Introduction
Breast carcinoma is a common malignant tumor among
women, the incidence and mortality rates continue to rise rapidly
worldwide along with lifestyle changes in diet and environmental
factors (1). In Asian countries,
the incidence of breast cancer increase doubled or even tripled in
the past few decades, similarly, in China ∼20–30% of the new cases
of breast cancer occurred in the past decade (2). Currently, most breast cancer patients
receive chemotherapy, but the long-term stimulation of cytotoxic
agents accelerate the occurrence of severe acquired resistance,
especially multidrug resistance (MDR) has become an important cause
of tumor recurrence after surgery and chemotherapy has failed
(3). However, the molecular
mechanisms of MDR are rather complicated, it involves in increased
drug efflux, drug metabolic biotransformation and alteration of
repair ability for DNA damage (4).
Therefore, investigating the other potential mechanisms and
screening the molecular targets of drug resistance have become key
factors in the study of MDR of breast cancer.
Methotrexate (MTX), an antifolate antineoplastic,
mainly prevents tumor cells growth and proliferation through potent
inhibition of dihydrofolate reductase (DHFR) for purine and
pyrimidine nucleotide biosynthesis and blockage of DNA replication.
As a classical anticancer drug, it possesses strong efficacy and
low price, which is widely used for the treatment of human
leukemia, lymphoma, breast cancer, head and neck cancers and other
solid tumors, both alone and in combination with other
chemotherapeutics (5). Although
antifolates are important and effective components of different
chemotherapeutic regimens currently used for various cancer
therapies, drug resistance frequently arises from the long-term use
for treatment in clinic, and the molecular mechanism of
antifolate-resistance in cancer cells is believed to be a
multifactorial process, it relates to the loss of folate carrier
(RFC) function, overexpression and mutation of DHFR and thymidylate
synthase (TS), increased ATP-driven MDR efflux transporters,
decreased polyglutamate synthase, the high expression of γ-glutamyl
hydrolase and tetrahydrofolic acid (THF) (6). However, the MTX resistance is still
an extremely complex process, and the mechanism of action may
involve changes in a variety of pathways and expressions of
different resistant proteins.
With the aim of gaining further insight into the
molecular mechanisms of MDR and to discover new biological targets
of MTX, the functional proteomic analysis using two-dimensional gel
electrophoresis (2-DE) and matrix-assisted laser desorption
ionization time of flight mass spectrometry (MALDI-TOF-MS) were
performed to identify the differential protein profiles between the
MTX-resistant breast cancer cell line MCF-7/MTX and drug sensitive
cell line MCF-7/S.
Materials and methods
Establishment of MCF-7/MTX cell line
The parental human breast cancer cell line MCF-7 was
obtained from the Chinese Academy of Science (Shanghai, China).
These cells were cultured in RPMI-1640 medium containing 10% fetal
bovine serum (Gibco), 1% penicillin and streptomycin, at 37°C in a
humidified atmosphere of 5% CO2. The MTX-resistant cell
line MCF-7/MTX was established from its sensitive cell line MCF-7/S
by progressively increasing methotrexate (Shanxi Pude
Pharmaceutical, China) over a period of 12 months. The culture
condition was the same as the MCF-7/S cell line. The drug
concentration was increased by 1.5-fold at each step of resistance,
from 4 to 220 nM. The cells were cultured in MTX of each step for
at least 2 weeks before each experiment.
Drug sensitivity assay
The MTT assay (Sigma) was used to measure the drug
sensitivity. Cells were seeded at a density of 4×104
cells per well in 96-well plate with each well containing 100
μl medium. The cells were grown for 24 h, then the culture
medium was replaced with 200 μl fresh medium containing MTX
at different concentrations for 72 h. The medium was removed and
added with 180 μl of RPMI-1640 and 20 μl MTT (5 mg
MTT/ml) and incubated for 4 h. Following medium removal, 150
μl of DMSO was added to solubilize the formazan. The
absorbance was read at 490 nm on a microplate reader (BioTek
ELx808, USA). The 50% growth inhibitory concentration
(IC50) of drug was determined to evaluate the drug
sensitivity. Resistance factor (RF) was calculated by the ratio of
the IC50 values of MCF-7/MTX to MCF-7/S cells.
MDR detection
The sensitivity of MCF-7/S and MCF-7/MTX cells to
doxorubicin (Zhejiang Haizheng Pharmaceutical, China), paclitaxel
(Nanjing Sike Pharmaceutical, China), 5-fluorouracil (Shanghai
Xudonghaipu Pharmaceutical, China), mitoxantrone (Shandong Luoxin
Pharmaceutical, China), vinorelbine (Hangzhou Minsheng
Pharmaceutical, China), gemcitabine (Jiangsu Haosen Pharmaceutical,
China), cisplatin, docetaxel and pemetrexed (Shandong Qilu
Pharmaceutical, China), were determined by MTT assay as described
above, the IC50 value and RF for each drug were
calculated.
Analysis of cell cycle distribution
MCF-7/S and MCF-7/MTX cells were seeded at a density
of 4×105 cells per well into 6-well plates and incubated
for 24 h. Cells were trypsinized, washed twice with cold PBS and
fixed by 70% ethanol overnight at 4°C, then washed and re-suspended
with PBS containing RNase and propidium iodide. After 30 min in the
dark, the cell cycle was analyzed using a flow cytometer
(FACSCanto™II, BD Biosciences, USA).
Preparation of proteins
MCF-7/S and MCF-7/MTX cells were harvested, washed
with PBS and then lysed in the lysis buffer (8 M urea, 2 M
thiourea, 4% CHAPS, 40 nM Tris, 50 mM DTT, 1 mM PMSF, 0.5% (w/v)
Bio-lyte and 0.5% IPG buffer) at 4°C for 30 min. After
centrifugation at 14,000 rpm for 40 min at 4°C, the supernatants
were purified by ReadyPrep2-D Cleanup kit (Bio-Rad, Hercules, CA,
USA) according to the manufacturer’s instructions and stored at
−80°C for use. The protein concentration was measured with Bradford
method.
2-DE and image analysis
Each protein sample (800 μg) was applied to
pH 3.0–10.0 (non-linear 17 cm) IPG strip (Bio-Rad) using a passive
rehydration method. After rehydration for 16 h, IEF was performed
successively for 1 h at 250 V, 1 h at 1,000 V, and 5 h at 10,000 V
to give a total of 60 kvh on an IPGphor. Then, the gel strip was
equilibrated for 15 min with DTT and iodoacetamide, respectively.
The second dimension was performed using 12% SDS-PAGE until
bromophenol blue reached the bottom of the gel. The gel was stained
with sensitive colloidal Coomassie blue G-250 and scanned with UMax
Powerlook 2110XL (Umax). Three independent experiments were made
for each cell line to ensure the accuracy of the analyses. The
images were analyzed by Imagemaster 2D Platinum (GE Amersham). The
gel spot pattern of each gel was normalized and matched. The
differentially expressed proteins between MCF-7/S and MCF-7/MTX in
a given spot were calculated as the ratio of all the normalized
spot values. Only spots that showed significant differences (±
>1.5-fold, P<0.05) were selected for further analysis.
In-gel digestion
Differential protein spots were excised from the
stained gels, destained in 30 mM potassium ferri-cyanide/100 mM
sodium thiosulfate (1:1 v/v) for 20 min and washed with water. Then
the spots were incubated in 0.2 M NH4HCO3 for
20 min and freeze-dried. Each spot was digested with 12.5 ng/ml
trypsin in 25 mM NH4HCO3 at 37°C overnight.
The peptides were extracted three times with 60% ACN/0.1% TFA, and
the extracts were pooled and dried in a vacuum centrifuge to
complete desiccation at room temperature.
MALDI-TOF-MS analysis
The mass spectra were analyzed using MALDI-TOF-TOF
instrument (4800 proteomics analyzer; Applied Biosystems). For
protein identification, combined peptide mass fingerprinting (PMF)
and the MS/MS queries were performed automatically by searching the
EBI database using the MASCOT search engine 2.2 (Matrix Science,
UK). The database search was carried out with the following
parameters: enzyme, trypsin; the allowance of one missed cleavage;
carbamidomethylation was set as fixed modification; oxidation of
methionine was allowed as variable modification; the peptide and
fragment mass tolerance were set at 100 ppm 0.4 Da, respectively. A
GPS Explorer protein confidence index ≥95% was used for further
manual validation. Proteins with probability based MOWSE scores
exceeding their threshold (P<0.05) were considered to be
positively identified.
Western blot analysis
Western blot assay was performed after 10% SDS-PAGE
separation, transfer to a PVDF membrane (Millipore) and saturation
with 5% non-fat dry milk for 2 h at room temperature. Followed by
incubation with primary rabbit polyclonal antibodies including
anti-P-gp (1:500 dilution; GeneTex), anti-MRP1 (1:600 dilution;
GeneTex), anti-RFC (1:1,500 dilution; Abcam), anti-hnRNP C1/C2
(1:600 dilution; GeneTex), rabbit monoclonal antibodies including
anti-BCRP (1:800 dilution; Epitomics), anti-DHFR (1:20,000
dilution; Epitomics), anti-NPM (1:1,000 dilution; Epitomics),
anti-ENO1 (1:2,500 dilution; Epitomics), anti-VIM (1:600 dilution;
Epitomics), anti-PGAM1 (1:1,000 dilution; Epitomics), anti-PSMA2
(1:2,000 dilution; Epitomics), or anti-β-actin (1:800 dilution;
Beijing Biosynthesis Biotechnology, China) as a loading control
overnight at 4°C, then the membranes were incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgG (1:20,000
dilution; CWbiotech, China) for 2 h at room temperature. Protein
bands were detected by ECL detection system. Images were analyzed
using a quantitative analysis system (Image-Pro Plus).
Quantitative PCR analysis
Total RNA was extracted using RNAfast2000 kit
(Fastagen) according to the manufacturer’s instructions.
Quantitative PCR was performed with PrimeScript RT Master Mix
Perfect Real Time Kit (Takara DRR036A) and SYBR Premix Ex Taq II
(Takara). The primer sequences and product length are listed in
Table I. The experiments were run
on the Bio-Rad CFX96™ Real-time system (Bio-Rad): pre-degeneration
for 95°C, 30 sec, 1 cycle, and PCR reaction, 95°C 5 sec followed by
60°C, 30 sec, 40 cycles, and 95°C for 15 sec, 60°C for 30 sec, 95°C
for 15 sec for dissociation. β-actin was used as an internal
control gene to normalize expression levels.
 | Table I.Primer sequences for quantitative
PCR. |
Table I.
Primer sequences for quantitative
PCR.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Product size
(bp) |
|---|
| MDR1 |
GAGCCCATCCTGTTTGACTG |
GCTGCCCTCACAATCTCTTC | 92 |
| MRP1 |
AAGGTGGACGAGAACCAGAA |
AACAGGGCAGCAAACAGAAC | 110 |
| BCRP |
AGCAGGGACGAACAATCATC |
GCCAATAAGGTGAGGCTATCA | 82 |
| RFC |
TCCTGTCCATCATCTACTTCTTG |
AGTGCCTGTGCTGCCTTCT | 130 |
| DHFR |
TCTCCAAGACCCCAACTGAG |
ATGTGAAAAGCCCGACAAT | 109 |
| NPM |
TGGCAGTGGAGGAAGTCTCT |
ATCAAACACGGTAGGGAAAGTT | 141 |
| ENO1 |
TCCCTTTGACCAGGATGACT |
GACTTTGAGCAGGAGGCAGTT | 151 |
| VIM |
GAAGAGAACTTTGCCGTTGAAG |
GAAGGTGACGAGCCATTTC | 100 |
| hnRNP
C1/C2 |
GCTTTGCCTTCGTTCAGTATG |
CCCGTTGAAAGTCATAGTCCA | 240 |
| PGAM1 |
GCTCCCCTTATCCAACAGAGTT |
TTGCTTCTCCTCACTGGTCAT | 114 |
| PSMA2 |
CCAAGCAGAATGATGAAATGAC |
CTTTATGGAAACACAGGCAACA | 103 |
| β-actin |
TGACGTGGACATCCGCAAAG |
CTGGAAGGTGGACAGCGAGG | 205 |
Small interfering RNA (siRNA)
transfection
MCF-7/MTX cells were seeded in a 6-well plate at a
density of 5×105 cells per well in RPMI-1640 without
antibiotics. After 24 h, the NPM or non-specific control siRNA
(Shanghai GenePharma, China) was transfected with Lipofectamine
2000 (Invitrogen) according to the manufacturer’s instructions. The
concentration of NPM siRNA was 50 nM. The efficiency of RNA
interference was checked by quantitative PCR and western blot
analysis, respectively. The transfected cells were used for
proliferation assay, other genes and proteins detection.
Statistical analysis
All experiments were carried out independently three
times. All data, unless stated otherwise, are shown as means ±
standard deviations. Statistical analyses of differences between
the groups were performed by Student’s t-test. A P-value of
<0.05 was considered statistically significant.
Results
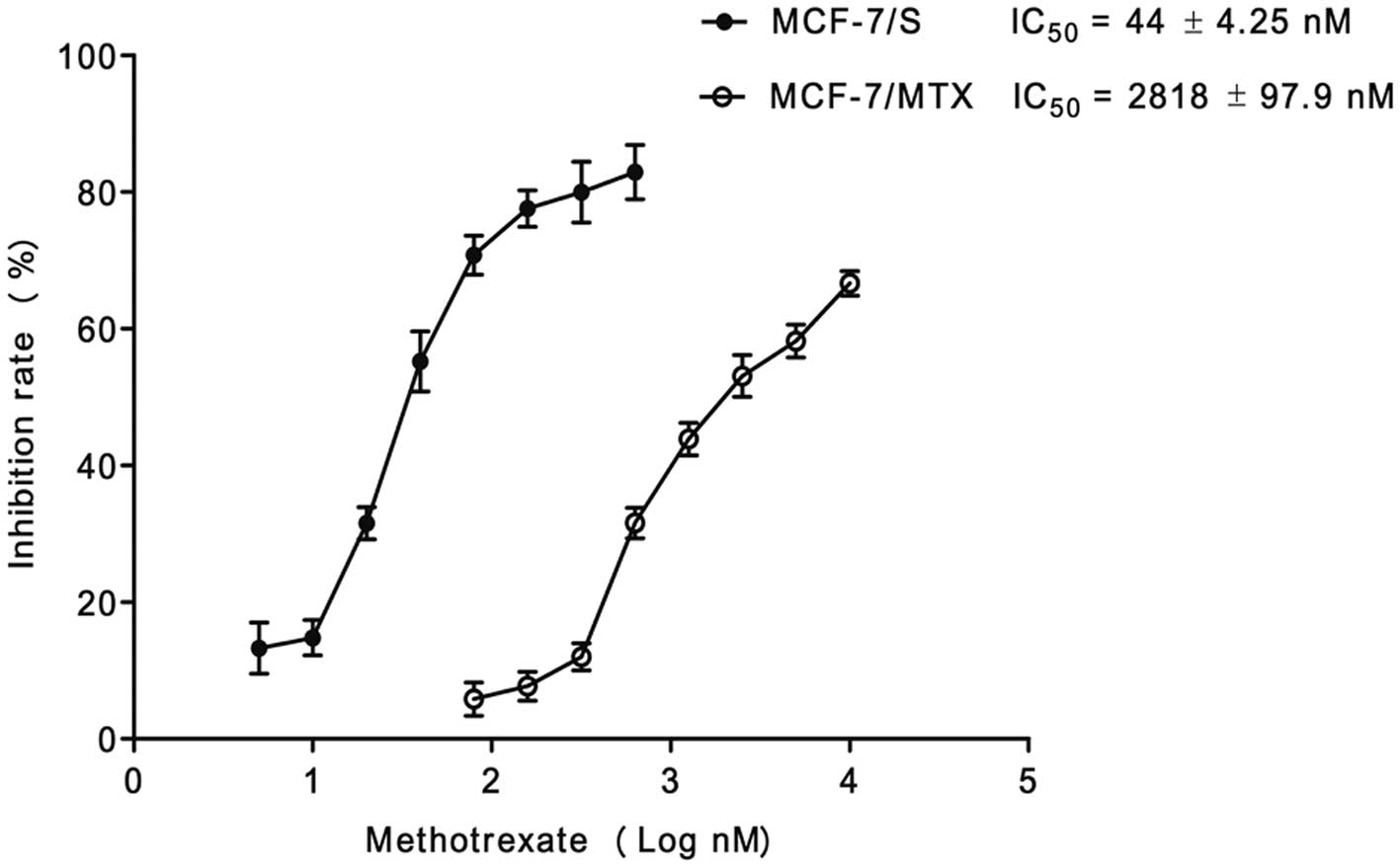
Methotrexate-resistant cell line
After a continuous induction with low concentration
of MTX in a gradually increasing manner for 12 months, the
MCF-7/MTX-resistant cell line grew steadily in the culture medium
with 220 nM MTX. The IC50 values of MTX for MCF-7/S
cells and MCF-7/MTX cell lines were 44±4.25 nM and 2818±97.9 nM,
respectively (Fig. 1). The
resistance of MCF-7/MTX cells to MTX was 64-fold higher than that
of MCF-7/S cells, which implied the MTX-resistant cell line was
successfully established.
MDR phenotype of MCF-7/MTX cells
Both MCF-7/S and MCF-7/MTX cell lines were treated
with different concentrations of chemotherapeutic drugs, and the
IC50 and RF values are summarized in Table II. The cross-resistance of
MCF-7/MTX cells to doxorubicin, paclitaxel, 5-fluorouracil,
mitoxantrone, vinorelbine, cisplatin, docetaxel, pemetrexed and
gemcitabine significantly increased compared with that of MCF-7/S
cells, which indicated the typical MDR phenotype of MCF-7/MTX
cells.
 | Table II.IC50 values (nM) of
different cells against selected anticancer drugs (mean ± SD). |
Table II.
IC50 values (nM) of
different cells against selected anticancer drugs (mean ± SD).
| Drugs | MCF-7/S | MCF-7/MTX | MCF-7/MTX RNAi | IC50
MCF-7/MTX/IC50 MCF-7/S
| IC50 MCF-7/MTX
RNAi/IC50 MCF-7/S
|
|---|
| RF | RF |
|---|
| Methotrexate | 44±4.25 | 2,818±97.9a | 1,288±29.0b | 64.0 | 29.3 |
| Doxorubicin | 71±2.57 | 245±7.39a | 174±5.42b | 3.45 | 2.45 |
| Paclitaxel | 20±0.45 | 48±1.62a | 33±1.05b | 2.40 | 1.65 |
| 5-fluorouracil | 3,890±178 | 7,943±55.3a | 6,892±57.9b | 2.04 | 1.77 |
| Mitoxantrone | 186±16.0 | 295±24.8a | 285±21.0 | 1.58 | 1.53 |
| Vinorelbine | 51±7.18 | 1,023±91.6a | 256±10.4b | 20.1 | 5.02 |
| Cisplatin | 2,042±74.2 | 5,495±66.2a | 3,987±187b | 2.69 | 1.95 |
| Docetaxel | 4.27±0.19 | 22±5.42a | 14±1.87b | 5.15 | 3.28 |
| Pemetrexed | 3,631±216 | 9,550±111a | 6,424±245b | 2.63 | 1.77 |
| Gemcitabine | 955±47.0 | 6,026±568a | 2,698±109b | 6.31 | 2.83 |
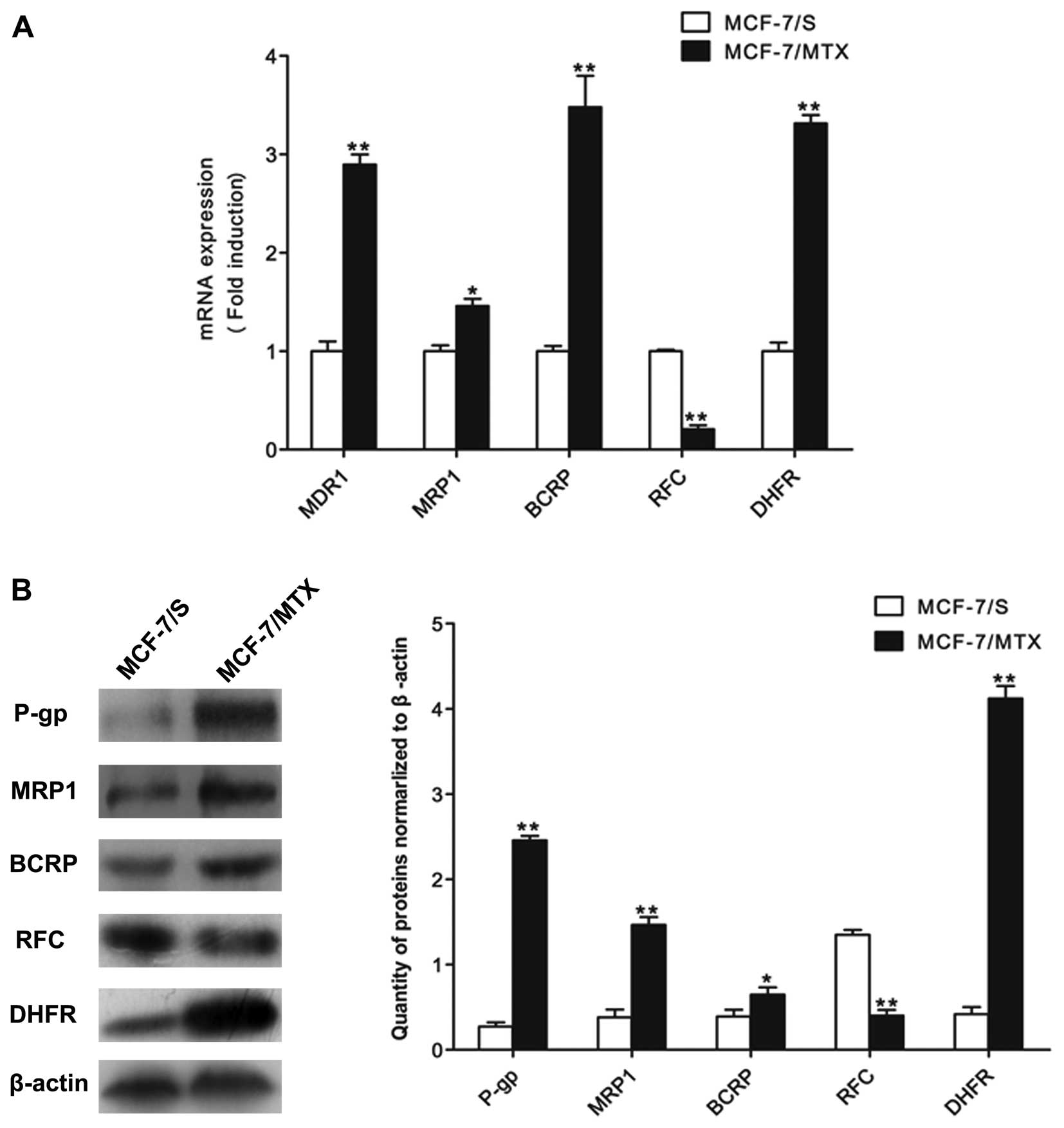
In addition, in order to detect whether the MDR of
MCF-7/MTX cells was associated with the classic resistance-related
proteins and resistance mechanism of MTX, we checked the mRNA and
protein levels of MDR1, MRP1, BCRP, RFC and DHFR by quantitative
PCR and western blot analyses. The mRNA levels of MDR1, MRP1, BCRP
and DHFR in MCF-7/MTX cells were higher than those in MCF-7/S
cells, while the mRNA expression of RFC was significant decreased
in MCF-7/MTX cell line (Fig. 2A).
The protein levels of these molecules were consistent with gene
expressions in both cell lines (Fig.
2B). These results demonstrated that the established MCF-7/MTX
cell line is a valuable model to study MDR mechanism.
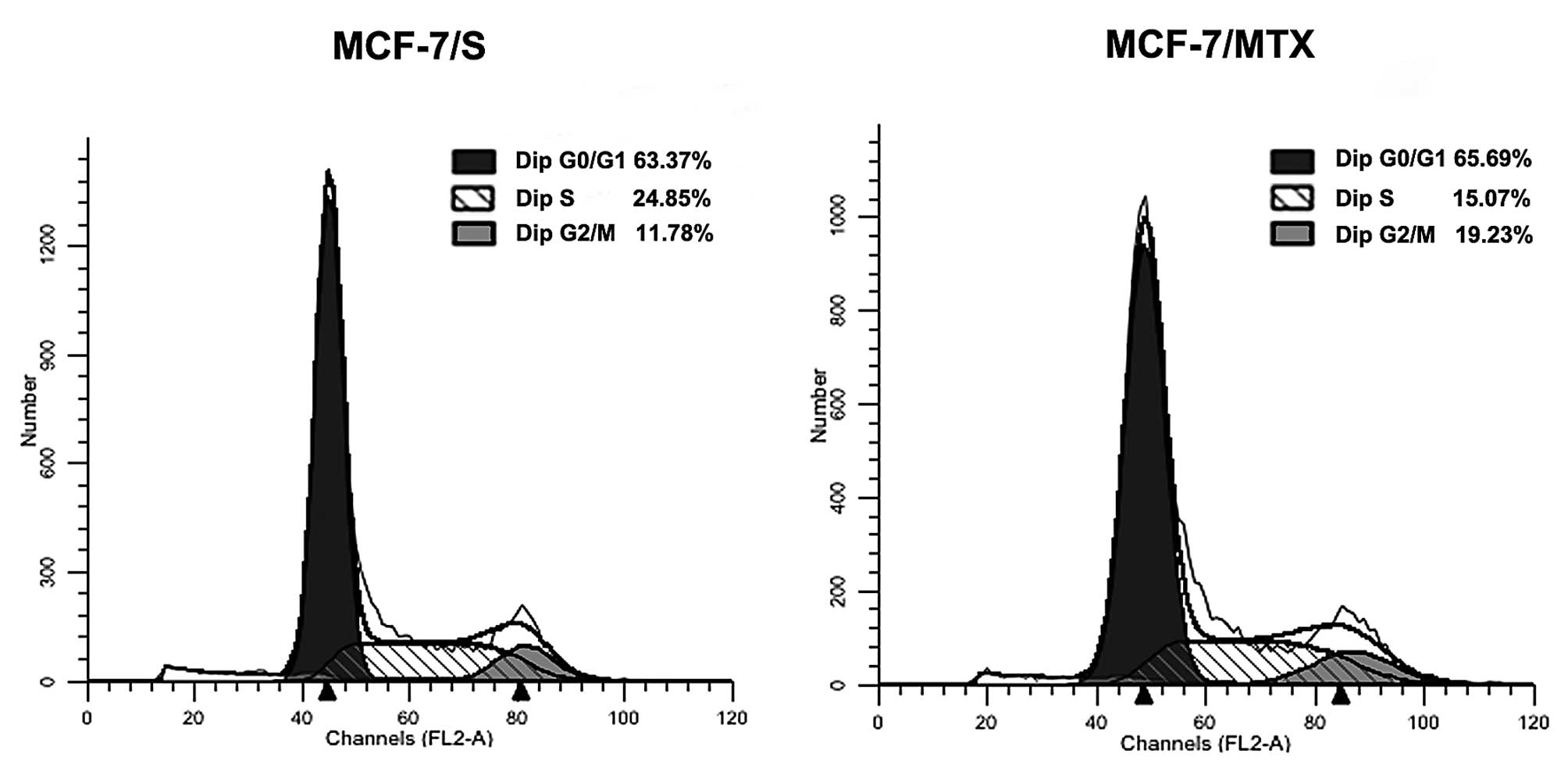
Cell cycle distribution
In order to further explore the effect of MTX on
cells, the cell cycle profiles of MCF-7/S and MCF-7/MTX was
performed using flow cytometry analysis. We found that the
proportion of MCF-7/MTX cells in G0/G1 and G2/M phases showed a
gradual increasing trend, from 63.37 to 65.69% and from 11.78 to
19.23%, respectively, while the proportion of S phase dropped from
24.85 to 15.07% (Fig. 3). These
results suggested that MTX could block the cell cycle at the G0/G1
phase of progression.
The proteome comparison of MCF-7/MTX and
MCF-7/S cells
To compare the proteome between MCF-7/MTX and
MCF-7/S cells, we performed 2-DE to separate the global protein
lysates of these two cell lines. The analysis was carried out in
six gels made from three independent protein samples of each cell
line. The images of protein spots were assessed using PDQuest6.0
analysis software. We selected 25 differentially expressed protein
spots (≥1.5-fold) between these two cell lines for further
identification using MALDI-TOF-MS, and 17 proteins that were
successively identified were marked with arrows in Fig. 4A, among which 7 proteins were
upregulated and 10 proteins were downregulated in the MCF-7/MTX
cell line (Table III).
 | Table III.MS identification of differentially
expressed proteins between MCF-7/MTX and MCF-7/S cells. |
Table III.
MS identification of differentially
expressed proteins between MCF-7/MTX and MCF-7/S cells.
| Spot no. | Accession no. | Protein name | MW (kDa)/PI | Peptide
matched | Score | Sequence coverage
(%) | Expression in
MCF-7/MTX/MCF-7/S |
|---|
| 1 | IPI00554788 | Keratin, type I
cytoskeletal 18 | 48.03/5.34 | 26 | 661 | 49 | 4.43↑ |
| 2 | IPI00645031 | Isoform 2 of
λ-crystallin homolog | 33.79/5.68 | 10 | 66 | 24 | 4.15↑ |
| 3 | IPI00658013 | Nucleophosmin
isoform 3 | 28.50/4.56 | 8 | 168 | 33 | 4.05↑ |
| 4 | IPI00893990 | NDUFA10
protein | 15.90/5.14 | 9 | 73 | 46 | 3.58↑ |
| 5 | IPI00022465 | Isoform 1 of citron
Rho-interacting kinase | 233.34/6.16 | 22 | 65 | 11 | 2.61↑ |
| 6 | IPI00465248 | Isoform α-enolase
of α-enolase | 47.48/7.01 | 16 | 277 | 39 | 2.43↑ |
| 7 | IPI00418471 | Vimentin | 53.68/5.06 | 23 | 663 | 53 | 2.08↑ |
| 8 | IPI00016610 | Poly(rC)-binding
protein 1 | 37.99/6.66 | 13 | 450 | 50 | 3.48↓ |
| 9 | IPI00216592 | Isoform C1 of
heterogeneous nuclear ribonucleoproteins C1/C2 | 32.37/4.94 | 14 | 624 | 43 | 3.34↓ |
| 10 | IPI00554648 | Keratin, type II
cytoskeletal 8 | 53.67/5.52 | 24 | 665 | 49 | 3.10↓ |
| 11 | IPI00024911 | Endoplasmic
reticulum resident protein 29 | 29.03/6.77 | 7 | 173 | 33 | 2.92↓ |
| 12 | IPI00549725 | Phosphoglycerate
mutase 1 | 28.90/6.67 | 15 | 895 | 62 | 2.79↓ |
| 13 | IPI00219622 | Proteasome subunit
α type-2 | 26.00/6.92 | 10 | 222 | 41 | 2.69↓ |
| 14 | IPI00739539 | POTE ankyrin domain
family member F | 123.02/5.83 | 13 | 386 | 47 | 2.37↓ |
| 15 | IPI00465439 |
Fructose-bisphosphatealdolase A | 39.85/8.30 | 14 | 386 | 44 | 2.12↓ |
| 16 | IPI00797270 | Isoform 1 of
triosephosphateisomerase | 26.94/6.45 | 19 | 682 | 90 | 1.83↓ |
| 17 | IPI00163187 | Fascin | 55.12/6.84 | 13 | 361 | 34 | 1.81↓ |
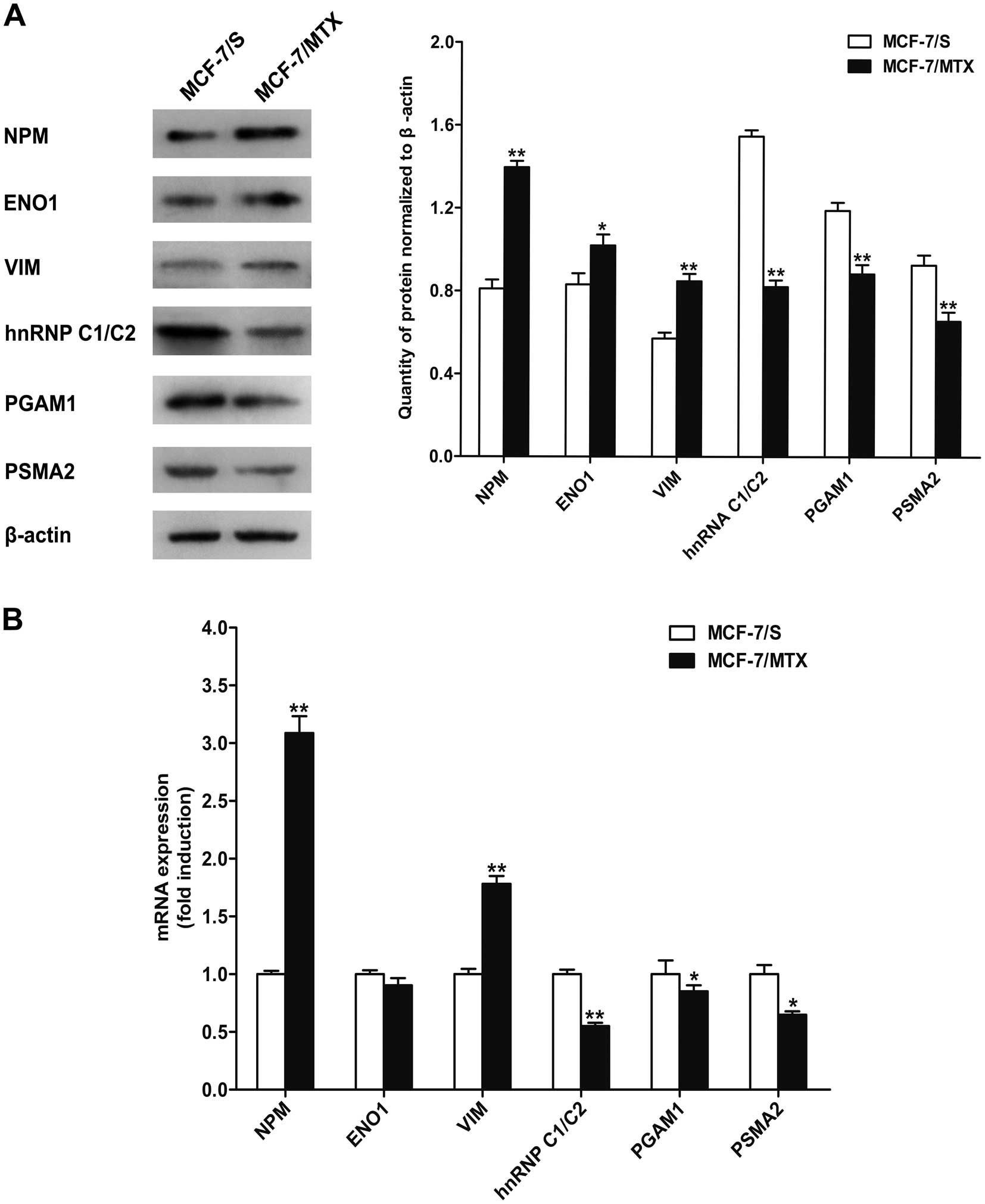
Validation of protein expression
To further confirm the expression of identified
proteins, western blotting was performed to validate six
differentially expressed proteins (Fig. 4B). Consistent with the results of
2-DE analysis, three proteins nucleophosmin (NPM), α-enolase
(ENO1), and vimentin (VIM) were significantly upregulated, while
other three proteins including heterogeneous nuclear
ribonucleoprotein (hnRNP C1/C2), phosphoglycerate mutase 1 (PGAM1),
and proteasome subunit α type-2 (PSMA2) were obviously
downregulated in MCF-7/MTX cells (Fig.
5A).
Validation of mRNA expression
To further validate whether the mRNA levels of these
six differentially expressed proteins had variations, quantitative
PCR assay was applied to detect the mRNA expression of NPM,
ENO1, VIM, hnRNP C1/C2, PGAM1 and PSMA2 (Fig. 5B). Compared with MCF-7/S cells, the
mRNA levels of NPM and VIM were increased while
hnRNP C1/C2, PGAM1 and PSMA2 genes were decreased in
MCF-7/MTX cell line, which was consistent with the alteration of
the protein levels. Unexpectedly, the mRNA expression of ENO1 was
slightly downregulated in the MCF-7 MTX cells, which was different
from the protein level.
Reversal of MDR in MCF-7/MTX cells by
knockdown of NPM
We found the protein and mRNA expressions of NPM
were both significantly upregulated in the MCF-7/MTX cells. To
study the functions of the increased expression of NPM in the MDR
of MCF-7/MTX cells, MTT assay was performed to determine the
sensitivity of MCF-7/MTX cells to MTX and other antitumor drugs
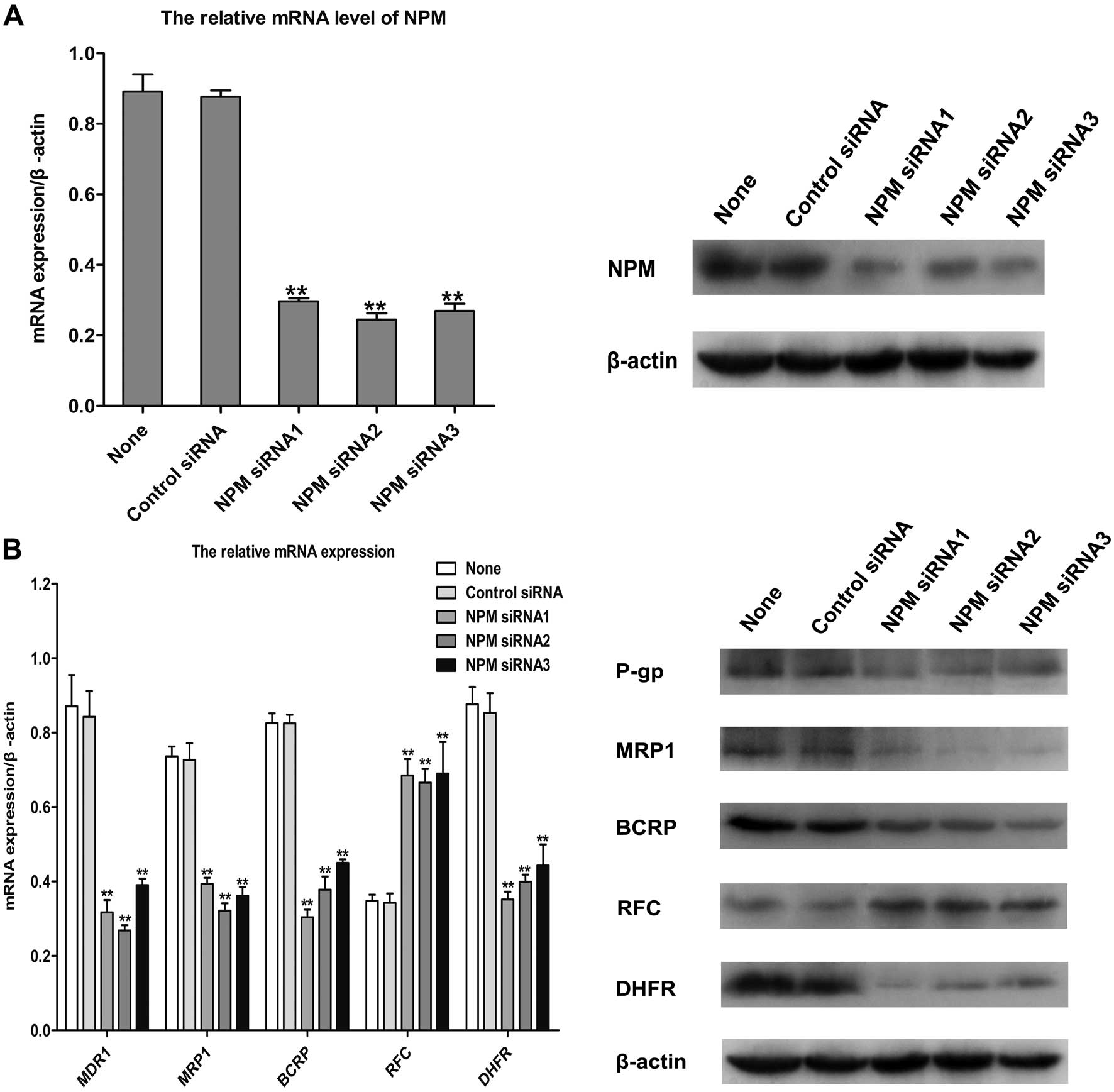
following knockdown of NPM by siRNA. Our results showed that
siRNA significantly down-regulated the gene and protein of NPM in
the MCF-7/MTX cells (Fig. 6A).
Knockdown of NPM by siRNA in MCF-7/MTX cells obviously
enhanced the inhibitory effect of MTX on proliferation. In
addition, the IC50 value and RF of MCF7/MTX cells
exposed to doxorubicin, paclitaxel, 5-fluorouracil, vinorelbine,
cisplatin, docetaxel, pemetrexed and gemcitabine were significantly
decreased after downregulating the expression of NPM by
siRNA (Table II). Surprisingly,
the protein and mRNA levels of drug resistance-related molecules,
MDR1, MRP1, BCRP and DHFR were also significantly decreased while
the protein and gene expressions of RFC were increased after
knockdown of NPM in MCF-7/MTX cells (Fig. 6B). These observations demonstrate
that NPM has a prominent role in the multidrug resistance of
MCF-7/MTX cells.
Discussion
The antifolate MTX is applied to the treatment of a
wide variety of cancers, it is usually given in combination with
cyclophosphamide and 5-flurouracil to cure breast cancer. However,
drug resistance is frequently observed upon treatment with MTX for
cancer patients, thus compromising the drug effectiveness. Although
the alteration of gene expressions also contribute to MTX
resistance, such as increase in S100A4, UGT1A6, caveolin-1,
enolase-2, PRKCA and the decrease of miR-224 or E-cadherin
(7–10), the resistance mechanism of MTX,
especially the MDR caused by MTX is not fully understood. In the
present study, we have successfully established the MTX-resistant
human breast cancer cell line MCF-7/MTX, which displays
cross-resistance to a variety of commonly used chemotherapy drugs
and multidrug resistance phenotype. Using the proteomic assay, we
integrally analyzed the protein expression differences between
resistant cells and parental cells to identify potential molecular
targets for MDR of breast cancer.
In this study, 17 differentially expressed proteins
were identified, seven proteins were upregulated and 10 proteins
were downregulated in MCF-7/MTX cells. Moreover, six significantly
expressed proteins with distinct functions were validated by
western blot and quantitative PCR assays at protein and mRNA
levels, respectively. These proteins are involved in tumorigenesis,
metabolism, glycolysis, cell proliferation, apoptosis and the
invasion process. Their roles in the formation of drug resistance
and molecular mechanism are discussed below.
α-enolase (ENO1), known as 2-phosphate-D-glycerate
hydrolase, catalyzes the formation of phosphoenolpyruvate from
2-phosphoglycerate in the process of glycolysis, which was a key
role in cellular energy metabolism (11). ENO1 has been recognized as a
conserved and single function protein, but recent studies found
that the effect of this enzyme is involved in transcription,
regulation of apoptosis and cell differentiation processes.
Research has shown that the expression of ENO1 significantly
increased in certain tumor cells, including liver cancer, lung
cancer, head and neck cancer, which could be used as prognostic
indicators for clinical treatment (12–14).
It was reported that the elevated expression of ENO1 was closely
related to tamoxifen and adriamycin resistance in breast cancer
(15,16). The expression of ENO1 was regulated
by ERK1/2 appearing to be mediated by c-Myc, which changed the
level of extracellular ATP, thus affecting cell survival (17). In our study, the protein level of
ENO1 in MCF-7/MTX cells are significantly increased, but its gene
level was slightly lower in the resistant cells, which indicates
the mRNA level is not fully present in the protein level, due to
the mRNA possessed storage, transport, degradation, translational
regulation and post-translational processing of the product, which
affects the quality and quantity of the protein (18). We have found the correlation
between ENO1 and MDR in breast cancer, which may be regulated by
activating ERK1/2 pathway, further affecting tumor cell
proliferation.
Vimentin (VIM) is a member of intermediate filament
protein family, which constitutes the cytoskeleton with
microtubules and actin filaments, it is involved in cell adhesion,
migration, apoptosis and cell signal transduction (19). In prostate cancer and
triple-negative breast cancer cells, the high expression of VIM was
significantly associated with cell invasiveness, moreover, the
activation of ERK signaling pathway promotes the overexpression of
VIM and increased cell migration and invasion in head and neck
cancer (20–22). In recent research, VIM was
intimately associated with the resistance to chemotherapeutic
drugs, it was significantly higher expression in resistant cancer
cells including paclitaxel-resistant prostate cancer,
temozolomide-resistant malignant glioma and tamoxifen-resistant
breast cancer (23–25). Our results show that the protein
and mRNA levels of VIM are significantly increased in the
MTX-resistant cells, which may be related to the variation of cell
adhesion, thereby possibly influencing cell metastasis and
invasion, and its resistance mechanisms need to be further
explored.
Heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNP
C1/C2) is a member of hnRNPs family, as an RNA-binding protein, it
participates in the regulation of pre-mRNA splicing and
post-translational modification (26). In addition, studies have shown that
hnRNPs were associated with tumor development and prognosis
(27). Previously, hnRNP C1/C2 has
been considered as an apoptosis-related protein, and the protein
level of hnRNP C1/C2 was p53-dependently upregulated, but its gene
transcription did not depend on the p53 role in colon cancer cells
after mitomycin C treatment (28,29).
Furthermore, in A549 lung cancer cells of wild-type p53, the
expression of hnRNP C1/C2 was significantly reduced in the process
of apoptosis, it regulated the synthesis of p53 isoforms with p53
IRES interaction and affected tumor cell death (30,31).
On the contrary, hnRNP C1/C2 was associated with DNA repair
function, knockdown of hnRNP C1/C2 by RNA interference could
increase the sensitivity of HeLa cells to chemotherapeutic drugs
(32). In the present study, both
protein and mRNA expression of hnRNP C1/C2 are downregulated in
MCF-7/MTX cells, which may be connected with mutations of apoptosis
regulatory factors and affects the normal process of cell
apoptosis, while the exact mechanism needs further
investigation.
Phosphoglycerate mutase 1 (PGAM1), as a key enzyme
of glycolysis and gluconeogenesis process, catalyzes the conversion
of 3-phosphoglycerate (3-PG) to 2-phosphoglycerate (2-PG). It was
found that PGAM1 was overexpressed in certain tumor cells including
liver cancer, breast cancer, and glioma, its upregulation may
facilitate the proliferation and metabolism of cancer cells, which
could be used as a potential therapeutic target for tumors
(33–36). Moreover, PGAM1 was also negatively
correlated with the tumor suppressor p53, which further illustrates
the interaction between PGAM1 and cell apoptosis (37). However, we discovered that the
protein and mRNA levels of PGAM1 are significantly reduced in
MTX-resistant cells, abnormal glucose metabolism may be involved in
mediating the role of MDR in breast cancer, and the resistance
mechanism is still unclear and requires further study.
Proteasomes are macromolecular complexes with a
variety of proteolytic functions, acting as catalysts in the
ubiquitin-protea-some pathway of intracellular protein degradation.
However, the ubiquitin-proteasome pathway may control numerous
important physiological functions, such as cell cycle, signal
transduction, DNA damage repair and cell apoptosis, additionally,
abnormal alteration of this pathway is closely bound with prognosis
and development of malignant tumor. Increased expression of
proteasome subunit protein can lead to formation of tumors,
especially to protect anti-apoptotic effects of cancer cells,
therefore, the proteasome subunit proteins could be recognized as
potential antitumor targets. Some studies demonstrated that PSMA7
could inhibit proliferation and invasion in A549 cells (38), it regulated cell transcription,
cell cycle and tumor development with other important proteins
interaction (39). It was found
that PSMA5 was highly expressed in 5-fluorouracil-resistant
colorectal cancer cells, which showed certain anti-apoptotic
activity (40). Whereas, in our
established MTX-resistant cell line, we first discover that the
protein and gene levels of PSMA2 are significantly downregulated,
which reflects a correlation between PSMA2 and MDR in breast
cancer, this provides the basis for future investigation of
resistance mechanisms.
Nucleolar phosphoprotein (NPM) is abundant and
highly conserved phosphoprotein which shuttles rapidly between
nucleus and cytoplasm, it participates in the assembly and
synthesis of ribosome, replication of chromosomes and centrosomes
and intracellular signal transduction, which plays an important
role in cell growth, proliferation and transformation. An
increasing number of studies manifested that there may exist a
connection between NPM and the occurence of tumors, high expression
of the proto-oncogene c-Myc could lead to upregulation of NPM
transcriptional level; its overexpression could inactivate the
function of p53 and enhance the anti-apoptosis effect in cancer
cells; NPM was able to regulate tumor progression by activating the
phosphorylation of MAPK/ERK and c-Myc signaling pathway (41). The overexpression of NPM defended
the p53-mediated cellular senescence and growth arrest in
colorectal cancer cells, which showed its role in promoting tumor
growth (42), conversely, reducing
the expression of NPM by shRNA sensitized the resistant leukemia
cells to chemotherapy (43). There
is also evidence indicating that NPM has a tumor suppressor
property maintaining the gene stability and regulating the ARF
tumor suppressor (44). Similarly,
in invasive breast cancer MDA-MB-231 cells, the overexpression of
NPM inhibited cell growth, which could be used as a tumor
suppressor factor and predictor of poor prognosis (45). Although the above demonstrated
functions of NPM are controversial, in our study, the protein and
gene levels of NPM are both significantly increased in MCF-7/MTX
cells. We discovered the connection between NPM and MTX resistance
mechanism, which may induce MDR through acting on MAPK/ERK
signaling pathway in breast cancer. Subsequently, we reduced the
expression of NPM by siRNA interference, which attenuates the
resistance of MCF-7/MTX cells to various drugs and the expressions
of cell factors related to MDR and MTX resistance mechanism. These
results indicate that NPM contributes to MTX resistance in breast
cancer cells and suggest that NPM may play a significant role in
the occurrence of MDR of cancer cells.
In conclusion, we have successfully established the
MTX-resistant human breast cancer cell line and identified 17
differentially expressed proteins. NPM, involving in tumorigenesis
and signal transduction, plays a prominent role in the MDR of
MCF-7/MTX cells. This study investigated the resistance mechanisms
of breast cancer and provided a theoretical basis for the clinical
diagnosis of MDR.
Acknowledgements
This study was supported by National
Natural Science Foundation of China (nos. 30973673 and
30973578).
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Bhoo-Pathy N, Yip CH, Hartman M, et al:
Breast cancer research in Asia: adopt or adapt Western knowledge?
Eur J Cancer. 49:703–709. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Longley DB and Johnston PG: Molecular
mechanisms of drug resistance. J Pathol. 205:275–292. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Baguley BC: Multiple drug resistance
mechanisms in cancer. Mol Biotechnol. 46:308–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Olsen EA: The pharmacology of
methotrexate. J Am Acad Dermatol. 25:306–318. 1991. View Article : Google Scholar
|
|
6.
|
Assaraf YG: Molecular basis of antifolate
resistance. Cancer Metastasis Rev. 26:153–181. 2007. View Article : Google Scholar
|
|
7.
|
de Almagro MC, Selga E, Thibaut R, Porte
C, Noe V and Ciudad CJ: UDP-glucuronosyltransferase 1A6
overexpression in breast cancer cells resistant to methotrexate.
Biochem Pharmacol. 81:60–70. 2011.PubMed/NCBI
|
|
8.
|
Selga E, Morales C, Noe V, Peinado MA and
Ciudad CJ: Role of caveolin 1, E-cadherin, Enolase 2 and PKCalpha
on resistance to methotrexate in human HT29 colon cancer cells. BMC
Med Genomics. 1:352008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Mencia N, Selga E, Noé V and Ciudad CJ:
Underexpression of miR-224 in methotrexate resistant human colon
cancer cells. Biochem Pharmacol. 82:1572–1582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Mencia N, Selga E, Rico I, et al:
Overexpression of S100A4 in human cancer cell lines resistant to
methotrexate. BMC Cancer. 10:2502010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Subramanian A and Miller DM: Structural
analysis of alpha-enolase. Mapping the functional domains involved
in down-regulation of the c-myc protooncogene. J Biol Chem.
275:5958–5965. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Takashima M, Kuramitsu Y, Yokoyama Y, et
al: Overexpression of alpha enolase in hepatitis C virus-related
hepatocellular carcinoma: association with tumor progression as
determined by proteomic analysis. Proteomics. 5:1686–1692. 2005.
View Article : Google Scholar
|
|
13.
|
Chang GC, Liu KJ, Hsieh CL, et al:
Identification of alpha-enolase as an autoantigen in lung cancer:
its overexpression is associated with clinical outcomes. Clin
Cancer Res. 12:5746–5754. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Tsai ST, Chien IH, Shen WH, et al: ENO1, a
potential prognostic head and neck cancer marker, promotes
transformation partly via chemokine CCL20 induction. Eur J Cancer.
46:1712–1723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Tu SH, Chang CC, Chen CS, et al: Increased
expression of enolase alpha in human breast cancer confers
tamoxifen resistance in human breast cancer cells. Breast Cancer
Res Treat. 121:539–553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Chuthapisith S, Layfield R, Kerr ID,
Hughes C and Eremin O: Proteomic profiling of MCF-7 breast cancer
cells with chemoresistance to different types of anti-cancer drugs.
Int J Oncol. 30:1545–1551. 2007.PubMed/NCBI
|
|
17.
|
Mizukami Y, Iwamatsu A, Aki T, et al:
ERK1/2 regulates intracellular ATP levels through alpha-enolase
expression in cardiomyocytes exposed to ischemic hypoxia and
reoxygenation. J Biol Chem. 279:50120–50131. 2004. View Article : Google Scholar
|
|
18.
|
de Sousa Abreu R, Penalva LO, Marcotte EM
and Vogel C: Global signatures of protein and mRNA expression
levels. Mol Biosyst. 5:1512–1526. 2009.PubMed/NCBI
|
|
19.
|
Ivaska J, Pallari HM, Nevo J and Eriksson
JE: Novel functions of vimentin in cell adhesion, migration, and
signaling. Exp Cell Res. 313:2050–2062. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Singh S, Sadacharan S, Su S, Belldegrun A,
Persad S and Singh G: Overexpression of vimentin: role in the
invasive phenotype in an androgen-independent model of prostate
cancer. Cancer Res. 63:2306–2311. 2003.PubMed/NCBI
|
|
21.
|
Karihtala P, Auvinen P, Kauppila S,
Haapasaari KM, Jukkola-Vuorinen A and Soini Y: Vimentin, zeb1 and
Sip1 are up-regulated in triple-negative and basal-like breast
cancers: association with an aggressive tumour phenotype. Breast
Cancer Res Treat. 138:81–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Tseng YH, Chang KW, Yang CC, et al:
Association between areca-stimulated vimentin expression and the
progression of head and neck cancers. Head Neck. 34:245–253. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Kim JJ, Yin B, Christudass CS, et al:
Acquisition of paclitaxel resistance is associated with a more
aggressive and invasive phenotype in prostate cancer. J Cell
Biochem. 114:1286–1293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Sun S, Wong TS, Zhang XQ, et al: Protein
alterations associated with temozolomide resistance in subclones of
human glioblastoma cell lines. J Neurooncol. 107:89–100. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Liu H, Zhang HW, Sun XF, et al:
Tamoxifen-resistant breast cancer cells possess cancer stem-like
cell properties. Chin Med J (Engl). 126:3030–3034. 2013.PubMed/NCBI
|
|
26.
|
Krecic AM and Swanson MS: hnRNP complexes:
composition, structure, and function. Curr Opin Cell Biol.
11:363–371. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Carpenter B, MacKay C, Alnabulsi A, et al:
The roles of heterogeneous nuclear ribonucleoproteins in tumour
development and progression. Biochim Biophys Acta. 1765:85–100.
2006.PubMed/NCBI
|
|
28.
|
Brockstedt E, Rickers A, Kostka S, et al:
Identification of apoptosis-associated proteins in a human Burkitt
lymphoma cell line. Cleavage of heterogeneous nuclear
ribonucleoprotein A1 by caspase 3. J Biol Chem. 273:28057–28064.
1998. View Article : Google Scholar
|
|
29.
|
Rahman-Roblick R, Johannes Roblick U,
Hellman U, et al: p53 targets identified by protein expression
profiling. Proc Natl Acad Sci USA. 104:5401–5406. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Koryllou A, Patrinou-Georgoula M, Troungos
C and Pletsa V: Cell death induced by N-methyl-N-nitrosourea, a
model SN1 methylating agent, in two lung cancer cell lines of human
origin. Apoptosis. 14:1121–1133. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Grover R, Sharathchandra A, Ponnuswamy A,
Khan D and Das S: Effect of mutations on the p53 IRES RNA
structure: Implications for de-regulation of the synthesis of p53
isoforms. RNA Biol. 8:132–142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Hossain MN, Fuji M, Miki K, Endoh M and
Ayusawa D: Downregulation of hnRNP C1/C2 by siRNA sensitizes HeLa
cells to various stresses. Mol Cell Biochem. 296:151–157. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Ren F, Wu H, Lei Y, et al: Quantitative
proteomics identification of phosphoglycerate mutase 1 as a novel
therapeutic target in hepatocellular carcinoma. Mol Cancer.
9:812010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Evans MJ, Saghatelian A, Sorensen EJ and
Cravatt BF: Target discovery in small-molecule cell-based screens
by in situ proteome reactivity profiling. Nat Biotechnol.
23:1303–1307. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Gao H, Yu B, Yan Y, et al: Correlation of
expression levels of ANXA2, PGAM1, and CALR with glioma grade and
prognosis. J Neurosurg. 118:846–853. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Jiang X, Sun Q, Li H, Li K and Ren X: The
role of phosphoglycerate mutase 1 in tumor aerobic glycolysis and
its potential therapeutic implications. Int J Cancer. Nov
28–2013.(Epub ahead of print). View Article : Google Scholar
|
|
37.
|
Chaneton B and Gottlieb E: PGAMgnam style:
a glycolytic switch controls biosynthesis. Cancer Cell. 22:565–566.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Tan JY, Huang X and Luo YL: PSMA7 inhibits
the tumorigenicity of A549 human lung adenocarcinoma cells. Mol
Cell Biochem. 366:131–137. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Du H, Huang X, Wang S, Wu Y, Xu W and Li
M: PSMA7, a potential biomarker of diseases. Protein Pept Lett.
16:486–489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Sakai A, Otani M, Miyamoto A, Yoshida H,
Furuya E and Tanigawa N: Identification of phosphorylated serine-15
and -82 residues of HSPB1 in 5-fluorouracil-resistant colorectal
cancer cells by proteomics. J Proteomics. 75:806–818. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Yung BY: Oncogenic role of
nucleophosmin/B23. Chang Gung Med J. 30:285–293. 2007.
|
|
42.
|
Wong JC, Hasan MR, Rahman M, et al:
Nucleophosmin 1, upregulated in adenomas and cancers of the colon,
inhibits p53-mediated cellular senescence. Int J Cancer.
133:1567–1577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Lin M, Hu J, Liu T, Li J, Chen B and Chen
X: Knockdown of nucleophosmin by RNA interference reverses
multidrug resistance in resistant leukemic HL-60 cells.
Immunobiology. 218:1147–1154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Grisendi S, Mecucci C, Falini B and
Pandolfi PP: Nucleophosmin and cancer. Nat Rev Cancer. 6:493–505.
2006. View Article : Google Scholar
|
|
45.
|
Karhemo PR, Rivinoja A, Lundin J, et al:
An extensive tumor array analysis supports tumor suppressive role
for nucleophosmin in breast cancer. Am J Pathol. 179:1004–1014.
2011. View Article : Google Scholar : PubMed/NCBI
|