stock solution in
phosphate-buffered saline (PBS) was added to each well, and the
cells were incubated for a further 4 h at 37°C. The medium was
removed, the cells were solubilized in 150 μl of
dimethylsulfoxide, and colorimetric analysis was performed
(wavelength, 490 nm). One plate was analyzed immediately after the
cells adhered (∼4 h after plating), and the remaining plates were
assayed every day for the next 4 consecutive days.
Flow cytometric analysis of apoptotic
cells
C4-2 cells were transfected for 48 h with no siRNA
(parental), specific siRNA (si-Linc00963-1 or si-Linc00963-2) and
control siRNA (si-scramble), then cells were suspended in
incubation buffer at a density of 1×106 cells/ml.
Apoptotic cells were analyzed by flow cytometry using a CYTOMICS FC
500 flow cyto-meter (Beckman Coulter), after incubating the cells
with a reagent containing Annexin V-FITC and Propidium Iodide (BD
Bioscience, CA, USA) for 15 min in darkness at room
temperature.
Analysis of invasiveness and mobility
(migration and invasion assays)
Cell invasion and migration potentials were measured
by Transwell assays (Millipore, Billerica, MA, USA). C4-2 cells
were transfected for 24 h with no siRNA (parental), specific siRNA
(si-Linc00963-1 or si-Linc00963-2) and control siRNA (si-scramble);
the cells were suspended in RPMI-1640 with 8 g/l BSA, 10 mM HEPES,
1.0 mM sodium bicarbonate at a density of 50 cells/μl; 200
μl cell suspensions were seeded into the upper chambers of
the Transwells whose porous membrane was coated with (for Transwell
invasion assay) or without (for migration assay) Matrigel (BD
Bioscience). RPMI-1640 (500 μl) with 8% FBS, 10 mM HEPES,
1.0 mM sodium bicarbonate was added to the bottom chamber as a
chemoattractant. After migration for 24 h, or invasion for 48 h,
the cells that had penetrated the filters were fixed in methanol,
and stained in 4 g/l crystal violet. The numbers of migrated and
invasive cells were determined from five random fields under an
Olympus microscope (Olympus) at ×10 magnification.
Western blot analysis
C4-2 cells which transfected for 48 h with no siRNA
(parental), specific siRNA (si-Linc00963-1, or si-Linc00963-2) and
control siRNA (si-scramble) were harvested in
radioimmunoprecipitation (RIPA) buffer. Protein concentration was
determined using the BCA protein assay. Proteins were resolved
using 10% SDS-PAGE gradient gels, and 30 mg/well was loaded.
Proteins were transferred electrophoretically to PVDF membrane
(Bio-Rad) at 25 V for 2 h. The membrane was blocked 2 h at room
temperature in PBS containing 5% nonfat dry milk. Antibodies AKT
and p-AKT (Bioworld Technology) were diluted in PBS-T (0.1%
Tween-20, Fisher) at 1:1000 working concentration, incubated
overnight at 4°C. The second antibody was HRP-conjugated
anti-rabbit, at 1:5000 in PBS-T, and incubated for 2 h at room
temperature. Following this, and all other incubations, membranes
were washed in PBS-T 3×5 min. HRP activity was detected using ECL
western blotting detection reagents. The bands were visualized by
chemiluminescence (New England Nuclear, Boston, MA, USA).
Statistical analysis
All statistical data were analyzed by Statistical
Program for Social Sciences (SPSS) 20.0 software (SPSS 20.0
software, IBM, USA) and GraphPad Prism 5.0 (GraphPad Software, La
Jolla, CA, USA). One-way analysis of variance test, two-tailed
Student’s t-test and rank-sum test were used as appropriate.
P<0.05 was considered statistically significant.
Results
Identification of the LncRNAs
differentially expressed in the LNCaP and C4-2 cell lines
In order to gain better understanding of the
androgen-independent and metastasis progression of prostate cancer
at the lncRNA level, we generated a comprehensive lncRNA expression
profile for LNCaP and C4-2. Total RNA was extracted from the cell
lines and analyzed using microarray from Aglient to characterize
the expression patterns of lncRNAs. In addition, the microarray
analyses were conducted three times. The relative expression of
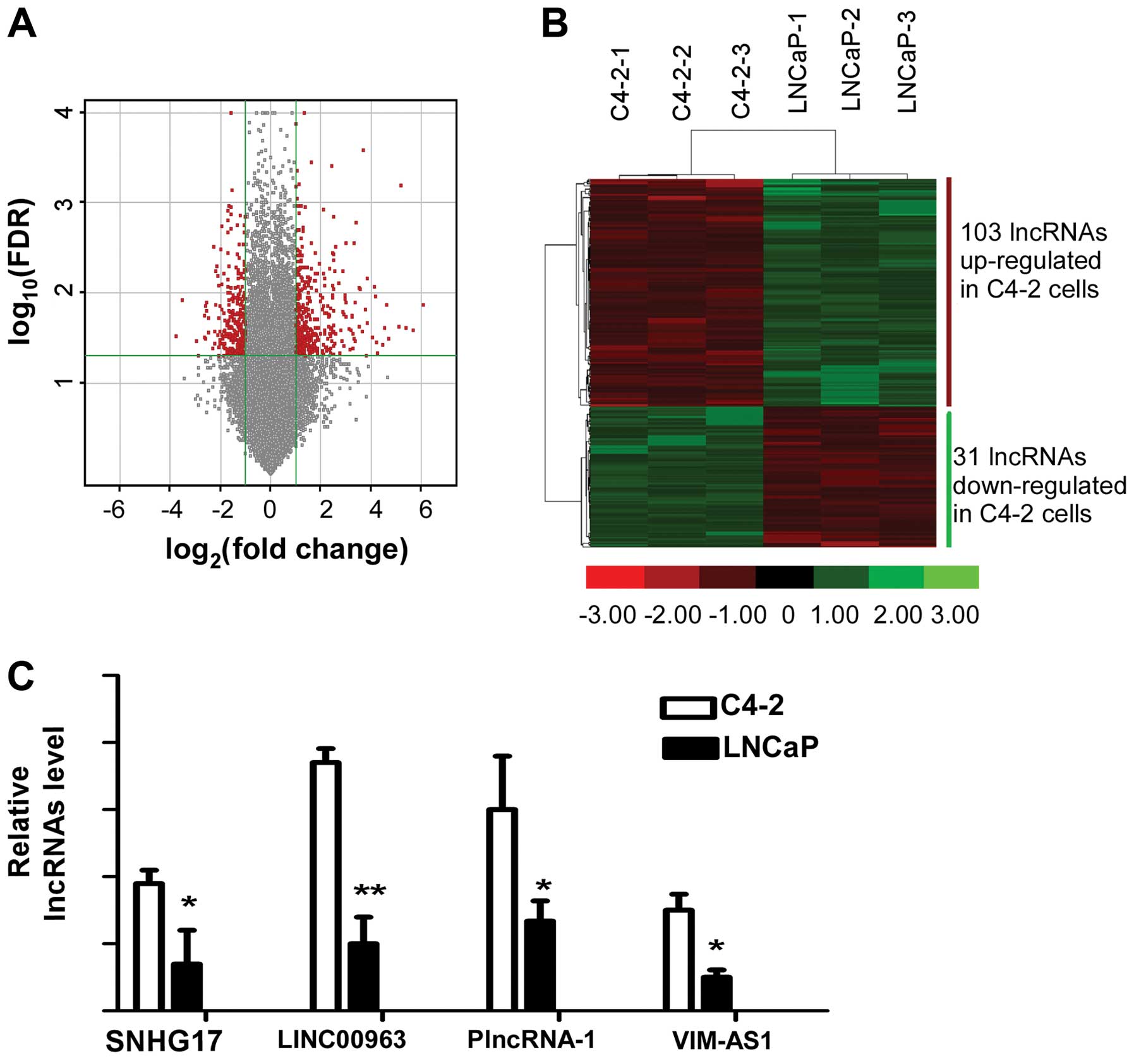
lncRNAs expressed by LNCaP and C4-2 is shown in Fig. 1A, representatively. We picked out
134 lncRNAs which were differentially expressed (FDR <0.001 and
fold change ≥2) at three times between LNCap and C4-2 from 46506
lincRNAs represented on the chips. From the 134 lncRNAs, we
identified 4 lncRNAs which were differentially expressed in LNCaP
and C4-2 to the most significant extent (Fig. 1B), including the well-known lncRNA
in prostate cancer, PlncRNA-1. Using RNA isolated from LNCaP and
C4-2, we performed qRT-PCR to validate the expression level of the
four identified lncRNAs. We found that lncRNA Linc00963 was
expressed differentially most significantly compared to the others
in LNCaP and C4-2 cell lines (Fig.
1C). As LNCaP is a hormone-sensitive but C4-2 was
hormone-insensitive prostate cell line derived from LNCaP, the
lncRNAs differentially expressed in LNCaP and C4-2 may be involved
in the transition from AD to AI.
Effect of Linc00963 knockdown on cell
viability
C4-2 is a hormone-insensitive prostate cell line and
possesses the capacity of metastasizing to lymph node and bone, and
our preliminary results indicated that lncRNA Linc00963 was
differentially expressed in C4-2 and LNCaP significantly.
Therefore, in order to investigate biological function of Linc00963
in PCa cell line C4-2, Linc00963 was suppressed by siRNAs in C4-2.
The effective knockdown of Linc00963 was confirmed by qRT-PCR.
Compared to cells transfected with scrambled siRNA, cells
transfected with siRNAs to Linc00963 showed significantly reduced
Linc00963 expression (each experiment was performed three times,
and a typical result is presented in Fig. 2A. After confirming the knockdown
efficiency of the siRNAs targeting Linc00963, we determined the
effect of a reduced Linc00963 level on cell viability using an MTT
assay. C4-2 cells that were transfected with siRNAs targeting
Linc00963 showed significant decrease in cell viability compared to
the parental or scrambled siRNA-transfected cells (Fig. 2B). This result demonstrated that
the Linc00963 had a direct effect on cell viability in C4-2
cells.
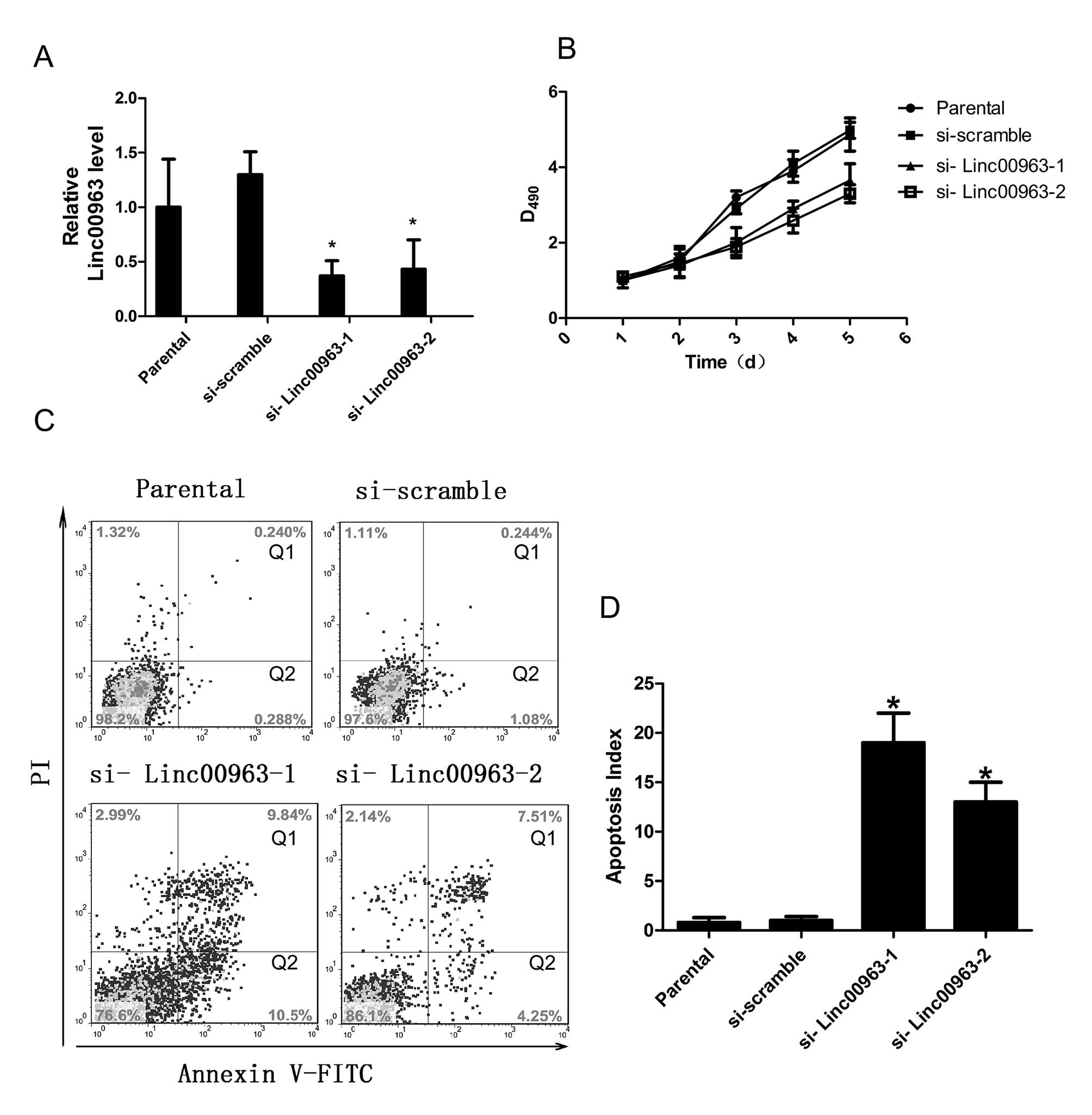 | Figure 2.The Linc00963 knockdown affects cell
viability. (A) QRT-PCR analysis of cell lines with reduced
Linc00963 level after transfection with siRNAs: parental C4-2 cells
carrying no siRNA (parental); scramble siRNA (si-scramble); siRNA
to Linc00963 (si-Linc00963-1, si-Linc00963-2). Compared to cells
transfected with scrambled siRNA, cells transfected with siRNAs
targeting Linc00963 showed significantly reduced Linc00963
expression. (B) MTT cell growth assays of control cells (parental,
si-scramble); cells with reduced Linc00963 expression
(si-Linc00963-1, si-Linc00963-2). The results show that cells
transfected with siRNAs targeting Linc00963 showed significant
decrease in cell viability. Data are presented as mean ± SD, N=3,
P<0.05. (C and D) Flow cytometric analyses of cells stained with
Annexin V-FITC and PI: cells carrying no siRNA (parental), scramble
siRNA (si-scramble), siRNA targeting Linc00963 (si-Linc00963-1,
si-Linc00963-2). Data are presented as mean ± SD, N=3,
*P<0.05. The results indicate that silencing
Linc00963 expression increased cell apoptosis. |
Effect of the Linc00963 knockdown on cell
apoptosis
To explore the mechanism by which Linc00963 affected
cell vitality of C4-2, we tested whether the inhibited cell
viability may be caused by increased cell apoptosis in Linc00963
knockdown cells. C4-2 cells transfected with scramble siRNA or
siRNAs targeting Linc00963 for 48 h were analyzed for apoptosis.
The results indicated that compared with the parental or scrambled
siRNA-transfected cells, cells transfected with siRNAs targeting
Linc00963 had increased apoptosis index which was calculated by
adding the cells in the Q1 and the cells in the Q2 (15.1±4.8% and
13.2±5.2% vs. 1.07±0.8%, P<0.05; 15.1±4.8% and 13.2±5.2% vs.
0.89±0.75%, P<0.05; each experiment was performed three times,
and a typical result is presented as Fig. 2C and D. Collectively, these results
suggested that the expression of Linc00963 in C4-2 cells was
important for both cell viability and apoptosis.
Effect of the Linc00963 knockdown on cell
migration and invasion
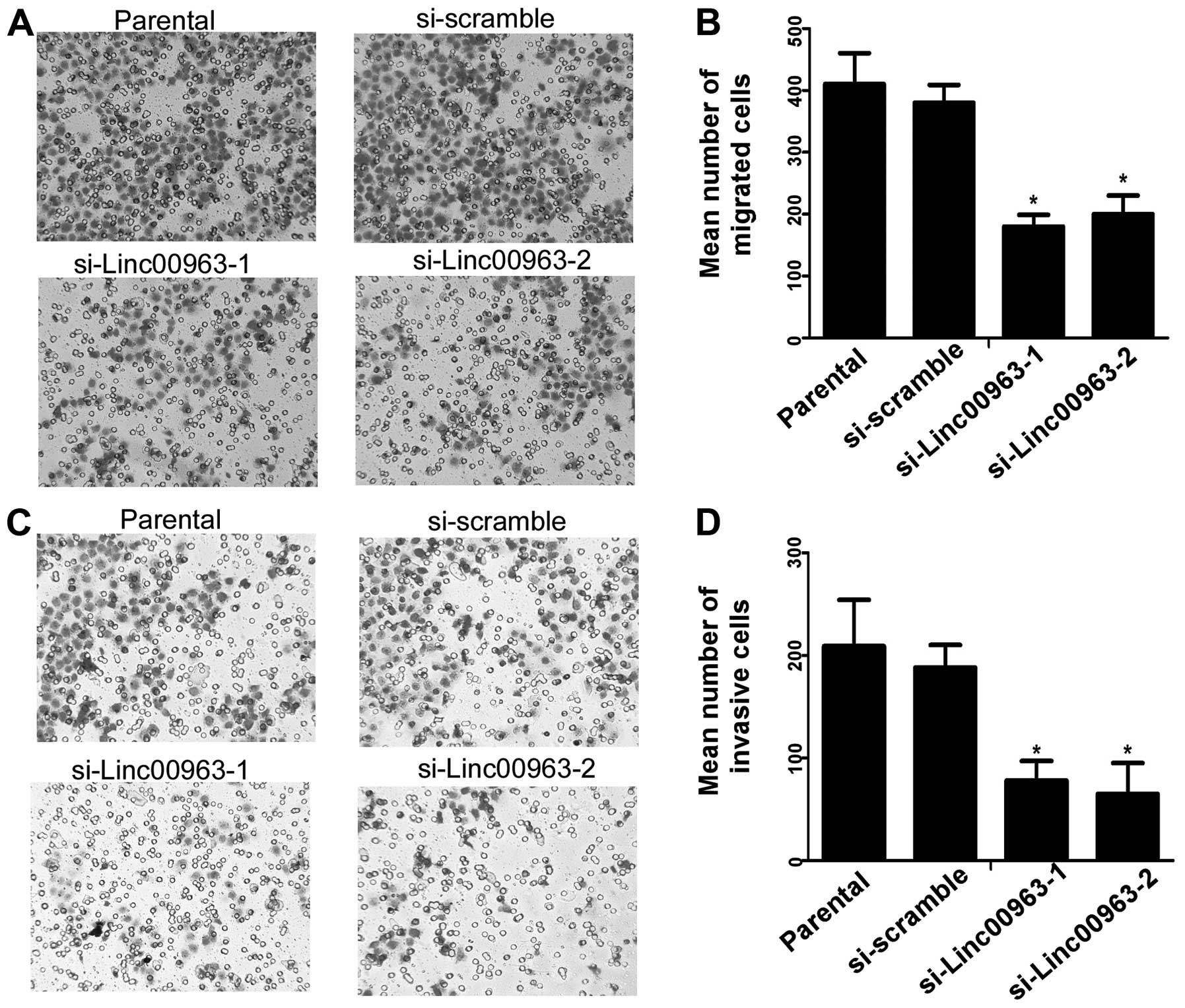
To test the effects of linc00963 on migration and
invasion of C4-2, we used standard Matrigel-coated or uncoated
transwell chamber assays. We found that compared with the scrambled
siRNA-transfected cells, C4-2 cells transfected with siRNAs
targeting Linc00963 had reduced migration and invasion ability
(Fig. 3A and C), and a reduced
invasive index (invasion cell number/migration cell number,
Fig. 3B and D). Thus, our results
indicated that Linc00963 was correlated with cell migration and
invasion in prostate cancer cell line C4-2.
Identification of the target molecule for
Linc00963 in prostate cancer cell lines
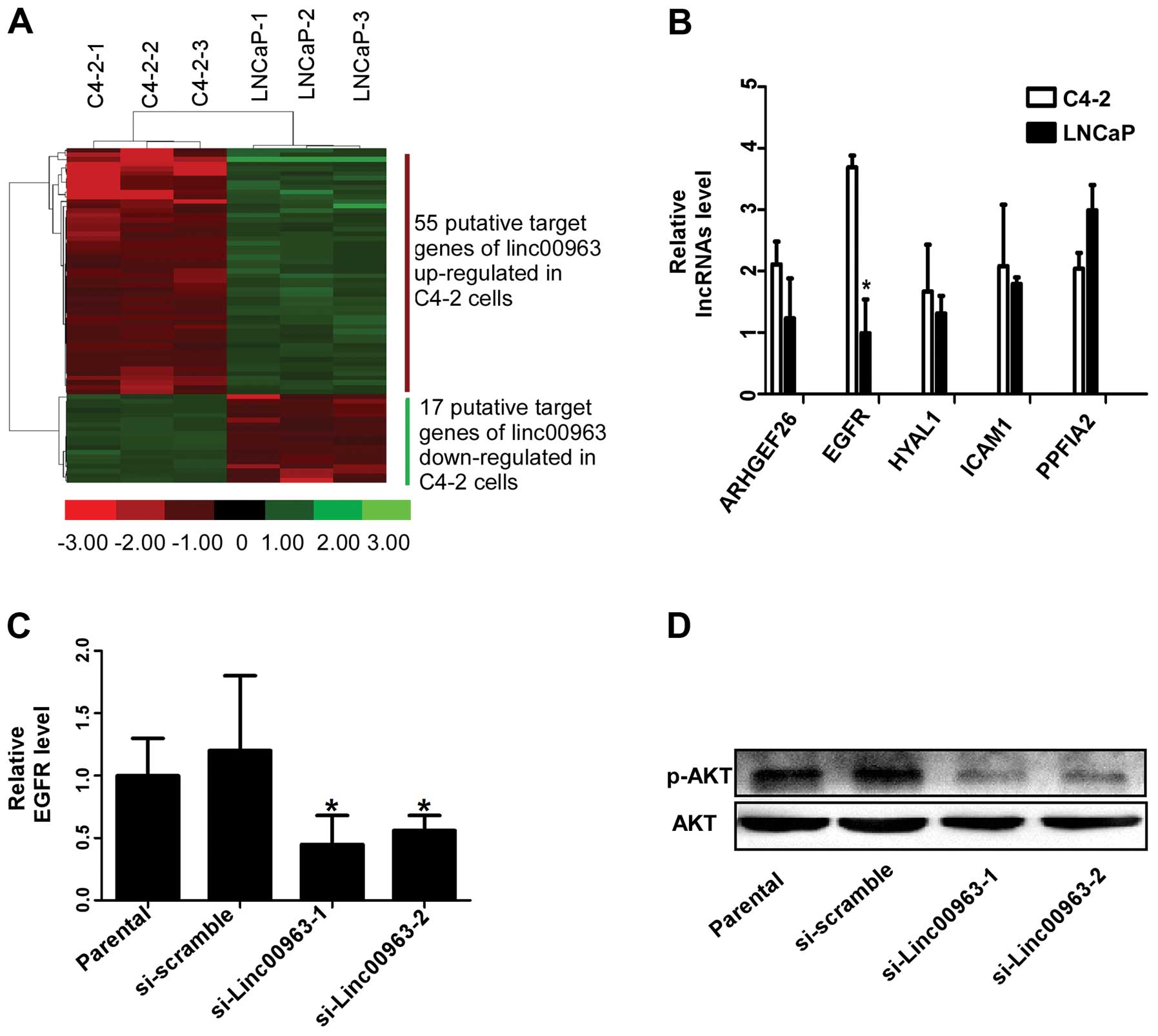
To understand the biological function of Linc00963,
the putative target sites were identified by 3 steps. Step 1: we
sought to determine whether Linc00963 act in cis or in
trans to regulate target gene expression. We use genome
annotation, genome browser and RNAplex to find these putative
targets (Fig. 4A). Step 2:
unsupervised hierarchical clustering was used to analyze the
differential mRNA expression profiles. Among these candidate
targets, we identified 5 genes, ARHGEF26, EGFR, HYAL1, ICAM1 and
PPFIA2 as the putative targets, which were differentially expressed
in C4-2 and LNCaP cells to the most significant extent and may be
regulated by Linc00963 through transcriptional interference
(Table I). Step 3: after
identification of these targets, we further validated our results
in the prostate cell lines LNCaP and C4-2. From the results of
qRT-PCR, we found EGFR was expressed most differentially in LNCaP
and C4-2 (Fig. 4B). To investigate
whether EGFR was the putative target for Linc00963, we performed
qRT-PCR to examine the expression level of EGFR in C4-2 cells
transfected with Linc00963 siRNAs. In addition, we tested the
phosphorylation level of its downstream molecule AKT which has been
proven to correlate with castration resistant cell growth and
androgen receptor level in CRPC (25). Our results showed knockdown of
Linc00963 significantly decreases EGFR, and the phosphorylation
level of AKT (Fig. 4C and D),
indicating Linc00963 involved in the transactivation of EGFR in
hormone-insensitive prostate cancer cells. In conclusion, our
result indicated that EGFR was the target molecule of Linc00963 in
prostate cancer cells.
 | Table I.The potential target molecular for
Linc00963 in prostate cancer cell lines. |
Table I.
The potential target molecular for
Linc00963 in prostate cancer cell lines.
| Gene name | Description | Fold change
expression C4-2/LNCaP |
|---|
| ATG9B | Autophagy related
9B | 3.123 |
| ADAMTS1 | ADAM metallopeptidase
with thrombospondin type 1 motif, 1 | 3.735 |
| ANKRD18A | Ankyrin repeat domain
18A | 3.178 |
| ARHGEF26 | Rho guanine
nucleotide exchange factor (GEF) 26 | 4.416 |
| AZGP1 | α-2-glycoprotein 1,
zinc-binding | 1/3.576 |
| GPR158 | G protein-coupled
receptor 158 | 3.186 |
| HES1 | Hes family bHLH
transcription factor 1 | 1/3.314 |
| HYAL1 |
Hyaluronoglucosaminidase 1 | 5.643 |
| ICAM1 | Intercellular
adhesion molecule 1 | 4.176 |
| KLK2 | Kallikrein-related
peptidase 2 | 1/3.591 |
| KLK3 | Kallikrein-related
peptidase 3, prostate specific antigen | 1/3.332 |
| PIK3R1 |
Phosphoinositide-3-kinase, regulatory
subunit 1 (α) | 3.611 |
| PPFIA2 | Protein tyrosine
phosphatase, receptor type, f polypeptide (PTPRF), interacting
protein (liprin), α2 | 1/4.872 |
| PRSS2 | Protease, serine, 2
(trypsin 2) | 3.331 |
| S100A10 | S100 calcium
binding protein A10 | 1/3.012 |
| Nrp1 | Neuropilin 1 | 3.277 |
| NUCB2 | Nucleobindin 2 | 3.638 |
| MYLK | Myosin light chain
kinase | 1/3.547 |
| EGFR | Epidermal growth
factor receptor | 5.347 |
| ELF3 | E74-like factor 3
(ets domain transcription factor, epithelial-specific) | 1/3.326 |
| ELOVL5 | ELOVL fatty acid
elongase 5 | 3.936 |
| GDF11 | Growth
differentiation factor 11 | 1/3.176 |
| GNAI1 | Guanine nucleotide
binding protein (G protein), α inhibiting activity polypeptide
1 | 1/3.195 |
| GAS6 | Growth
arrest-specific 6 | 3.079 |
Discussion
Progression to androgen resistance and metastasis of
PCa, involve alterations in gene expression and dysregulation of
signaling pathways, and remains both an intensive and elusive area
of investigation. In addition to protein coding genes and miRNAs,
dysregulatory of lncRNAs is emerging as a ubiquitous component in
the gene regulatory networks of cancer progression (11,26).
In the current study, for the first time, lncRNA Linc00963
(GeneBank accession ID: 100506190) is found to be involved in the
progression from AD to AI of prostate cancer. We characterized
Linc00963, which is more frequently overexpressed in
hormone-insensitive prostate cell line C4-2 than hormone-sensitive
prostate cell line LNCaP, and its overexpression correlated with
cell viability, cell apoptosis, cell migration and cell invasion of
C4-2, suggesting an important role of Linc00963 in the transition
from AD to AI.
Previous studies showed that lincRNAs could act in
cis (on neighboring genes) by transcriptional interference
and/or function in trans (on distant located genes) though
targeting epigenetic modifiers (27,28).
We identified that Linc00963 and EGFR were located in different
chromosomes and Linc00963 had a functional relationship with EGFR,
so we supposed Lin00963 might act in trans with EGFR. We
further confirmed that EGFR and the phosphorylation level of its
downstream gene AKT decreased following knockdown of Linc00963.
These results demonstrated that EGFR was the putative target
molecule of Linc00963 and Linc00963 involved in the transactivation
activity of EGFR in prostate cancer cells. However, further studies
are required to clarify the molecular mechanism underlying the
regulation of Linc00963 and EGFR.
LncRNAs are being recognized at every level of gene
expression in various physiological processes, and alteration of
the expression of lncRNAs in cancer is considered as one of the
main driving forces during tumorigenesis (26,29,30).
In prostate cancer, the function of lncRNAs is more complex than
previously believed. More and more lncRNAs were found dysregulated
in prostate cancer, and most of them exhibit oncogenic function,
including prostate cancer antigen 3 (PCA3) (31), PCGEM1, PRNCR1, MALAT-1, PlncRNA-1
(32), and CTBP1-AS (33). Most of these identified lncRNAs was
proven to be associated with androgen receptor (AR), but no
previous studies concerning lncRNAs paid attention to the mechanism
that would account for the transition from AD to AI. Thus, we used
microarray technologies to delineate the differential expression
profiles of cancer-related lncRNAs in hormone-sensitive and
hormone-insensitive prostate cancer cell lines. RNA-seq revealed
that 134 lncRNAs were expressed differentially in LNCaP and C4-2,
and Linc00963 was upregulated to the most significant extent in
C4-2 among these dysregulated lncRNAs. These results led us to
believe that Linc00963 is potentially involved in the progeression
of AD PCa to the lethal AI phenotype. To our knowledge, this is the
first study using microarray technologies to delineate the lncRNA
profiles between LNCaP and C4-2 cells.
Castration-resistant prostate cancer (CRPC) tends to
progress and metastasize, and shows short survival. The prostate
cancer cell line C4-2 acquires the phenotypes of
androgen-independence and osseous metastases, and it is commonly
used in models of castration-resistant and aggressive prostate
cancer (22). The second part of
our study identified the role of Linc00963 in AI prostate cancer
cells. Our results indicated that Linc00963 is significantly
associated with the cell viability, cell motility and cell
invasiveness. Although a more detailed mechanism must be discovered
to explain our results, these results provide evidence that
Linc00963 may function as an oncogenic molecule and linc0096 is a
ubiquitous component in the gene regulatory networks of prostate
cancer development and progression. Though previous studies had
identified some lncRNAs aberrantly expressed exhibiting oncogenic
function in PCa, such as inhibiting apoptosis or promoting cell
proliferation (10,16,17,21),
no direct evidence was provided indicating the role of lncRNAs in
AI prostate cancer development. Our lncRNA expression profiling
results and the effect of Linc00963 on tumor metastasis in LNCaP
and C4-2 partially illustrate the role lnRNAs played in the
transition from AD to AI and the progress of AI prostate cancer
metastasis.
The third part of our study identified EGFR as a
putatively functional target of Linc00963 in the
hormone-insensitive prostate cancer cell line C4-2. Enhanced
expression of EGFR/ErbB1 had been proven to correlate with high
grades of prostate cancer malignancies and contribute to the
progression from localized and AD prostate cancer to more
metastatic and AI state (34,35),
and the phosphorylation level of AKT which was the downstream gene
of EGFR had been proven to correlate with androgen receptor level
and promote castration- resistant cell growth (25). Importantly, AR was observed to be
slightly downregulated in C4-2 (36). Taken together, all these studies
indicated EGFR might play a more important role in the process from
AD to AI than AR. However, most of the well characterized prostate
cancer-related lncRNAs were proved to be closely associated with
AR, which indicated these lncRNAs played secondary role in the
tansition from AD to AI. Whereas, we infer that lncRNAs, which were
correlated with EGFR, might play a pivotal role in the progression
of PCa.
Although we identified lncRNAs as a ubiquitous
component in the gene regulatory networks of prostate cancer
progression, more work need to be done to eventually clarify the
underlying mechanism mediating the transition from AD to AI. The
most important aspect is to make clear the interaction between
lncRNAs and EGFR. Our results indicated that the lncRNA Linc00963
could affect the expression level of EGFR at transcriptional level,
but we did not obtain any evidence to support their direct
interaction between EGFR protein complex and Linc00963. The second
limitation of our study is we confirmed the role of lincRNAs in
hormone-sensitive and hormone-insensitive prostate cancer cell line
LNCaP and C4-2, but we did not further validate our result in the
tissues obtained from AD and AI prostate cancer patients. Although
use of LNCaP and C4-2 cell line is an accepted model for studying
the mechanism underlying the progression of PCa from AD to AI, the
confirmation from experiments in clinical specimens will make our
results more conclusive and convincing. These limitations indicate
that more detailed work is required to clarify the relationship
between lincRNAs and EGFR, to further improve our knowledge
concerning the transition from AD to AI in prostate cancer.
In summary, we have identified that the long
intergenic non-protein coding RNA 00963 is upregulated in
hormone-insensitive prostate cancer cell line C4-2 but
downregulated in hormone-sensitive prostate cancer cell line LNCaP.
The knockdown of Linc00963 in hormone-insensitive prostate cancer
cells inhibits cell viability, motility and invasiveness. Linc00963
physically associates with EGFR and could be an important regulator
for the transition in prostate cancer from AD to AI. Collectively,
our data provide insight into molecular characteristics of AI and
metastatic prostate cancer and provide clues for finding new
strategies to prevent PCa metastasis.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (no. 81272200,
81072117).
References
|
1.
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2.
|
Lee AK: Radiation therapy combined with
hormone therapy for prostate cancer. Semin Radiat Oncol. 16:20–28.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Molina A and Belldegrun A: Novel
therapeutic strategies for castration resistant prostate cancer:
inhibition of persistent androgen production and androgen receptor
mediated signaling. J Urol. 185:787–794. 2011. View Article : Google Scholar
|
|
4.
|
Mlcochova J, Faltejskova P, Nemecek R,
Svoboda M and Slaby O: MicroRNAs targeting EGFR signalling pathway
in colorectal cancer. J Cancer Res Clin Oncol. 139:1615–1624. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Shuch B, Mikhail M, Satagopan J, et al:
Racial disparity of epidermal growth factor receptor expression in
prostate cancer. J Clin Oncol. 22:4725–4729. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Syed S: Combination chemotherapy for
hormone-refractory prostate carcinoma: progress and pitfalls.
Cancer. 98:2088–2090. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Traish AM and Morgentaler A: Epidermal
growth factor receptor expression escapes androgen regulation in
prostate cancer: a potential molecular switch for tumour growth. Br
J Cancer. 101:1949–1956. 2009. View Article : Google Scholar
|
|
8.
|
Antonarakis ES, Carducci MA and
Eisenberger MA: Novel targeted therapeutics for metastatic
castration-resistant prostate cancer. Cancer Lett. 291:1–13. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Guttman M, Amit I, Garber M, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Reis EM, Nakaya HI, Louro R, et al:
Antisense intronic non-coding RNA levels correlate to the degree of
tumor differentiation in prostate cancer. Oncogene. 23:6684–6692.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Prensner JR, Iyer MK, Balbin OA, et al:
Transcriptome sequencing across a prostate cancer cohort identifies
PCAT-1, an unannotated lincRNA implicated in disease progression.
Nat Biotechnol. 29:742–749. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Ji P, Diederichs S, Wang W, et al:
MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Petrovics G, Zhang W, Makarem M, et al:
Elevated expression of PCGEM1, a prostate-specific gene with cell
growth-promoting function, is associated with high-risk prostate
cancer patients. Oncogene. 23:605–611. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Srikantan V, Zou Z, Petrovics G, et al:
PCGEM1, a prostate-specific gene, is overexpressed in prostate
cancer. Proc Natl Acad Sci USA. 97:12216–12221. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Chung S, Nakagawa H, Uemura M, et al:
Association of a novel long non-coding RNA in 8q24 with prostate
cancer susceptibility. Cancer Sci. 102:245–252. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Cui Z, Ren S, Lu J, et al: The prostate
cancer-up-regulated long noncoding RNA PlncRNA-1 modulates
apoptosis and proliferation through reciprocal regulation of
androgen receptor. Urol Oncol. 31:1117–1123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
de Kok JB, Verhaegh GW, Roelofs RW, et al:
DD3(PCA3), a very sensitive and specific marker to detect prostate
tumors. Cancer Res. 62:2695–2698. 2002.
|
|
21.
|
Hessels D, Klein Gunnewiek JM, van Oort I,
et al: DD3(PCA3)-based molecular urine analysis for the diagnosis
of prostate cancer. Eur Urol. 44:8–16. 2003. View Article : Google Scholar
|
|
22.
|
Xie BX, Zhang H, Yu L, et al: The
radiation response of androgen-refractory prostate cancer cell line
C4-2 derived from androgen-sensitive cell line LNCaP. Asian J
Androl. 12:405–414. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Horoszewicz JS, Leong SS, Kawinski E, et
al: LNCaP model of human prostatic carcinoma. Cancer Res.
43:1809–1818. 1983.
|
|
24.
|
Thalmann GN, Anezinis PE, Chang SM, et al:
Androgen-independent cancer progression and bone metastasis in the
LNCaP model of human prostate cancer. Cancer Res. 54:2577–2581.
1994.PubMed/NCBI
|
|
25.
|
Chen L, Mooso BA, Jathal MK, et al: Dual
EGFR/HER2 inhibition sensitizes prostate cancer cells to androgen
withdrawal by suppressing ErbB3. Clin Cancer Res. 17:6218–6228.
2011. View Article : Google Scholar
|
|
26.
|
Gutschner T and Diederichs S: The
hallmarks of cancer: a long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar
|
|
28.
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Roberts LD, Murray AJ, Menassa D, Ashmore
T, Nicholls AW and Griffin JL: The contrasting roles of PPARdelta
and PPARgamma in regulating the metabolic switch between oxidation
and storage of fats in white adipose tissue. Genome Biol.
12:R752011. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Cheng W, Zhang Z and Wang J: Long
noncoding RNAs: New players in prostate cancer. Cancer Lett.
339:8–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Bussemakers MJ, van Bokhoven A, Verhaegh
GW, et al: DD3: a new prostate-specific gene, highly overexpressed
in prostate cancer. Cancer Res. 59:5975–5979. 1999.PubMed/NCBI
|
|
32.
|
Bu D, Yu K, Sun S, et al: NONCODE v3.0:
integrative annotation of long noncoding RNAs. Nucleic Acids Res.
40:D210–D215. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Takayama K, Horie-Inoue K, Katayama S, et
al: Androgen-responsive long noncoding RNA CTBP1-AS promotes
prostate cancer. EMBO J. 32:1665–1680. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Mimeault M and Batra SK: Recent advances
on multiple tumorigenic cascades involved in prostatic cancer
progression and targeting therapies. Carcinogenesis. 27:1–22. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Nickerson T, Chang F, Lorimer D, Smeekens
SP, Sawyers CL and Pollak M: In vivo progression of LAPC-9 and
LNCaP prostate cancer models to androgen independence is associated
with increased expression of insulin-like growth factor I (IGF-I)
and IGF-I receptor (IGF-IR). Cancer Res. 61:6276–6280.
2001.PubMed/NCBI
|
|
36.
|
Xie BX, Zhang H, Wang J, et al: Analysis
of differentially expressed genes in LNCaP prostate cancer
progression model. J Androl. 32:170–182. 2011. View Article : Google Scholar : PubMed/NCBI
|