Ovarian cancer is the most lethal cancer of the
female reproductive system, with high rate of mortality worldwide.
Likewise, ovarian cancer is one of the five leading types of cancer
death in women (1,2). In the last 80 years (1930–2010), the
overall death rate of ovarian cancer patients declined very
slightly (2). The main obstacle to
a successful treatment for ovarian cancer is the development of
drug resistance to combined chemotherapy (3). Drug resistance results from a variety
of factors including decreased cell-associated drugs, altered drug
inactivation, increased DNA damage tolerance/repair, increased
anti-apoptotic regulator activity and growth factor receptor
deregulation (4,5). Besides, abnormal expressions of genes
mediated by microRNA regulation also play critical roles in
development of drug resistance in ovarian cancer (6,7).
However, regardless of mechanisms, abnormal expression of drug
resistance-related genes often play important roles in drug
resistance (8). Thus, mining and
exploring of potentially drug resistance-related genes would be a
feasible and reasonable way to meet the challenge of the drug
resistance in ovarian cancer (9).
NEK [NIMA (never in mitosis gene A)-related
expressed kinase] family is the never-in-mitosis A (NIMA) protein
of Aspergillus nidulans, which was first identified by
Oakley and Morris in a genetic screen for cell division cycle
mutants (10). NEKs have now been
identified in a wide range of organisms, and eleven genetically
distinct proteins named NEK1 to NEK11 are expressed in humans. The
functions of NEKs mainly are involved in the regulation of cell
cycle progression, in particular, NEK2, NEK6, NEK7 and NEK9
contribute to the establishment of the microtubule-based mitotic
spindle, whereas NEK1, NEK10 and NEK11 have been implicated in the
DNA damage response (11). NEKs
have been proven to be associated with cancer development and
progression, in a cell cycle-dependent manner, and thus the NEKs
are considered as potential therapeutic targets in cancer (11–13).
The association of NEKs with drug resistance in
cancer is limited, with only a few studies. For example, NEK2
downregulation may improve the sensitivity of breast cancer cells
during chemotherapy treatments (14), and its upregulation may induce drug
resistance mainly through activation of efflux drug pumps in
myeloma and other cancers (15).
NEK4 is downregulated at 8 and/or 24 h in colon cancer cell lines
in response to 5-fluorouracil (16), and its suppression sensitize cancer
cells to taxol and vincristine, via regulation on mitosis and
microtubule homeostasis (17). In
ovarian cancer, we revealed that the upregulation of NEK2 is
associated with drug resistance in ovarian cancer, via its direct
or indirect interaction with a number of genes, proteins, microRNAs
and biological processes (18),
and further analysis indicated that the NEK2 expression is
regulated by NR2F2 (19).
Additionally, NEK1 expression can be induced by paclitaxel
(20), NEK4 is identified as a
candidate gene as potentiators of cisplatin (21), NEK6 can be affected by paclitaxel
through preventing phosphorylation of Thr389 (22), and NEK8 is significantly
downregulated in DDP-resistant ovarian cell line IGROV-1 (23). These results indicated that the
NEK1, 4, 6 and NEK8 were chemo-treatment related
proteins/genes.
However, among all the NEKs, the association of
NEK11 with cancer is rare, with one study considering it as a
potential tumor suppressor gene (24), and it is significantly
downregulated in ovarian cancer, mediated by methylation or
mutation (25,26), but its association with drug
resistance in cancer has not been reported. In this study, on the
basis of comprehensive bioinformatics analyses, we aimed to
illustrate the association of NEK11 with drug resistance in ovarian
cancer.
The microarray data of NEK11 in ovarian cancer
tissues was retrieved from the TCGA Ovarian array data deposited in
the Oncomine online database (https://www.oncomine.org/resource/main.html) (27). The microarray data of NEK11 in
ovarian cancer cells was retrieved from the Gene Expression Omnibus
(GEO) (http://www.ncbi.nlm.nih.gov/geoprofiles/) (28,29).
There were three probe sets targeting NEK11, while only two probe
sets with significant variability in statistics were kept.
Unpaired, two-tailed t-test assuming homogeneity of the variances
was performed with Excel software.
The protein/gene interaction analysis was performed
using GeneMANIA online tool (http://www.genemania.org/) (30–32).
Protein-small molecule/chemical interaction analysis was performed
using STITCH 4.0 beta (http://stitch-beta.embl.de/) (33–35)
and DrugBank online database (36,37).
Annotation of biological process was performed by Coremine Medical
online database (http://www.coremine.com/medical/) (38). The microRNAs targeted to the gene
were predicted by miRWalk online tool which including 10 prediction
tools (DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid, PICTAR4,
PICTAR5, PITA, RNA22, Targetscan) (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)
(39).
The mRNA expression of NEK11 in ovarian cancer
tissues and cells was retrieved from TCGA ovarian array and DataSet
Record GDS3754 which have been deposited in Oncomine and GEO online
database, respectively. As shown in Fig. 1A and C, compared with the
expression in 8 cases of normal ovaries, the NEK11 mRNA was
significantly downregulated in 586 cases of ovarian serous
cystadenocarcinomas, with a 3.05-fold change (P=3.45E-5).
Similarly, the expression of the NEK11 in cisplatin-resistant
ovarian cancer cells was measured (Fig. 1B and D). As shown in the figure,
the mRNA expression of NEK11 was decreased in cisplatin-resistant
ovarian cancer cells compared with expression in its sensitive
counterpart, with at least a 1.8-fold change (P<0.001). These
results suggested that the downregulation of NEK11 might
potentially play crucial roles in development of ovarian cancer and
drug resistance.
Protein/gene-protein/gene interactions performed
with GeneMANIA was used to reveal the drug resistance-related
functions of NEK11. As shown in Fig.
2, NEK11 had direct interactions with NEK2, MET, JUN, ERBB2,
EGFR, AKT1, AKT2, CHEK2, WWOX, IKBKE, ELF3 and MAST2, among those,
10 proteins/genes have been proven to be involved in the regulation
of drug resistance in ovarian cancer. NEK2, MET, JUN, ERBB2, EGFR,
AKT1, AKT2 and IKBKE are oncogenes associated with drug resistance
in ovarian cancer (18,41–50).
For example, IKBKE is found to be upregulated in ovarian, breast
and prostate cancer, and its upregulation biologically and
clinically relevant to the cancer development and progression, as
well as the chemoresistance (50).
Downregulation of the AKT2 sensitizes the ovarian cancer cells to
paclitaxel-induced apoptosis, and inhibits the survivin expression
which can induce drug-resistance to paclitaxel (49). NEK2 shared protein domains and had
very strong physical interaction with NEK11, and has been proven to
be responsible for the development of drug resistance not only in
ovarian cancer (18), but also in
breast (14) and colorectal cancer
(51). CHEK2 and WWOX are the
tumor suppressor genes which directly interacted with NEK11. CHEK2
is one of the critical kinases governing cell apoptosis, cell cycle
checkpoint and DNA damage repair. In ovarian cancer cells, CHEK2 is
degraded at the protein level in response to cisplatin through the
ubiquitin-proteasome pathway, suggesting that degradation or
decreased expression of CHEK2 is partially responsible for
chemoresistance (52). WWOX
suppression by RNA interference reverses platinum resistance in
DDP-resistant SKOV3 ovarian cancer cells (53). In addition to the direct
interactions, NEK11 indirectly interacted with 15 proteins/genes in
the network. Of which, 10 including BRCA1, BRCA2, PTEN, CDKN2A,
MLH1, KRAS, MYC, HGF, PMF2 and MLH3 are associated with drug
resistance in ovarian cancer (54–63).
Taken together, based on the protein/gene
interaction network, total of 28 proteins/genes were identified to
be directly/indirectly interacted with NEK11, and of which, 22 were
contributed to drug resistance in ovarian cancer. These results
strongly supported the idea that NEK11 might associate with drug
resistance in ovarian cancer.
The protein-small molecule/chemical interaction was
performed to further explain the drug resistance-related functions
of NEK11 in ovarian cancer. As shown in Table I, five chemicals including CC 243,
staurosporine, dasatinib, flavin mononucleotide and dabrafenib were
found to interact with NEK11. Except for the CC 243, all the
chemicals are associated with drug resistance in ovarian and other
cancers. For example, treatment of ovarian cancer cells with
staurosporine induces apoptosis in a time-dependent manner
(64), and reduces P-glycoprotein
expression and modulates multidrug resistance (65). Dasatinib is an antitumor agent for
many solid tumors (66), and can
significantly enhance the sensitivity to carboplatin in ovarian
cancer cells (67). As for the
flavin mononucleotide, a previous study showed that the
overexpression of riboflavin kinase increases the levels of flavin
mononucleotide and render cell resistance not only to cisplatin but
also to hydrogen peroxide and diamide (68), suggesting a role of flavin
mononucleotide in regulation of cisplatin resistance. Dabrafenib is
an inhibitor of BRAF leading to constitutive activation of the MAPK
signaling pathway (69,70), while the BRAF and MAPK signaling
have been proven to play important roles in drug resistance in
ovarian cancer (43,71–73).
MicroRNA-mediated post-transcriptional gene
regulation is considered as a significant regulator of many
cellular processes, both physiological and pathological (77,78).
The microRNAs perform their functions through the regulation on
their target genes, and it has been well established that microRNAs
represent a class of genes with a great potential for use in
diagnostics, prognosis and therapy (79). Therefore, the function of a gene
can be predicted based on functionality of the microRNAs that
target it (9). A microRNA-mRNA
interaction analysis was performed with miRWalk to predict the
microRNAs that target NEK11. Using this tool, we identified 443
microRNAs predicted to transcriptionally target NEK11. These
results suggesting that the microRNAs might be an important
regulation mechanism on NEK11. As shown in Table II, among the top 8 microRNAs most
strongly targeted NEK11, 3 microRNAs including miR-21, -590-5p and
-149 were shown to be involved in drug resistance in ovarian and
other cancers (80–84). For example, the expression of
miR-21 is notably upregulated in the paclitaxel-resistant ovarian
cancer A2780/taxol cells compared with the parental A2780 cells.
Inhibition of miR-21 in A2780/taxol cells decreased the expression
of P-gp and HIF-1α proteins, and increased the sensitivity of the
A2780/taxol cells to paclitaxel. These results suggested that
miR-21 may be involved in the development of drug resistance and
the regulation of MDR1/P-gp expression, at least in part, by
targeting HIF-1α in ovarian cancer cells (80). The other microRNAs did not indicate
direct involvement in drug resistance, however, except for the
miR-876-5p, they all play important roles in cancer development and
progression (as shown in Table
II), and might also be involved in the drug resistance. For
example, miR-376b controls mTOR inhibition-related autophagy
(85), while the autophagy plays
an important role in wt p53 and mutant p53-immediated multidrug
resistance in ovarian cancer (86).
Functional annotation of proteins/genes is a
fundamental problem in the post-genomic era, and determining the
functions of proteins encoded in genome sequences represents a
major challenge in current biology (92). Experimental determination of
protein function is expensive and time-consuming. Thus,
computational approaches based on the diverse genomic and proteomic
datasets can facilitate more rapid annotation of protein function
and guide laboratory experiments (92,93).
The computational approaches to gene function prediction have
relied on a variety of genomic and proteomic data, at least
including usage of microarray expression data (94), protein-protein interaction networks
(95), protein-small
molecule/chemical interactions (33–35)
and the annotation of gene with biological processes (93). Computational methods for inferring
protein function, which exploit the context of a protein in
cellular networks, can provide both a first hand hint into the
functional role of a protein and offer complementary insights to
understanding the function of proteins (96). Thus, on the basis of many
large-scale databases and networks, gene function prediction based
on bioinformatics analysis is a potential, feasible and valuable
way for gene function prediction (92). Using the comprehensive
bioinformatics analyses, we identified that two genes CCL21 and
SPARCL1 are associated with drug resistance in ovarian cancer
(9). Similarly, we identified that
the upregulation of NEK2 and E2F3 are associated with drug
resistance in ovarian cancer and poor prognosis in hepatocellular
carcinoma, respectively (18,97).
The association of NEK11 with drug resistance in
ovarian cancer was analyzed, on the basis of comprehensive
bioinformatics analysis (Fig. 4),
including microarray data retrieving, protein/gene interaction,
protein-small molecule/chemical interaction, biological process
annotation and microRNA-mRNA interaction. The
database/tool/software used in the analysis included GeneMANIA
online tool (30–32), Coremine Medical (38), Oncomine online database (27), GEO profiles (28,29),
STITCH 4.0 beta (33–35), DrugBank online database (36,37)
and miRWalk (39), which all are
regularly used and reliable databases/tools. For example, GeneMANIA
is a web-based database and a tool for prediction of gene functions
on the basis of multiple networks derived from different genomic or
proteomic data/sources (30). With
a query gene, GeneMANIA can find a small set of genes that are most
likely to share function with that gene based on their interactions
with it (32). The GEO at the
National Center for Biotechnology Information (NCBI) has emerged as
the leading fully public repository for gene expression data,
predominantly gene expression data generated by DNA microarray
technology (28), and about a
billion individual gene expression measurements are stored,
submitted by over 1500 laboratories, addressing a wide range of
biological phenomena (98). Thus,
the protein/gene functions predicted by these online
tools/databases/software were accurate and reliable.
NEK11 mRNA was significantly downregulated in both
ovarian cancer tissues and cisplatin-resistant cells (Fig. 1), indicating the potential roles of
NEK11 in regulation of drug resistance in ovarian cancer.
Protein/gene interaction indicated that among the total 28
proteins/genes which interacted with NEK11, 22 were contributed to
drug resistance in ovarian cancer (Fig. 2). Protein-small molecule
interaction suggested that among the 5 small molecules/chemicals
which interacted with NEK11, 4 were directly/indirectly associated
with drug resistance in ovarian and other cancers (Table I). Annotation of biological process
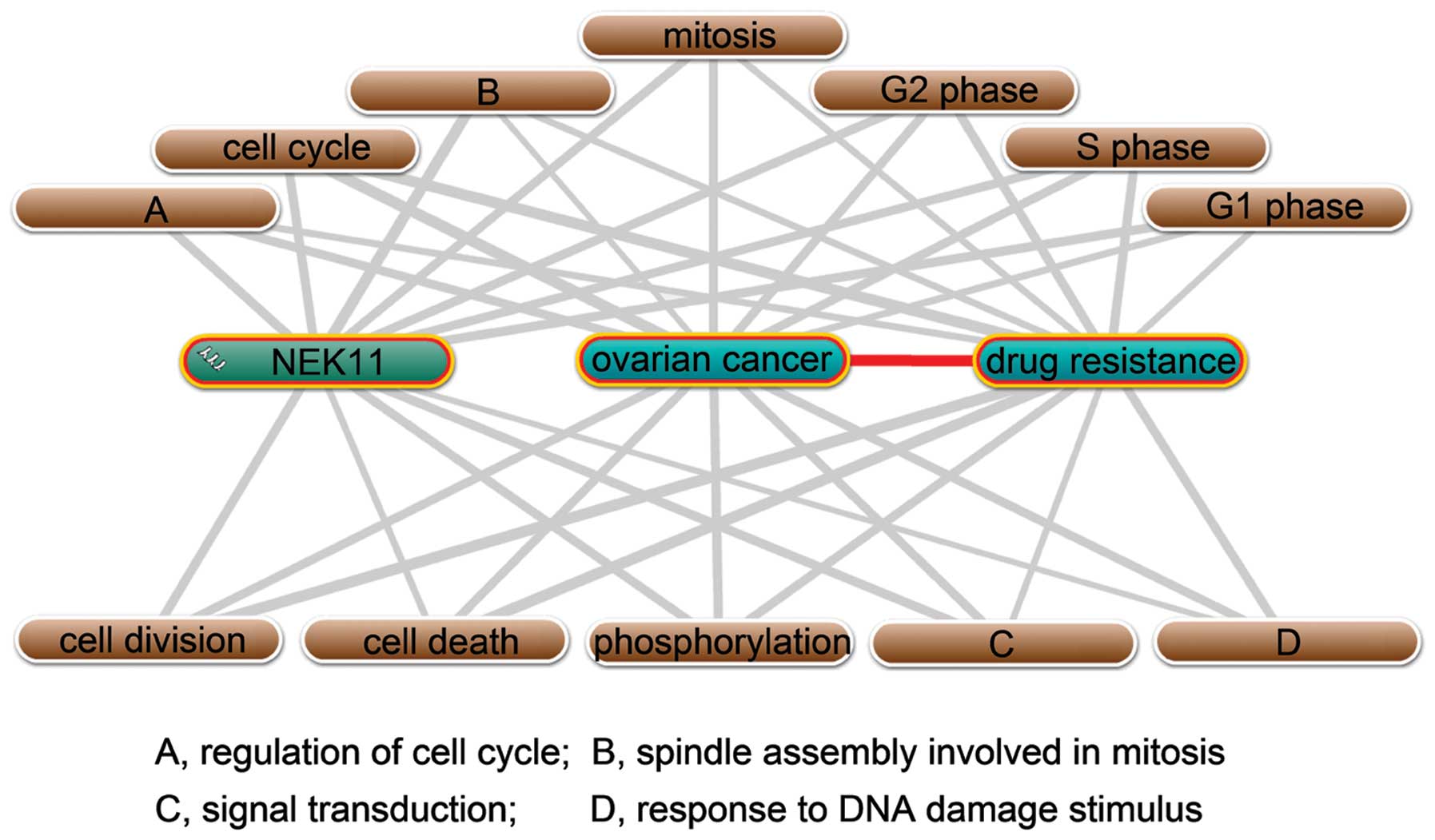
indicated that NEK11 probably were involved in the regulation of
drug resistance in ovarian cancer via its regulatory roles in 12
biological processes (Fig. 3).
MicroRNA-mRNA interaction analysis indicated that among the eight
microRNAs most strongly targeting NEK11, the majority were involved
in drug resistance in ovarian and other cancers (Table II). All these results provide a
very strong possibility that NEK11 is involved in drug resistance
in ovarian cancer, via its interactions with a number of genes,
proteins, small molecules, microRNAs and related biological
processes.
Cell cycle-mediated drug resistance is best
described as a relative insensitivity to a chemotherapeutic agent
because of the position of the cells in the cell cycle. Cell cycle
is closely involved in chemosensitivity for combination
chemotherapy, and the chemotherapeutic agents correlated with cell
cycle events include taxanes, platinum, camptothecin and
fluorouracil (99). Cell cycle is
closely related to drug resistance in ovarian cancer. For instance,
integration of DNA methylation and gene expression reveals specific
platinum resistance related signaling pathways in ovarian cancer,
which include cell growth-promoting pathways PI3K/Akt and cell
cycle progression (40). Besides,
curcumin and cisplatin or oxaliplatin can induce cell cycle
inhibition (100), and cell cycle
synchronization can reverse taxol resistance of human ovarian
cancer cell lines (101). In
addition, comprehensive bioinformatics analysis indicates that 15
TSGs perform their drug resistance-related functions through 5
pathways including cell cycle (8).
Annotation of NEK11 with ovarian cancer and drug resistance
generated 12 biological processes, of which, 7 were cell
cycle-related processes. Thus, given the important roles of NEK11
in regulation of cell cycle (11,12),
the important roles of the cell cycle in drug resistance (99), and the close linkages between NEK11
and drug resistance (Fig. 3), we
concluded that the NEK11 might be associated with drug resistance
in ovarian cancer via regulation of the cell cycle.
Taken together, for the first time, we illustrated
that downregulation of NEK11 in drug resistant cells might
contribute to drug resistance, via their interactions with a number
of drug resistance-related genes, proteins, small molecules and
microRNAs, and probably through regulation of the cell cycle. This
study set the stage for further investigation into the drug
resistant-related functions of NEK11, in ovarian and other
cancers.
The project was supported by the National Natural
Science Foundation of China (Grant No. 81302283), Youth Science
Foundation of Guangxi Medical University (No. GXMUYSF201205), and
China Postdoctoral Science Foundation (No. 2014M552535XB).
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar
|
|
3
|
Cannistra SA: Cancer of the ovary. N Engl
J Med. 351:2519–2529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson SW, Ozols RF and Hamilton TC:
Mechanisms of drug resistance in ovarian cancer. Cancer.
71:644–649. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parikh A, Lee C, Peronne J, Marchini S,
Baccarini A, Kolev V, Romualdi C, Fruscio R, Shah H, Wang F,
Mullokandov G, Fishman D, D’Incalci M, Rahaman J, Kalir T, Redline
RW, Brown BD, Narla G and DiFeo A: microRNA-181a has a critical
role in ovarian cancer progression through the regulation of the
epithelial-mesenchymal transition. Nat Commun. 5:29772014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li H, Xu H and Shen H: microRNA-106a
modulates cisplatin sensitivity by targeting PDCD4 in human ovarian
cancer cells. Oncol Lett. 7:183–188. 2014.PubMed/NCBI
|
|
8
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Tumor suppressor genes associated with drug resistance in
ovarian cancer (review). Oncol Rep. 30:3–10. 2013.PubMed/NCBI
|
|
9
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Bioinformatic analysis of chemokine (C-C motif) ligand 21 and
SPARC-like protein 1 revealing their associations with drug
resistance in ovarian cancer. Int J Oncol. 42:1305–1316.
2013.PubMed/NCBI
|
|
10
|
Oakley BR and Morris R: A mutation in
Aspergillus nidulans that blocks the transition from
interphase to prophase. J Cell Biol. 96:1155–1158. 1983.
|
|
11
|
Fry AM, O’Regan L, Sabir SR and Bayliss R:
Cell cycle regulation by the NEK family of protein kinases. J Cell
Sci. 125:4423–4433. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Malumbres M and Barbacid M: Cell cycle
kinases in cancer. Curr Opin Genet Dev. 17:60–65. 2007. View Article : Google Scholar
|
|
13
|
Bou Zgheib N, Xiong Y, Marchion DC, Bicaku
E, Chon HS, Stickles XB, Sawah EA, Judson PL, Hakam A and
Gonzalez-Bosquet J: The O-glycan pathway is associated with in
vitro sensitivity to gemcitabine and overall survival from ovarian
cancer. Int J Oncol. 41:179–188. 2012.PubMed/NCBI
|
|
14
|
Lee J and Gollahon L: Nek2-targeted ASO or
siRNA pretreatment enhances anticancer drug sensitivity in
triplenegative breast cancer cells. Int J Oncol. 42:839–847.
2013.PubMed/NCBI
|
|
15
|
Zhou W, Yang Y, Xia J, Wang H, Salama ME,
Xiong W, Xu H, Shetty S, Chen T and Zeng Z: NEK2 induces drug
resistance mainly through activation of efflux drug pumps and is
associated with poor prognosis in myeloma and other cancers. Cancer
Cell. 23:48–62. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De Angelis PM, Svendsrud DH, Kravik KL and
Stokke T: Cellular response to 5-fluorouracil (5-FU) in
5-FU-resistant colon cancer cell lines during treatment and
recovery. Mol Cancer. 5:202006.PubMed/NCBI
|
|
17
|
Doles J, Hemann MT, Doles J and Hemann M:
Nek4 status differentially alters sensitivity to distinct
microtubule poisons. Cancer Res. 70:10332010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu X, Gao Y, Lu Y, Zhang J, Li L and Yin
F: Upregulation of NEK2 is associated with drug resistance in
ovarian cancer. Oncol Rep. 31:745–754. 2014.PubMed/NCBI
|
|
19
|
Hawkins SM, Loomans HA, Wan YW,
Ghosh-Choudhury T, Coffey D, Xiao W, Liu Z, Sangi-Haghpeykar H and
Anderson ML: Expression and functional pathway analysis of nuclear
receptor NR2F2 in ovarian cancer. J Clin Endocrinol Metab.
98:E1152–E1162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bani MR, Nicoletti MI, Alkharouf NW,
Ghilardi C, Petersen D, Erba E, Sausville EA, Liu ET and Giavazzi
R: Gene expression correlating with response to paclitaxel in
ovarian carcinoma xenografts. Mol Cancer Ther. 3:111–121. 2004.
|
|
21
|
Arora S, Bisanz KM, Peralta LA, Basu GD,
Choudhary A, Tibes R and Azorsa DO: RNAi screening of the kinome
identifies modulators of cisplatin response in ovarian cancer
cells. Gynecol Oncol. 118:220–227. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Le XF, Hittelman WN, Liu J, McWatters A,
Li C, Mills GB and Bast RC Jr: Paclitaxel induces inactivation of
p70 S6 kinase and phosphorylation of Thr421 and Ser424 via multiple
signaling pathways in mitosis. Oncogene. 22:484–497. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng L, Lu W, Kulkarni B, Pejovic T, Yan
X, Chiang JH, Hood L, Odunsi K and Lin B: Analysis of chemotherapy
response programs in ovarian cancers by the next-generation
sequencing technologies. Gynecol Oncol. 117:159–169. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bozic I, Antal T, Ohtsuki H, Carter H, Kim
D, Chen S, Karchin R, Kinzler KW, Vogelstein B and Nowak MA:
Accumulation of driver and passenger mutations during tumor
progression. Proc Natl Acad Sci USA. 107:18545–18550. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moniz L, Dutt P, Haider N and Stambolic V:
Nek family of kinases in cell cycle, checkpoint control and cancer.
Cell Div. 6:182011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kashuba V, Dmitriev AA, Krasnov GS,
Pavlova T, Ignatjev I, Gordiyuk VV, Gerashchenko AV, Braga EA,
Yenamandra SP, Lerman M, Senchenko VN and Zabarovsky E: NotI
microarrays: novel epigenetic markers for early detection and
prognosis of high grade serous ovarian cancer. Int J Mol Sci.
13:13352–13377. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: a cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barrett T and Edgar R: Mining microarray
data at NCBI’s Gene Expression Omnibus (GEO)*. Methods
Mol Biol. 338:175–190. 2006.
|
|
29
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: a real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9(Suppl 1): S42008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD and
Morris Q: The GeneMANIA prediction server: biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zuberi K, Franz M, Rodriguez H, Montojo J,
Lopes CT, Bader GD and Morris Q: GeneMANIA prediction server 2013
update. Nucleic Acids Res. 41:W115–W222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kuhn M, Szklarczyk D, Franceschini A, von
Mering C, Jensen LJ and Bork P: STITCH 3: zooming in on
protein-chemical interactions. Nucleic Acids Res. 40:D876–D880.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuhn M, Szklarczyk D, Franceschini A,
Campillos M, von Mering C, Jensen LJ, Beyer A and Bork P: STITCH 2:
an interaction network database for small molecules and proteins.
Nucleic Acids Res. 38:D552–D556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuhn M, von Mering C, Campillos M, Jensen
LJ and Bork P: STITCH: interaction networks of chemicals and
proteins. Nucleic Acids Res. 36:D684–D688. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: a
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36:D901–D906. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Law V, Knox C, Djoumbou Y, Jewison T, Guo
AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A,
Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y and Wishart DS:
DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids
Res. 42:D1091–D1097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, et
al: Diagnostic interpretation of array data using public databases
and internet sources. Hum Mutat. 33:930–940. 2012.PubMed/NCBI
|
|
39
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011.
|
|
40
|
Li M, Balch C, Montgomery JS, Jeong M,
Chung JH, Yan P, Huang TH, Kim S and Nephew KP: Integrated analysis
of DNA methylation and gene expression reveals specific signaling
pathways associated with platinum resistance in ovarian cancer. BMC
Med Genomics. 2:342009. View Article : Google Scholar
|
|
41
|
Tang MK, Zhou HY, Yam JW and Wong AS:
c-Met overexpression contributes to the acquired apoptotic
resistance of nonadherent ovarian cancer cells through a cross talk
mediated by phosphatidylinositol 3-kinase and extracellular
signal-regulated kinase 1/2. Neoplasia. 12:128–138. 2010.
|
|
42
|
Pan B, Yao KS, Monia BP, Dean NM, McKay
RA, Hamilton TC and O’Dwyer PJ: Reversal of cisplatin resistance in
human ovarian cancer cell lines by a c-jun antisense
oligodeoxynucleotide (ISIS 10582): evidence for the role of
transcription factor overexpression in determining resistant
phenotype. Biochem Pharmacol. 63:1699–1707. 2002. View Article : Google Scholar
|
|
43
|
Mansouri A, Ridgway LD, Korapati AL, Zhang
Q, Tian L, Wang Y, Siddik ZH, Mills GB and Claret FX: Sustained
activation of JNK/p38 MAPK pathways in response to cisplatin leads
to Fas ligand induction and cell death in ovarian carcinoma cells.
J Biol Chem. 278:19245–19256. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Smith V, Hobbs S, Court W, Eccles S,
Workman P and Kelland LR: ErbB2 overexpression in an ovarian cancer
cell line confers sensitivity to the HSP90 inhibitor geldanamycin.
Anticancer Res. 22:1993–1999. 2002.PubMed/NCBI
|
|
45
|
Zhou BP, Hu MC, Miller SA, Yu Z, Xia W,
Lin SY and Hung MC: HER-2/neu blocks tumor necrosis factor-induced
apoptosis via the Akt/NF-kappaB pathway. J Biol Chem.
275:8027–8031. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qiu L, Di W, Jiang Q, Scheffler E, Derby
S, Yang J, Kouttab N, Wanebo H, Yan B and Wan Y: Targeted
inhibition of transient activation of the EGFR-mediated cell
survival pathway enhances paclitaxel-induced ovarian cancer cell
death. Int J Oncol. 27:1441–1448. 2005.PubMed/NCBI
|
|
47
|
Sen T, Sen N, Brait M, Begum S, Chatterjee
A, Hoque MO, Ratovitski E and Sidransky D: DeltaNp63alpha confers
tumor cell resistance to cisplatin through the AKT1 transcriptional
regulation. Cancer Res. 71:1167–1176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xing H, Weng D, Chen G, Tao W, Zhu T, Yang
X, Meng L, Wang S, Lu Y and Ma D: Activation of
fibronectin/PI-3K/Akt2 leads to chemoresistance to docetaxel by
regulating survivin protein expression in ovarian and breast cancer
cells. Cancer Lett. 261:108–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Weng D, Song X, Xing H, Ma X, Xia X, Weng
Y, Zhou J, Xu G, Meng L, Zhu T, Wang S and Ma D: Implication of the
Akt2/survivin pathway as a critical target in paclitaxel treatment
in human ovarian cancer cells. Cancer Lett. 273:257–265. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guan H, Zhang H, Cai J, Wu J, Yuan J, Li
J, Huang Z and Li M: IKBKE is over-expressed in glioma and
contributes to resistance of glioma cells to apoptosis via
activating NF-κB. J Pathol. 223:436–445. 2011.PubMed/NCBI
|
|
51
|
Suzuki K, Kokuryo T, Senga T, Yokoyama Y,
Nagino M and Hamaguchi M: Novel combination treatment for
colorectal cancer using Nek2 siRNA and cisplatin. Cancer Sci.
101:1163–1169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang P, Gao W, Li H, Reed E and Chen F:
Inducible degradation of checkpoint kinase 2 links to
cisplatin-induced resistance in ovarian cancer cells. Biochem
Biophys Res Commun. 328:567–572. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu YY, Li L, Li DR, Zhang W and Wang Q:
Suppression of WWOX gene by RNA interference reverses platinum
resistance acquired in SKOV3/SB cells. Zhonghua Fu Chan Ke Za Zhi.
43:854–858. 2008.(In Chinese).
|
|
54
|
Yang D, Khan S, Sun Y, Hess K, Shmulevich
I, Sood AK and Zhang W: Association of BRCA1 and BRCA2 mutations
with survival, chemotherapy sensitivity, and gene mutator phenotype
in patients with ovarian cancer. JAMA. 306:1557–1565. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou C, Smith JL and Liu J: Role of BRCA1
in cellular resistance to paclitaxel and ionizing radiation in an
ovarian cancer cell line carrying a defective BRCA1. Oncogene.
22:2396–2404. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lee S, Choi EJ, Jin C and Kim DH:
Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA
amplification contributes to cisplatin resistance in an ovarian
cancer cell line. Gynecol Oncol. 97:26–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wu H, Cao Y, Weng D, Xing H, Song X, Zhou
J, Xu G, Lu Y, Wang S and Ma D: Effect of tumor suppressor gene
PTEN on the resistance to cisplatin in human ovarian cancer cell
lines and related mechanisms. Cancer Lett. 271:260–271. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kawakami Y, Hama S, Hiura M, Nogawa T,
Chiba T, Yokoyama T, Takashima S, Tajiri H, Eguchi K, Nagai N,
Shigemasa K, Ohama K, Kurisu K and Heike Y: Adenovirus-mediated p16
gene transfer changes the sensitivity to taxanes and Vinca
alkaloids of human ovarian cancer cells. Anticancer Res.
21:2537–2545. 2001.PubMed/NCBI
|
|
59
|
Plumb JA, Strathdee G, Sludden J, Kaye SB
and Brown R: Reversal of drug resistance in human tumor xenografts
by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene
promoter. Cancer Res. 60:6039–6044. 2000.
|
|
60
|
Ratner ES, Keane FK, Lindner R, Tassi RA,
Paranjape T, Glasgow M, Nallur S, Deng Y, Lu L, Steele L, Sand S,
Muller RU, Bignotti E, Bellone S, Boeke M, Yao X, Pecorelli S,
Ravaggi A, Katsaros D, Zelterman D, Cristea MC, Yu H, Rutherford
TJ, Weitzel JN, Neuhausen SL, Schwartz PE, Slack FJ, Santin AD and
Weidhaas JB: A KRAS variant is a biomarker of poor outcome,
platinum chemotherapy resistance and a potential target for therapy
in ovarian cancer. Oncogene. 31:4559–4566. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Vogt U, Falkiewicz B, Bielawski K, Bosse U
and Schlotter CM: Relationship of c-myc and erbB oncogene family
gene aberrations and other selected factors to ex vivo
chemosensitivity of ovarian cancer in the modified
ATP-chemosensitivity assay. Acta Biochim Pol. 47:157–164.
2000.PubMed/NCBI
|
|
62
|
Bardella C, Dettori D, Olivero M, Coltella
N, Mazzone M and Di Renzo MF: The therapeutic potential of
hepatocyte growth factor to sensitize ovarian cancer cells to
cisplatin and paclitaxel in vivo. Clin Cancer Res. 13:2191–2198.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xiao X, Melton DW and Gourley C: Mismatch
repair deficiency in ovarian cancer - molecular characteristics and
clinical implications. Gynecol Oncol. 132:506–512. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gregory-Bass RC, Olatinwo M, Xu W,
Matthews R, Stiles JK, Thomas K, Liu D, Tsang B and Thompson WE:
Prohibitin silencing reverses stabilization of mitochondrial
integrity and chemoresistance in ovarian cancer cells by increasing
their sensitivity to apoptosis. Int J Cancer. 122:1923–1930. 2008.
View Article : Google Scholar
|
|
65
|
Sampson KE, Wolf CL and Abraham I:
Staurosporine reduces P-glycoprotein expression and modulates
multidrug resistance. Cancer Lett. 68:7–14. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gnoni A, Marech I, Silvestris N, Vacca A
and Lorusso V: Dasatinib: an anti-tumour agent via Src inhibition.
Curr Drug Targets. 12:563–578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jinawath N, Vasoontara C, Jinawath A, Fang
X, Zhao K, Yap KL, Guo T, Lee CS, Wang W, Balgley BM, Davidson B,
Wang TL and Shih Ie M: Oncoproteomic analysis reveals
co-upregulation of RELA and STAT5 in carboplatin resistant ovarian
carcinoma. PLoS One. 5:e111982010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hirano G, Izumi H, Yasuniwa Y, Shimajiri
S, Ke-Yong W, Sasagiri Y, Kusaba H, Matsumoto K, Hasegawa T,
Akimoto M, Akashi K and Kohno K: Involvement of riboflavin kinase
expression in cellular sensitivity against cisplatin. Int J Oncol.
38:893–902. 2011.PubMed/NCBI
|
|
69
|
Hertzman Johansson C and Egyhazi Brage S:
BRAF inhibitors in cancer therapy. Pharmacol Ther. 142:176–182.
2014.
|
|
70
|
Bucheit AD and Davies MA: Emerging
insights into resistance to BRAF inhibitors in melanoma. Biochem
Pharmacol. 87:381–389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Janku F, Tsimberidou AM, Garrido-Laguna I,
Wang X, Luthra R, Hong DS, Naing A, Falchook GS, Moroney JW,
Piha-Paul SA, Wheler JJ, Moulder SL, Fu S and Kurzrock R: PIK3CA
mutations in patients with advanced cancers treated with
PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 10:558–565. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Janku F, Wheler JJ, Westin SN, Moulder SL,
Naing A, Tsimberidou AM, Fu S, Falchook GS, Hong DS, Garrido-Laguna
I, Luthra R, Lee JJ, Lu KH and Kurzrock R: PI3K/AKT/mTOR inhibitors
in patients with breast and gynecologic malignancies harboring
PIK3CA mutations. J Clin Oncol. 30:777–782. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fister S, Gunthert AR, Aicher B, Paulini
KW, Emons G and Grundker C: GnRH-II antagonists induce apoptosis in
human endometrial, ovarian, and breast cancer cells via activation
of stress-induced MAPKs p38 and JNK and proapoptotic protein Bax.
Cancer Res. 69:6473–6481. 2009. View Article : Google Scholar
|
|
74
|
Gene Ontology consortium. http://www.geneontology.org.
|
|
75
|
Gamberoni G, Storari S and Volinia S:
Finding biological process modifications in cancer tissues by
mining gene expression correlations. BMC Bioinformatics. 7:62006.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lagreid A, Hvidsten TR, Midelfart H,
Komorowski J and Sandvik AK: Predicting gene ontology biological
process from temporal gene expression patterns. Genome Res.
13:965–979. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tili E, Michaille JJ, Gandhi V, Plunkett
W, Sampath D and Calin GA: miRNAs and their potential for use
against cancer and other diseases. Future Oncol. 3:521–537. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xie Z, Cao L and Zhang J: miR-21 modulates
paclitaxel sensitivity and hypoxia-inducible factor-1alpha
expression in human ovarian cancer cells. Oncol Lett. 6:795–800.
2013.PubMed/NCBI
|
|
81
|
Li B, Ren S, Li X, Wang Y, Garfield D,
Zhou S, Chen X, Su C, Chen M, Kuang P, Gao G, He Y, Fan L, Fei K,
Zhou C and Schmit-Bindert G: MiR-21 overexpression is associated
with acquired resistance of EGFR-TKI in non-small cell lung cancer.
Lung Cancer. 83:146–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yang SM, Huang C, Li XF, Yu MZ, He Y and
Li J: miR-21 confers cisplatin resistance in gastric cancer cells
by regulating PTEN. Toxicology. 306:162–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Watson JA, Bryan K, Williams R, Popov S,
Vujanic G, Coulomb A, Boccon-Gibod L, Graf N, Pritchard-Jones K and
O’Sullivan M: miRNA profiles as a predictor of chemoresponsiveness
in Wilms’ tumor blastema. PLoS One. 8:e534172013.PubMed/NCBI
|
|
84
|
Berghmans T, Ameye L, Willems L, Paesmans
M, Mascaux C, Lafitte JJ, Meert AP, Scherpereel A, Cortot AB,
Cstoth I, Dernies T, Toussaint L, Leclercq N and Sculier JP:
Identification of microRNA-based signatures for response and
survival for non-small cell lung cancer treated with
cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer.
82:340–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Korkmaz G, le Sage C, Tekirdag KA, Agami R
and Gozuacik D: miR-376b controls starvation and mTOR
inhibition-related autophagy by targeting ATG4C and BECN1.
Autophagy. 8:165–176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kong D, Ma S, Liang B, Yi H, Zhao Y, Xin
R, Cui L, Jia L and Liu X: The different regulatory effects of p53
status on multidrug resistance are determined by autophagy in
ovarian cancer cells. Biomed Pharmacother. 66:271–278. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Hidaka H, Seki N, Yoshino H, Yamasaki T,
Yamada Y, Nohata N, Fuse M, Nakagawa M and Enokida H: Tumor
suppressive microRNA-1285 regulates novel molecular targets:
aberrant expression and functional significance in renal cell
carcinoma. Oncotarget. 3:44–57. 2012.PubMed/NCBI
|
|
88
|
Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S,
Shao W, Cai J, Du Q, Zhu Y and Mao J: Up-regulation of
microRNA-1290 impairs cytokinesis and affects the reprogramming of
colon cancer cells. Cancer Lett. 329:155–163. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yungang W, Xiaoyu L, Pang T, Wenming L and
Pan X: miR-370 targeted FoxM1 functions as a tumor suppressor in
laryngeal squamous cell carcinoma (LSCC). Biomed Pharmacother.
68:149–154. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Fan C, Liu S, Zhao Y, Han Y, Yang L, Tao
G, Li Q and Zhang L: Upregulation of miR-370 contributes to the
progression of gastric carcinoma via suppression of FOXO1. Biomed
Pharmacother. 67:521–526. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Feng Y1, Wang L, Zeng J, Shen L, Liang X,
Yu H, Liu S, Liu Z, Sun Y, Li W, Chen C and Jia J: Fork head box M1
is overexpressed in Helicobacter pylori-induced gastric
carcinogenesis and is negatively regulated by hsa-miR-370. Mol
Cancer Res. 11:834–844. 2013.
|
|
92
|
Sharan R, Ulitsky I and Shamir R:
Network-based prediction of protein function. Mol Syst Biol.
3:882007. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Phuong T and Nhung N: Predicting gene
function using similarity learning. BMC Genomics. 14(Suppl 4):
S42013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Uetz P, Giot L, Cagney G, Mansfield TA,
Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart
P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T,
Vijayadamodar G, Yang M, Johnston M, Fields S and Rothberg JM: A
comprehensive analysis of protein-protein interactions in
Saccharomyces cerevisiae. Nature. 403:623–627. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Janga S, Díaz-Mejía JJ and
Moreno-Hagelsieb G: Network-based function prediction and
interactomics: the case for metabolic enzymes. Metab Eng. 13:1–10.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zeng X, Yin F, Liu X, Xu J, Xu Y, Huang J,
Nan Y and Qiu X: Upregulation of E2F transcription factor 3 is
associated with poor prognosis in hepatocellular carcinoma. Oncol
Rep. 31:1139–1146. 2014.PubMed/NCBI
|
|
98
|
Barrett T and Edgar R: Gene expression
omnibus: microarray data storage, submission, retrieval, and
analysis. Methods Enzymol. 411:352–369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Shah MA and Schwartz GK: Cell
cycle-mediated drug resistance: an emerging concept in cancer
therapy. Clin Cancer Res. 7:2168–2181. 2001.PubMed/NCBI
|
|
100
|
Montopoli M, Ragazzi E, Froldi G and
Caparrotta L: Cell-cycle inhibition and apoptosis induced by
curcumin and cisplatin or oxaliplatin in human ovarian carcinoma
cells. Cell Prolif. 42:195–206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wang X, Pan L, Mao N, Sun L, Qin X and Yin
J: Cell-cycle synchronization reverses Taxol resistance of human
ovarian cancer cell lines. Cancer Cell Int. 13:772013. View Article : Google Scholar : PubMed/NCBI
|